Summary
Dysregulated cardiac function after sepsis in intensive care unit is known to predict poor long-term outcome and increase mortality. Their pathological feature and molecular mechanism remain unclear. We observed that septic patients with depressed left ventricular ejection fraction (LVEF) have the highest in-hospital and 28 days mortality comparing to patients with hyperdynamic LVEF or with heart failure with preserved LVEF. Echocardiograms reveal that survivors post cecum ligation and puncture (CLP) on rodents have stable LVEF and non-survivors have fluctuated LVEF at CLP early phase. CLP-induced mice fall into three groups based on LVEF 24 h post-surgery: high-, low-, and normal-LVEF. Transcriptomic and proteomic analyses identify jointly and distinctively changed genes, proteins and biologically essential pathways in left ventricles from three CLP groups. Notably, transmission electron microscopy shows different mitochondrial and sarcomere defects associated with LVEF variances. Together, this study systematically characterizes the molecular, morphological, and functional alterations in CLP-induced cardiac injury.
Subject areas: Health sciences, Proteomics, Transcriptomics
Graphical abstract

Highlights
-
•
Uncovers the temporal dynamics of LVEF in sepsis animal model
-
•
CLP induction results in three different subtypes base on LVEF
-
•
Shared and distinct molecular signatures in three CLP group with different LVEF
-
•
Defects in mitochondria and sarcomere are associated with different LVEF
Health sciences; Proteomics; Transcriptomics
Introduction
Sepsis is a systemic condition with life-threatening organ dysfunction caused by infection and the consequent dysregulated host response.1 Sepsis induces a wide spectrum of clinical complications including cardiac dysfunction, which has been recognized as an essential pathological feature in sepsis. Sepsis-induced cardiac dysfunction is associated with increased mortality and influences the long-term outcome of septic patients.2,3 Given the key roles of myocardial circulation in supporting the function of multiple organs and the interplay among organs during septic pathogenies, cardiac dysfunction or failure contributes significantly to the organ abnormalities in sepsis, hence underscoring critical clinical importance.
Currently, the diagnostic criteria and precise definition of sepsis-induced cardiac dysfunction remain uncertain, posing great difficulties in clinical management and understanding of the disease pathogenesis. Left ventricular ejection fraction (LVEF) assessed by using echocardiograms has been widely employed to evaluate ventricular function in intensive care unit (ICU). A recent large cohort study reveals that patients with either lower or higher LVEF had higher mortality risks,4 suggesting that both depressed and hyperdynamic LVEF are prognostic of poor outcome in ICU. These results lead to a hypothesis that distinct change of LVEF might be related to Sepsis-induced cardiac dysfunction associated with multiple clinical presentations including left ventricular (LV) systolic or diastolic dysfunction, right ventricular (RV) impairment. Acutely depressed LVEF associated with ventricular dilation has been long viewed as critical characteristics in sepsis-induced cardiomyopathy.5,6,7 Nevertheless, preserved or hyperdynamic LVEF was also frequently observed in patients with cardiac dysfunction or heart failure in sepsis. Recent study reported that hyperdynamic LVEF was associated with mortality in septic patients.8 The complexity and heterogeneous of LVEF in septic patients, as well as different alterations in LV and RV structure and function, together implicate that different mechanisms are involved in the cardiac dysfunction or failure associated with sepsis, which however, are poorly understood.
In this study, we show that cecum ligation and puncture (CLP) induction of polymicrobial sepsis in rodents results in three different subtypes base on LVEF: HEF (high LVEF, LVEF ≥90%), LEF (low LVEF, LVEF < 65%), and NEF (normal LVEF, 65% ≤LVEF< 90%), recapitulating the clinical observations in septic patients by our multi-centered intensive care units data and other clinical reports.8 Moreover, by systematic transcriptomic and proteomic profiling, as well as gene set enrichment and protein coexpression network analysis, we reveal that these 3 types of LVEF response contain shared and distinct molecular signatures. Intriguingly, transmission electron microscopy (TEM) examination shows that all these 3 types of CLP groups display defects in mitochondria and sarcomere with unique characteristics. Thus, we provided a comprehensive catalog of differentially expressed genes and proteins, as well as enriched gene ontology terms and pathways in sepsis-induced cardiomyopathy with different LVEF.
Results
Clinical assessment of septic patients with different left ventricle ejection fraction
Totally, we evaluated clinical data of 939 patients that meet diagnostic criterion of Sepsis 3.0 from our multi-centered intensive care units (ICU) project (NCT number ChiCTR1800015056). All the patients were categorized into three groups for further analysis based on LVEF, including patients with hyperdynamic left ventricular ejection fraction (HDLVEF) (high LVEF group, LVEF higher than or equal to 70%), patients with heart failure with preserved ejection fraction (HFpEF) (normal LVEF group, LVEF higher than or equal to 50% and less than 70%), and patients with depressed LVEF (low LVEF group, LVEF less than 50%). Kaplan-Meier survival curve analysis shows that high LVEF group and normal LVEF group both have fair cumulative descending survival rate, and low LVEF group had the lowest cumulative survival rate in 30 days follow up (Figure 1). The three groups of septic patients all have considerable hospital death rate (8.6%, 7.1%, and 13.7%), and substantial 28 days mortality (13.2%, 13.2%, and 26.3%) (Table S1). Notably, patients with low LVEF have the highest in-hospital mortality, as well as 28 days mortality among all the 3 septic groups (Table S1).
Figure 1.
Survivor rate of septic patients with depressed, preserved, and hyperdynamic LVEF
(A) Kaplan-Meier survival curve analysis of three group of septic patients with different LVEF.
(B) Number of patients analyzed in the Kaplan-Meier survival curve analysis. 517 patients diagnosed with sepsis with complete follow-up data were included. Low LVEF group (n = 156), normal EF group (n = 284) and high EF group (n = 77). Significance threshold was set 0.05 for p value.
Meantime we assessed clinical parameters of patients from the three groups. Patients with low LVEF have lower SOFA score, APACHE II score, MV hours, vasoactive drug use time, lactate level, oxygen absorption concentration (FiO2), and higher age level than those with normal LVEF. Furthermore, patients with low LVEF have higher age level, APACHE II score, vasoactive drug use time, pH value level, heart rate (HR) level, norepinephrine (NE) use dosage, FiO2, but lower lactate level, oxygenation index than high LVEF group. By contrast, patients in high LVEF group have lower age level, NE use dosage and FiO2 than those in normal LVEF group (Table S1).
CLP induction in rodents recapitulates LVEF changes observed in septic patients
Next, we performed echocardiogram to measure LVEF of mice at different time point after CLP induction of polymicrobial sepsis. The experimental design and workflow are illustrated in Figure 2A. We found that the baseline LVEF of sham-treated mice is 76% ± 6.22% (n = 10), which is consistent with previous studies.9,10 Intriguingly, by assessing LVEF at different time point through 24 h after CLP (the early phase of sepsis), we observed that mean LVEF of CLP mice is comparable to that of sham mice at each time point after CLP with the exception that CLP mice had moderate higher LVEF than control mice at 3 h after induction (Figure 2B). In addition, in this early phase of sepsis, survived mice had stable LVEF ([77.25 ± 2.613] % at 6 h after CLP; [80.97 ± 1.567] % at 15 h after CLP), whereas LVEF of non-survivors changes markedly across time points (reduced to [67.00 ± 6.081] % at 6 h, increased to [92.42 ± 4.922] % at 15 h, and further reduced to 53.67% at 24 h) (Figure 2C). Notably, non-survivors exhibited decreased LVEF at 6 h and 24 h after CLP compared to survivors, but exhibited increased LVEF at 15 h and 3 days (Figures 2C and 2D). Together, these results suggest that stable LVEF might be associated with high survival rate in sepsis. Moreover, LVEF of non-survivors return to a range that is comparable to that of survivors at 6 days after CLP (Figure 2D).
Figure 2.
Temporal dynamic change of LVEF in mice after CLP and LPS challenge
(A) Schematic illustration of experimental design and workflow.
(B) Quantitative analysis of LVEF from mice at designated time point after CLP and sham treatment. n = 4–17 mice per group.
(C) Quantitative analysis of LVEF from survived and non-survived mice at designated time point through 24 h post CLP. n = 5–12 mice per group.
(D) Quantitative analysis of LVEF from survived and non-survived mice at designated time point through 30 days post CLP. n = 13–56 mice per group.
(E) Representative images of mice left ventricle measured by small animal ultrasound at 24 h post CLP and sham operation.
(F) Percentage of mice in HEF (LVEF > = 90%), LEF (LVEF< 65%), and NEF (65% = <LVEF < 90%) at different time point after CLP.
(G) Quantitative analysis of LVEF from mice of sham and three CLP groups according to LVEF measured at 24 h post treatment. n = 2–9 mice per group.
(H) Kaplan-Meier survival curve analysis of these 3 CLP groups with different LVEF. Sham: n = 11 mice, HEF: n = 17 mice; LEF: n = 4 mice; NEF: n = 26 mice.
(I) Quantitative analysis of LVEF from mice at designated time point through 24 h post LPS and saline administration. n = 4–17 mice per group.
(J) Quantitative analysis of LVEF from mice at designated time point through 40 days post LPS and saline administration. n = 4–17 mice per group. Data are shown as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Student’s t test.
The various trend of LVEF post CLP in mice recapitalize the heterogeneous LVEF observed in sepsis patients. Given hemodynamic changes normally come out at 24 h after sepsis onset in both clinical and animal research,6,11 we then subjected CLP mice into three categories based on LVEF assessed at 24 h post CLP: larger than or equal to 90% (HEF), less than 65% (LEF), less than 90% and larger than or equals to 65% (NEF) (Figures 2E and 2F). Notably, LVEF of all the three groups recovered to a range between 65% and 90% after 3 days and remained in this range in the following 30 days in mice CLP model, which is undistinguished from sham group (Figure 2G). We performed Kaplan-Meier survival curve analysis of these 3 groups. The results show that group of HEF and NEF had considerable death rate, while LEF group had the highest mortality compared to the other two groups (Figure 2H). To test whether LVEF before death is associated with the survival rate, we regrouped the mice according to LVEF last time measured before death. The two groups of LVEF < 65% and LVEF ≥90% occupied the maximal proportion of mortality, while 65% ≤ LVEF < 90% group had highest survival rate among three groups (Figure S1). Together, these data further suggest that keeping LVEF in an appropriate level might be beneficial for the long-term outcome of sepsis patients.
LPS induction leads to depressed LVEF in mice
We also examined the LVEF in LPS-induced endotoxemia model. Mice were intraperitoneal injected with a single dose of LPS (15 mg/kg) and monitored with echocardiogram at various time points after injection. Unlike CLP mice, the mean LVEF of LPS-treated mice is uniformly depressed to about 57% at 6 h and recovered to baseline range that is similar as saline-treated group at 15 h (Figures 2I and 2J). Therefore, CLP sepsis model likely involves more complicated feature and mechanism compared to LPS-induced endotoxemia model.
Evaluation of hemodynamics in CLP-induced rats
To better evaluate the hemodynamics parameters during sepsis, we performed CLP on rats, which are suitable to provide high resolution of hemodynamics parameters. In accordance with mice CLP model, mean LVEF is comparable in CLP and sham group, whereas non-survivors after CLP exhibit more fluctuant LVEF than survivors rat CLP model (Figures 3A and 3B). In addition, the CLP group falls into 3 subtypes based on LVEF at 24 h after CLP, which again manifests the LVEF heterogeneous in mice CLP model and sepsis patients. (Figure 3C). Furthermore, we conducted a serial of hemodynamics examination on rats at 24 h after CLP. The mean cardiac output (CO) is higher in CLP group than that in sham-treated control rats at 24 h after surgery (Figure 3D). In addition, non-survivors displayed lower CO and velocity time integral (VTI) when compared with survivors at 24 h after CLP (Figures 3E and 3G). However, mean blood pressure (MBP), end diastolic volume (EDV) and left ventricular diastolic dimension (LVIDd), HR, E/A ratio, end-systolic volume (ESV) and left ventricular systolic dimension (LVIDd), left ventricular outflow tract (LVOT) remained unchanged in non-survivors compared with survivors at 24 h after CLP (Figures S2A–S2U).
Figure 3.
Hemodynamics measurement of rats in CLP sepsis model
(A) Quantitative analysis of LVEF from rats at designated time point after CLP and sham treatment. n = 4–15 rats per group.
(B) Quantitative analysis of LVEF from survived and non-survived rats at designated time point post CLP. n = 4–11 rats per group.
(C) Quantitative analysis of LVEF from rats of sham and three CLP groups according to LVEF measured at 24 h post treatment.
(D) Quantitative analysis of cardiac output (CO) of rats at designated time point after CLP and sham treatment. n = 4–15 rats per group.
(E) Comparison of CO in survivors and non-survivors at designated time point after CLP. n = 4–11 rats per group.
(F) Quantitative analysis of velocity time integral (VTI) of rats at designated time point after CLP and sham treatment. n = 4–15 rats per group.
(G) Comparison of VTI in survivors and non-survivors at designated time point after CLP. n = 4–11 rats per group. Data are shown as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, Student’s t test.
RNA-seq and GSEA analysis reveal molecular signature in CLP mice with different LVEF
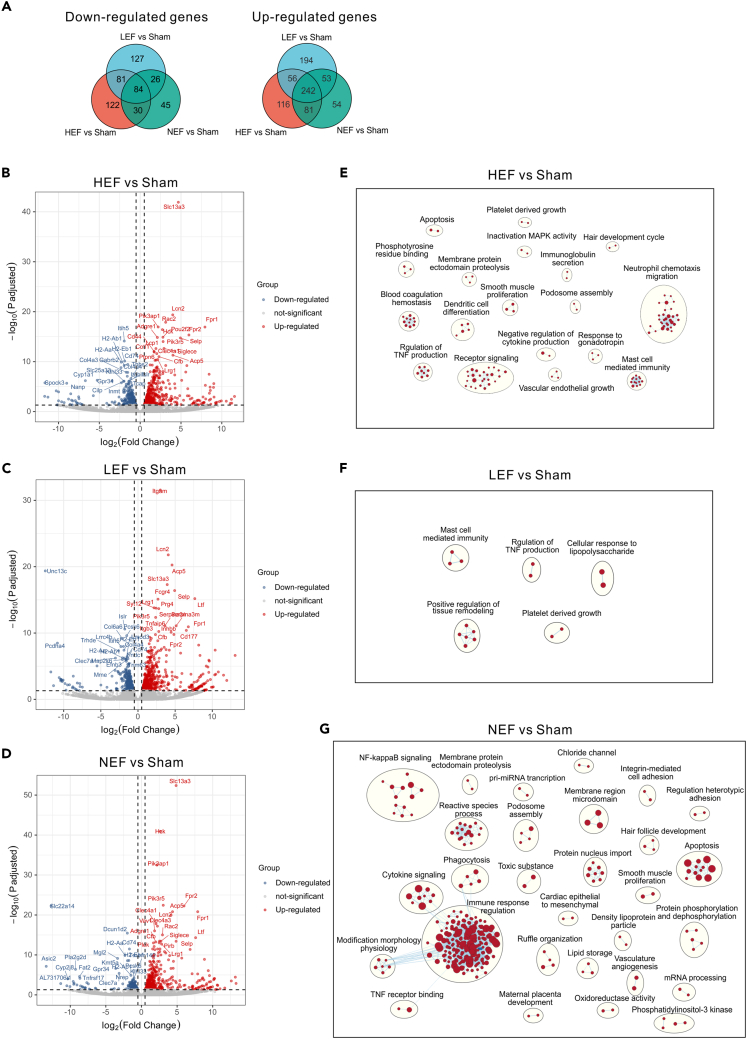
To systematic explore the molecular signatures of CLP-induced septic hearts, we characterized the transcriptomes of LV collected from mice 24 h after CLP and sham treatment. Accordingly, the CLP mice were divided into 3 groups (HEF, LEF, and NEF) for transcriptomic analysis based on LVEF measured at 24 h after CLP. Principal component analysis (PCA) showed the transcriptomes of three CLP groups were distinct from that of sham group (Figure S3A). We next analyzed the deferential expressed genes (DEGs) in comparisons of 3 CLP groups versus sham group, respectively. 3 CLP groups display profound transcriptional changes when compared to sham group (Figures 4A–4D). Gene Ontology and combined pathway (KEGG and Reactome) analysis by using metascape reveal that inflammatory response and the associated terms (such as regulation of cytokine production, leukocyte cell-cell adhesion) are the most over-representative molecular signatures among up-regulated genes shared in 3 CLP versus sham comparisons (Figures S3B–S3D), suggesting an increased infiltration and activation of immune cells during CLP-induced sepsis. Extracellular matrix related terms (such as collagen and basement membrane) are most significantly enriched in down-regulated genes in comparisons of 3 different CLP group versus sham (Figures S3B–S3D). This support the notion that extracellular matrix remodeling is essential pathological feature in many types of myocardial injury.12 Notably, we observed that down-regulated genes in HEF and LEF groups, but not in NEF are enriched for GO terms related to channel activity (sodium channel, potassium channel) (Figures S3B–S3D). These results suggest that disturbance of ion channel activity, critical for maintaining normal heart rhythm and function, might contribute to LVEF changes and cardiac dysfunction after sepsis-induced cardiac dysfunction.
Figure 4.
Differential expressed genes and enriched gene sets of left ventricles from mice 24 h post CLP
(A) Venn plot of significantly down-regulated and up-regulated genes (adjusted p < 0.05, fold change > 2) in comparisons of each CLP group versus sham. (HEF, LVEF > =90%), low LVEF (LEF, LVEF < 65%), and normal LVEF (NEF, 65% = < LVEF < 90%).
(B–D) Volcano plots illustrate the differentially expressed genes in comparisons of HEF versus sham (B), LEF versus sham (C), NEF versus sham (D). Top 20 differential expressed genes are labeled in the plots.
(E–G) Enrichment map of gene set enrichment analysis (GSEA) of HEF versus sham (E), LEF versus sham (F), NEF versus sham (G) node cutoff, p < 0.01 and FDR < 0.15; edge cutoff, overlap coefficient > 0.375, NSE < −1.7 and > 1.7. GSEA terms associated with upregulated (red) and downregulated (blue) genes are grouped into nodes with associated terms.
To archive a more comprehensive understanding of the pathways altered in response to CLP treatment, gene set enrichment analysis (GSEA) on transcriptomes from three CLP groups (HEF, LEF, and NEF) and sham group were performed. The gene sets are more pronouncedly enriched in upregulated than downregulated transcriptomes. Furthermore, we conducted clustering analysis of enriched gene sets using CytoScape EnrichmentMap.
The highly enriched clusters positive correlated to HEF transcripts included several immunity-related pathways such as neutrophil chemotaxis, mast cell immunity, dendritic cell differentiation, TNF and cytokine production, receptor signaling (Figure 4E). In addition, enrichment of clusters including blood coagulation, platelet growth, vascular endothelial growth, and smooth muscle proliferation suggested the microvascular endothelial dysfunction during HEF-related sepsis-induced cardiomyopathy (Figure 4E).
LEF transcripts contain similar enriched clusters as HEF transcripts, such as mast cell immunity, TNF production, and platelet growth (Figure 4F). Moreover, the largest enriched cluster positive associated with LEF transcripts is regulation of tissue remodeling (Figure 4F), indicating the regeneration of injured cardiac tissues during LEF-related cardiomyopathy. Notably, enrichment map nodes associated with upregulated genes in LEF transcripts include cellular response to LPS (Figure 4F). This result confirmed the molecular similarity in LEF group with LPS-injected mice, both of which exhibiting reduced LVEF (Figure 2).
Despite enrichment of many immune response-related gene sets (for example, cytokine signaling, TNF receptor binding, NF-κB signaling, phagocytosis) from upregulated genes in NEF group, reactive species, apoptosis, and nucleus import are also markedly enriched (Figure 4G). Increased mitochondrial-mediated production of reactive species and cell apoptosis might contribute to the cardiac dysfunction with preserved ejection fraction that has been observed in many cardiovascular diseases.13,14 It is also intriguing that nucleocytoplasmic transportation-related pathways are remarkedly upregulated in NEF transcripts (Figure 4G), which reveals the convergent mechanism also observed in ischemic and dilated human hearts.15,16
Define common transcriptomic alterations in three CLP groups
We noted that many genes are coordinately regulated in all the three CLP groups compared to sham group, respectively, resembling the common transcriptional changes of sepsis-induced cardiomyopathy. Totally, we identified 242 genes coordinately upregulated, and 84 genes coordinately downregulated in three CLP groups compared to sham group (Figure 4A). To probe the convergent biological alterations in three CLP groups, we conducted a pathway and protein-protein interaction (PPI) enrichment and clustering analysis. Similar to the shared enrichment analysis among three groups, the top represented pathways were cell activation and inflammatory response in co-upregulated transcriptome (Figure 5A), and collagen-containing extracellular matrix and MHC class 2 protein complex that is required for antigen processing and presentation, in downregulated transcriptome (Figure 5C). These results further confirmed that acute inflammatory response including innate immunity and adaptive immunity, as well as extracellular matrix homeostasis is key molecular feature at 24 h after CLP. In addition, lipid- (such as lipid transport and regulation of fatty acid biosynthetic process), ROS, and striated muscle contraction-related (such as myofibril assembly and sarcomere) pathways are enriched in down-regulated DEGs (Figure 5C), indicating the perturbation of lipid biosynthesis and transport and ROS, and defects in cardiomyocyte organization during sepsis-induced cardiac dysfunction. Together, these observations are in consistent with the transcriptome analysis results on human cardiac samples after septic shock that shows widespread dysregulation of genes associated with sarcomere contraction, extracellular matrix, and fatty acid and lipid metabolism.17
Figure 5.
Enriched pathways on coordinately changed transcriptomes from distinct CLP group
(A and C) Network shows enriched pathways (Gene Ontology, KEGG, and Reactome) of co-upregulated (A) or co-downregulated (C) transcriptome in CLP versus sham. Each color represents a cluster of enriched terms and pathways. Node indicates each term or pathway. The size of node is proportional to the number of genes under that term.
(B and D) Protein-protein interaction (PPI) analysis of co-upregulated (B), or co-downregulated genes (D). Center network represents the full protein-protein interactions. The surrounding networks include densely connected proteins identified according to their functions listed.
Next, we utilized the molecular complex detection algorithm (MCODE) to characterize protein-protein interaction (PPI) clusters associated with signaling pathways. We observed PPls including regulation of leukocyte degranulation, Natural killer cell mediated cytotoxicity, serine-type endopeptidase inhibitor activity, JAK-STAT signaling pathway, TNF signaling pathway, plasma lipoprotein particle in upregulated signaling pathways (Figure 5B), and MHC class II protein complex, basement membrane in downregulated signaling (Figure 5D). Most of the PPI clusters are known to be involved in immune system. Hence these results unveil various type of immune machinery included in the acute-phase immune response during sepsis-induced cardiomyopathy.
Proteomics responses to CLP-induced cardiomyopathy
Given that mRNA and protein levels are not well correlated, transcriptome alterations only reflect partial changes of protein levels. We thus performed proteomics analysis on left heart ventricles isolated from mice of three CLP groups and sham group, which are the same samples used in the transcriptomic analysis. We only observed minimums overlap in differential expressed proteins (DEPs) with DEGs identified in same comparisons of each CLP group versus sham (Figures 6A and 6B), suggesting a lag in changes in protein expression after mRNA expression changes during early phase of sepsis. Totally, 19, 19, and 20 genes are differentially expressed in both transcription and protein levels in each CLP group compared to sham group, respectively (Figure 6B; full catalog of genes and function are listed in Table S2, the expression profile of all the genes and proteins are listed in Tables S3 and S4, respectively). Consistently, quantitative real-time PCR analysis reveals that expression levels of Hcls1, Lbp, Lcn2, Mt1, and Mt2 are increased in LV samples from CLP mice compared to that from sham treated mice (Figure S4).
Figure 6.
Proteome and specific protein networks associated with distinct CLP group
(A) Volcano plots illustrate the differentially expressed proteins in comparisons of HEF, LEF, and NEF versus sham, respectively. Top 20 differential expressed proteins are labeled in the plots.
(B) Venn plots of DEGs and DEPs in comparisons of CLP versus sham.
(C–S) WGCNA analysis on proteome from HEF/Sham (C–G), LEF/Sham (H–N), and NEF/Sham group (O–S). Module-trait relationships indicate the protein modules (module eigengenes [MEs]) associated with HEF (C), LEF (H), and NEF (O) group. Only statistically significant modules are listed. p values are shown in parentheses. Red represents a positive correlation and blue represents a negative correlation to CLP group. D and F, ME expression in HEF and Sham samples; I, K, and M, ME expression in LEF and Sham samples; P and R, ME expression in NEF and Sham samples. Box in the plots displays 25 th and 75 th percentile values, the center line represents the median and the whiskers show minimum and maximum values. Pathway enrichment analysis in the proteins with high intramodular connectivity in the midnightblue (E) and skyblue3 (G) modules in the HEF/Sham group, green (J), black (L), and steelblue (N) modules in the LEF/Sham group, as well as green (Q) and brown (S) modules in the NEF/Sham group.
Notably, pathway enrichment analysis reveals that DEPs in comparisons of three CLP groups versus sham contain set of up-regulated proteins enriched for acute inflammatory responses, and down-regulated proteins enriched for high-density lipoprotein particle (Figure S5). These data suggest that signaling pathways related to inflammatory response and lipid metabolism and transport are commonly altered in mRNA and protein levels at 24 h after CLP despite the LVEF changes. Intriguingly, proteins related to ferroptosis (Fth1, Hmox1, Map1lc3a, Map1lc3b) and metal ion homeostasis (Fth1, Hmox1, LCN2, Mt, Mt2) are jointly upregulated in all the three CLP groups despite of LVEF changes (Figures 6B and S5), suggesting that metal ion disturbance-mediated cell death might contribute to pathology of sepsis-induced cardiac dysfunction.
Coexpression analysis identifies distinct protein network in response to different LVEF changes after CLP
To define biologically essential clusters of proteins according to their co-expression patterns, we conducted weighted gene co-expression network analysis (WGCNA) that helps us to identify which protein clusters is mostly influenced by CLP induction at different LVEF levels. We constructed three sets of coexpression networks separately for HEF/Sham, LEF/Sham, and NEF/Sham. These three networks are completely independent while module names are reused across networks that did not indicate module similarity.
In HEF/Sham network, we identified 39 modules, among which module midnightblue (p = 0.0002) is the most significantly positive-associated, and module skyblue3 (p = 0.0118) is the most significantly negative-associated with HEF group (Figure 6C). Eigengenes in midnightblue module are upregulated, while that in skyblue3 are downregulated in HEF group compared to sham group (Figures 6D and 6F). The striking feature of midnightblue module is particularly strong overrepresentation of acute response proteins (Figure 6E). Top hub proteins (Orm1, Orm2, Saa1, and Saa2), that are associated with acute inflammatory response are markedly upregulated in HEF group (Figures 6E and 7A), suggesting a rapid immune response to pathogen invasion after CLP induction. Module skyblue3 contains hub genes related to carbohydrate metabolic process (Rbp4, Mgat2), retinoid metabolism and cholesterol homeostasis (Apoa2, Ttr, Rbp4, Cav3) (Figures 6G and 7B), indicating that CLP might disrupt energy and lipid homeostasis in the heart.
Figure 7.
Protein-protein interaction analysis of module proteins from WGCNA
(A–G) Network plots of the proteins with high intramodular connectivity in the midnightblue (A) and skyblue3 (B) modules in the HEF/Sham group, green (C), black (D), and steelblue (E) modules in the LEF/Sham group, as well as green (F) and brown (G) modules in the NEF/Sham group. The size of node (named node degree), represents degree of association determined by the number of edges associated with this node, more the number of edges associated with this node, the bigger the node is.
In LEF/Sham network, WGCNA showed that green module (p = 0.0006) has a significant positive correlation, black (p = 0.0175) and steelblue module (p = 0.0200) have negative correlation with LEF group (Figure 6H). Green module eigengenes are upregulated in LEF group (Figure 6I). Functional annotation reveals that plasma lipid protein particle (Vldlr, Apoc4, Rbp4, Lpl, Saa1, Saa2) and mitophagy (Bnip3, Sqstm1, Mapk9, Foxo3, Gabarapl1, Map1lc3a, Map1lc3b) are highly enriched in green module (Figures 6J and 7C). Increased expression of mitophagy proteins indicates mitochondria dysregulation and clearing of damaged mitochondria during CLP-induced cardiomyopathy, which is in agreement with previous rat and human proteomic studies showing the mitochondria and energy production defects associated with sepsis-induced cardiac dysfunction.17,18 Black module is strikingly associated with complement cascade, endopeptidase regulator activity, collagen-containing extracellular matrix, and regulation of blood coagulation (Figures 6L and 7D). In particular, many complement proteins (for example, Serping1, C4b, C9, Cfi, F2, Hc, Cpn2, C8b, C8a) are downregulated in LEF group compared to sham (Figure 6L). Increased protein expression of distinct subset of components of complement cascade, including C1qb, C1qc, C3, has been reported in SARS-CoV-2-associated myocardial inflammation,19 suggesting the complication in regulation of complement cascade in different myocardial inflammation conditions. Steelblue is a small module containing 35 proteins, in which many proteins (for example, Apoa2, Pon1, Slc27a1, Agpat2) are related to glycerophospholipid metabolic process (Figures 6N and 7E), again suggesting the complicated disruption of lipid metabolism in sepsis-induced cardiomyopathy.
Moreover, WGCNA analysis on NEF/Sham network reveals that green (p = 0.0022) and brown (p = 0.0149) module show markedly positive association with NEF group, while lightyellow (p = 0.0323) and yellow (p = 0.0487) module show moderately negative association with NEF (Figure 6O). Strikingly, GO enrichment analysis reveals that terms like cellular component disassembly, nuclear membrane reassembly, are significantly overrepresented in green module (Figures 6Q and 7F). In particular, proteins related to nuclear membrane/nuclear pore complex, such as chmp2b, chmp7, Spast, show subtle, but significant upregulation in NEF group (Figures 6Q and 7F), implying that repair and homeostasis of nuclear pore complex are involved in pathology of sepsis-induced myocardiopathy. This result is also consistent with selective enrichment of nucleus import pathway revealed by GSEA analysis on NEF transcripts (Figure 4G). Some proteins related to mitochondrial envelope (for example, Acsl3, Eci1, Slc25a15) and fatty acid β-oxidation (for example, Acox1, Acsl3, Eci1, Sting1, Lpl) are upregulated in NEF group (Figures 6Q and 7F). Fatty acid β-oxidation is the major pathway utilized by mitochondria to degrade fatty acids, maintaining energy homeostasis.20 CLP induction may cause dysregulation of mitochondria and perturbation of energy homeostasis in heart even without LVEF changes. Brown module is a large module containing 525 proteins, which is enriched for terms including lipid, ribonucleoprotein complex biogenesis and translation, autophagy and mitophagy, ferroptosis (Figures 6S and 7G), suggesting diversified mechanism underlying sepsis-induced cardiac dysfunction. Notably, increased expression of ribosomal proteins in NEF group indicates that enhanced protein synthesis under sepsis-induced acute response.
Morphological abnormalities associated with CLP-induced cardiomyopathy
To characterize the morphological changes in heart after CLP, we performed HE staining on LV samples collected from mice of three different CLP group at 24 h after CLP. Results showed that focal pyknosis and necrosis occur in myocardial cells, and disorientated sarcomere and interstitial edema in three LVEF groups. These morphological changes are striking in HEF and LEF group, whereas are moderate in NEF group at 24 h after CLP induction (Figure 8A). As all subjects from LEF group were dead before 30 days after CLP, we were not able to assess the recovery of morphological changes for long-term post CLP. Notably, HE staining of left ventricle samples collected from HEF and LEF mice 30 days post CLP indicates that the morphological defects are alleviated at long-term period after sepsis (Figure 8A), suggesting a reversal of cardiac structural abnormalities that is in accordance with myocardial remission observed in patients with septic cardiomyopathy.21
Figure 8.
Defects in cardiac mitochondria and sarcomere in CLP mice
(A) HE staining of left ventricle samples from HEF, LEF, NEF, and Sham groups. The right image in each panel is in high-magnification. Scale bar = 50 μm in left image; Scale bar, 20 μm in right image.
(B) Representative images from TEM examination. Scale bar is labeled in each image.
(C–H) Quantitative analysis of TEM images from experiments in B. Mitochondria size (C), aspect (AR) ratio (D), circularity (E), number (F), solidity (G), and percentage of abnormal cristae (H) were quantified. n = 6–7 imaging fields from 2 mice per group (F and H), n = 194–435 mitochondria from 2 mice per group (C, D, E and G).
(I–K) Quantitative analysis of sarcomere at TEM images from experiments in B. Mitochondria size (C), aspect (AR) ratio (D), circularity (E), number (F), solidity (G), and percentage of abnormal cristae (H) were quantified. n = 9–12 images from 2 mice per group (I and J), n = 3–9 images from 2 mice per group (K).
(L) Z score normalized FPKM (Fragments Per Kilobase Million) of genes with significant association to sarcomere. Data are shown as mean ± SEM. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Student’s t test.
TEM reveals defects in mitochondria and cardiac myofibril in CLP mice
We next examined the ultrastructure of left ventricle cardiac tissues from mice of three CLP group by using TEM. Strikingly, we observed marked, but distinct morphological changes in mitochondria from LV cardiomyocytes in three CLP group, respectively (Figure 8A). All the three CLP group shows decreased mitochondrial area, increased AR ratio and abnormal mitochondrial circularity compared to sham group (Figures 8B–8E). However, number of cardiomyocytes mitochondria is increased in HEF group, but not in LEF and NFF group compared to sham group (Figure 8F), suggesting disturbance in mitochondrial fusion/fission dynamics. By contrast, cardiomyocytes in NEF and LEF group had considerably number of mitochondria compared to that in sham group, but their mitochondria are severely injured and the cristae structure is drastically broken (Figures 8G and 8H). The disruption of mitochondria structure in NEF and LEF group is in agreement with our WGCNA analysis showing specific association of mitophagy and mitochondria-related protein coexpression modules.
Moreover, we found remarkable abnormalities in cardiac myofibril and sarcomere with distinct alterations among three CLP group (Figures 8B and 8I–8K). HEF group shows dramatic cardiac myofibril disorientation, which is slightly changed in NEF group and remain intact in LEF group (Figures 8B and 8I). In addition, type 2,3 Z-bands are crushed into disorientated sarcomere in HEF group, but remain unchanged in other two groups (Figures 8B and 8J). In contrast, LEF and NEF, but not HEF group exhibited abnormal type 1 Z-band in cardiomyocyte (Figures 8B and 8K). Consistently, transcriptomic analysis indicated a genome-wide change in expression of myofibril and sarcomere-related genes in CLP-induced mice (Figure 8L).
Discussion
Measurement of LVEF has been widely conducted to assess cardiac function of septic patients in ICU. In this study, we performed echocardiogram in rodent sepsis model to examine the LVEF at different time after CLP and LPS challenge, with aim to delineate the temporal dynamics of LVEF after sepsis induction. LPS has been used to induce endotoxemia in rodents that mimic an acute inflammatory response to gram-negative bacteria infection. Our study shows that challenge with LPS leads to reduction of LVEF (% reduction compared to saline treated mice) in mice at 6 h after LPS injection, the depression of LVEF recovers to a level that is comparable to saline-treated mice at 15 h (Figure 2I). These results suggest that single-dose injection of LPS cause rapid, but reversible depression of LVEF in mice. Intriguingly, we unveil that CLP induction that models polymicrobial sepsis causes a much more complicated LVEF response than LPS treatment. Three different subtypes based on LVEF (HEF, LVEF > = 90%; LEF, LVEF < 65%; and NEF, 65% = < LVEF < 90%) assessed at 24 h after CLP were observed in both mice and rat model (Figures 2E–2G and 3C). With this strategy, we observed that fluctuation of LVEF toward high level or low level within 24 h post CLP brings poor prognosis, whereas, mice or rats with stable level of LVEF have better survival (Figures 2C, 2D and 3B). Hence, maintenance of a stable LVEF might be beneficial for the long-term outcome after sepsis. These data support the previous clinical assessment that both hyperdynamic or depressed LVEF in ICU have increased mortality or related to chronic cardiovascular disease.4,8,22
Here, by using combination of echocardiogram, transcriptomic and proteomic profiling, and histological and TEM examination, we further depict the molecular and structural alterations in CLP-induced septic mice model with different LVEF. Immune response is known to be involved in pathology of sepsis. We defined a serial of immune-related molecular interacting clusters in convergent upregulated transcriptome in all the CLP groups, such as regulation of leukocyte degranulation, natural killer cell mediated cytotoxicity, serine-type endopeptidase inhibitor activity, JAK-STAT signaling pathway, TNF signaling pathway, plasma lipoprotein particle (Figure 5B); whereas interacting cluster of MHC class II protein complex that is essential for adaptive immune response,23 is convergently down-regulated in transcriptomes of all the CLP mice (Figure 5D). These findings suggest that innate and adaptive immune systems are affected in different way during sepsis-induced cardiac dysfunction. In addition, we find down-regulated cluster of extracellular matrix organization and upregulated cluster of collagen catabolic process in transcriptomes in all the CLP mice LVEF changes (Figure 5D). These results are in line with previous reports that decreased collagen content in rat CLP model and dysregulated expression of extracellular matrix-related genes in postmortem heart from septic patients.17,24 Cardiac extracellular matrix provides essential mechanical support and transduce molecular signaling, and dynamics of extracellular matrix may trigger inflammation and repair upon myocardial injury.12 Our data hence suggest that disruption of extracellular matrix is key pathological event at early phase of cardiac dysfunction associated with sepsis.
By using TEM examination, we identify two prominent morphological changes after CLP induction. Mitochondria deficits have been implicated in many types of cardiac dysfunction.14,25,26,27,28,29 Transcriptional profiling on human postmortem heart also indicates that expression levels of many mitochondrial-related genes are changed in cardiac tissues from septic patients.17 Consistently, we observed that mitochondrial morphology is defective in CLP-induced cardiomyocytes. Notably, mitochondria become fragmented in HEF group (Figures 8B, 8C, and 8F), suggesting impaired mitochondria fission/fusion dynamics. By contrast, mitochondria crista is severe injured in NEF and LEF group (Figures 8B and 8H). Sarcomere dysregulation was reported in numerous cardiac disorders including sepsis-induced cardiomyopathy.17,30 Z-band is the structural backbone of cardiac sarcomere, providing a site for various signal transduction in cardiomyocytes.31,32 TEM examination reveals that cardiac myofibril is disorganized in HEF group of CLP mice, specifically, type 2, 3 Z-bands are disrupted (Figures 8B, 8I, and 8J). However, type 1 Z-band abnormality is observed in LEF and LEF group in CLP mice (Figures 8B and 8K). Hence, these results characterize the distinct ultrastructure changes in cardiomyocyte in different CLP subtypes.
Sepsis-induced cardiomyopathy lacks clear definition yet. Characteristics such as reduced LV contractility, LV dilation with not high filling pressure, RV dysfunction have been described in sepsis-associated acute syndrome of cardiac dysfunction.1 Animal studies reported that LVEF has decreased systemic vascular resistance (SVR), increased CO, DO2 (oxygen delivery), VO2 (oxygen consumption), and the inverse relationship of LVEF and SVR represents ventricular-arterial coupling in a porcine model.33,34 In this study, by assessing hemodynamics parameters from CLP-induced rat, we found that CLP induction leads to elevated mean CO. However, when compared to survivors after CLP, non-survivors had lower CO and velocity time integral (VTI) (Figure 3).
Our study represents a systematic analysis of transcriptional and translational variants in CLP-induced septic cardiac dysfunction with different LVEF. The strength of our study is that we uncover the temporal dynamics of LVEF with two well-established sepsis model, providing detailed information of LVEF changes associated with CLP- and LPS challenge-induced cardiomyopathy. The three subtypes of sepsis-induced cardiomyopathy based on LVEF are also in accordance with clinical practice and reports.8 Moreover, our study present, for the first time, a comprehensive list of changed genes and pathways in heart associated with different LVEF, providing new insight into the pathophysiology of sepsis-induced cardiomyopathy.
Limitations of the study
However, there are still some limitations of our study. The morphological changes and molecular mechanisms of the three subtypes need clinical validations. Moreover, current study reveals the changes in mRNA and protein levels at 24 h after CLP. The molecular alterations at early time point, such as 3 h or 6 h after CLP will be investigated in the future study to uncover the temporal dynamics of molecular signature during sepsis-induced cardiac dysfunction. In addition, given the diversified cell types in heart, single cell transcriptome analysis may help to understand the cell-type-specific contributions to pathology of sepsis-induced cardiomyopathy with different LVEF.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| LPS | Sigma | Cat#L2630-100mg |
| Electron microscope fixative fluid | Wuhan Servicebio Biotechnology | Cat#G1102-100ML |
| TransZol | TransGen Biotech | Cat#ET111-01-V2 |
| Critical commercial assays | ||
| BCA Protein Assay Kit | Bio-Rad | Cat#5000002 |
| High pH Reversed-Phase Peptide Fractionation Kit | Thermo Scientific | Cat#84868 |
| NovoScript Plus All-in-one 1st Strand cDNA Synthesis SuperMix | Novoprotein | Cat#E047 |
| PerfectStart Green qPCR SuperMix | TransGen Biotech | Cat#AQ611-01 |
| Deposited data | ||
| RNA-seq raw and analyzed data | This paper | GEO: GSE229925 |
| Proteomics raw and analyzed data | This paper | ProteomeXchange ID: PXD041530 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | Hunan SJA Laboratory Animal Co. Ltd | N/A |
| Rat: Sprague-Dawley | Hunan SJA Laboratory Animal Co. Ltd | N/A |
| Oligonucleotides | ||
| PCR primers (for full sequence, see Table S5) | This paper | Custom primer sets |
| Software and algorithms | ||
| MASCOT engine version 2.2 | Matrix Science | https://matrixscience.com |
| Proteome Discoverer | Thermo Scientific | Cat#OPTON-31101 |
| Broad GSEA software (3.0) | UC San Diego and Broad Institute | http://software.broadinstitute.org/gsea |
| EnrichmentMap Cytoscape App | Merico et al.35 | https://apps.cytoscape.org/apps/enrichmentmap |
| Metascape | Zhou et al.36 | https://metascape.org/gp/index.html#/main/step1 |
| Other | ||
| Easy nLC | Thermo Scientific | Cat#LC140 |
| Q exactive mass spectrometer | Thermo Scientific | Cat#IQLAAEGAAPFALGMAZR |
| Vivid IQ ultrasound | GE | https://www.gehealthcare.com/zh-cn/products/ultrasound/vivid/vivid-iq-ultra-edition |
| QuantStudio5 Real-Time PCR system | Applied Biosystems | Cat#A36320 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zhaoxin Qian (xyqzx@csu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Animals
6-8 weeks male C57BL/6 mice and 260-300g weight male SD rat (Hunan SJA Laboratory Animal Co. Ltd) were housed in 12 h light and 12 h dark cycle and temperature-controlled room. Our animal research was approval by Experimental Animal Ethics Committee, Xiangya Hospital, Central South University (202110109) and strictly abided by “3R” research animal principle.
We constructed well accepted animal model of sepsis: polymicrobial peritonitis induced by cecum ligation and puncture (CLP), and endotoxemia model by intraperitoneal injection of 15 mg/kg lipopolysaccharide (LPS). The CLP model was built as depicted previously.37 Briefly, after induced anesthesia by proper concentration of isoflurane maintained by anesthesia machine for small animals, the cecum was ligated at 1 cm from distal pole and then perforated single through-and-through puncture with 18-gauge needle, avoiding injuring blood vessels. After removing puncture needle, feces of mung bean size were squeezed out from one puncture hole. The ligation position of 1 cm from distal pole and the amount of excluded feces were strictly maintained the same in all animals to guarantee consistency. Sham group was operated all the same as experimental group except for ligation and puncture procedure. After surgery, animals were resuscitated with prewarmed 1 mL saline, and were wrapped in towel and placed on prewarmed heating pad.
Study participant details
From August 2017 to February 2019, critically ill patients’ data were collected from our multi-centered intensive care units (ICU) project (NCT number ChiCTR1800015056). Informed consent forms were signed consent by recruited patients or their family members. Patients in this project diagnosed with Sepsis 3.0 criterion were included for secondary analysis in our study.38 The inclusion criteria were age equal or greater than 18 years old and meeting diagnostic criterion of Sepsis 3.0 (n = 939). De-identification principle was used to protect personal privacy of patients. Continuous variables were stated as medians with interquartile ranges (IQR) or means with standard deviations (SD) depending on distribution of data. Categorical variables were stated as frequencies (percentages).
As there is no generally accepted criterion to distinguish different variation trend of cardiac function after stress of sepsis in clinical practice, we use left ventricular ejection fraction (LVEF) less than 50% as lower limit to define reduced or depressed left ventricular systolic function, that is low LVEF group in our research, according to heart failure guideline set by European Society of Cardiology (ESC).39 And LVEF higher than or equal to 70% was employed as upper limit to define hyperdynamic left ventricular ejection fraction (HDLVEF) and hyperdynamic shock, namely high LVEF group in our study, based on the American College of Cardiology (ACC) guidelines. And then LVEF higher than or equal to 50% and less than 70% was defined as heart failure with preserved ejection fraction (HFpEF) subtype, that is normal LVEF group in our research, according to heart failure guideline of ESC.39 On the basis of above-mentioned grouping criteria, 939 patients were classified into three groups: low LVEF group (n = 255), high LVEF group (n = 152) and normal LVEF group (n = 532).
Method details
Hemodynamic monitoring by ultrasound
Small animal ultrasound was proceeded under ultrasonic instrument of Vivid IQ (American GE Co. Ltd). Ultrasonic operation was executed by two experienced operators with more than 7 years rich experience of ultrasonic operation. And any ultrasonic measurement value was repeated three to four times by the two operators and calculated average value to diminish inter-operators’ error.
Histology
Mice myocardium was rapidly collected at 24 h and 30 days after CLP or sham operation and immersed into formalin for 24 h, and then ventricular section was paraffin-embedded. The sections were cut 4 μm and stained with hematoxylin and eosin.
Transmission electron microscope (TEM)
Mice myocardium was fixed in electron microscope fixative fluid (Wuhan Servicebio Biotechnology Co. Ltd) at 24 h after CLP or sham operation, then staying overnight under 4°C. Then after sequential steps of washing, fixation with osmic acid, washing, dehydration with acetone, resin embedding, baking, slicing by Leica EMUC7 ultramicrotome and staining with uranyl acetate and lead citrate. Images were taken under 1400plus JEOL TEM.
RNA sequencing and transcriptomic analysis
Mice myocardium tissue was rapidly collected at 24 h after CLP or sham operation and immersed into liquid nitrogen for quick freezing for following transcriptome and proteomics analysis. RNA sequencing was processed by Novogene Co., Ltd. Then mRNA was purified from total RNA by using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase. Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and dNTP. The library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then PCR amplification, the PCR product was purified by AMPure XP beads. After the library is qualified, the different libraries are pooling according to the effective concentration and the target amount of data off the machine, then being sequenced by the Illumina NovaSeq 6000. The end reading of 150 bp pairing is generated. Four fluorescent labeled dNTP, DNA polymerase and splice primers were added to the sequenced flow cell and amplified. When the sequence cluster extends the complementary chain, each dNTP labeled by fluorescence can release the corresponding fluorescence. The sequencer captures the fluorescence signal and converts the optical signal into the sequencing peak by computer software, so as to obtain the sequence information of the fragment to be tested. EdgeR R package was used to conduct significance analysis of expression difference. Adjusted p value (Padj) was obtained by Benjamini - Hochberg (BH) method to control false positive rate. Significantly differential expression genes were identified based on the following criteria: Adjusted P-value <0.05 and absolute foldchange >2.
Quantitative Proteomics
Quantitative Proteomics was processed by Applied Protein Technology Co., Ltd. After sample lysis and protein extraction, the amount of protein was quantified with the BCA method. Then protein was digested and the digest peptides of each sample were desalted on C18 Cartridges, concentrated by vacuum centrifugation and reconstituted in 40 μL of 0.1% (v/v) formic acid. Then samples were operated through filter-aided sample preparation (FASP Digestion) procedure. 20 μg of protein for each sample were mixed with 5X loading buffer respectively and boiled for 5 min. The proteins were separated on 12.5% SDS-PAGE gel (constant current 14 mA, 90 min). Protein bands were visualized by Coomassie Blue R-250 staining. 100 μg peptide mixture of each sample was labeled using TMT reagent according to the manufacturer’s instructions (Thermo Scientific). Labeled peptides were fractionated by High pH Reversed-Phase Peptide Fractionation Kit (Thermo Scientific). LC-MS/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Scientific) that was coupled to Easy nLC (Proxeon Biosystems, now Thermo Fisher Scientific) for 60/90 min. The MS raw data for each sample were searched using the MASCOT engine (Matrix Science, London, UK; version 2.2) embedded into Proteome Discoverer 1.4 software for identification and quantitation analysis.
Gene set enrichment analysis (GSEA)
GSEA of each comparison was performed with Broad GSEA software (3.0) (http://software.broadinstitute.org/gsea).40 GSEA results were further visualized with EnrichmentMap Cytoscape App,35,41 and enriched pathway clusters were manually curated.
Pathway enrichment and protein-protein interaction analysis
Pathway (Gene Ontology, KEGG, Reactome) and protein-protein interaction (PPI) enrichment were performed using Metascape (http://www.metascape.org/).36 Function-related sub-networks in PPI network were clustered by Molecular Complex Detection (MCODE) algorithm.42
Weighted gene co-expression network analysis (WGCNA)
WGCNA is a systematic biological method used to describe the gene association modes among different samples.43 It can be used to identify gene sets that are highly synergistic changed, and identify candidate biomarkers or therapeutic targets based on the coherence of gene sets and the correlation between gene sets and phenotypes. Our proteomics data of relative expression was used as input for WGCNA. We first assumed that the proteins network followed a scale-free distribution and defined the protein co-expression matrix and the adjacency function formed by protein network, and then calculated the different coefficients of different nodes, and construct the hierarchical clustering tree. The different branches of the cluster tree represented different protein modules. The coexpression degree of proteins in the same module was high, while the coexpression degree of proteins belonging to different modules was low. Then we identified modules that are significantly associated with the trait of each subgroup of HEF, LEF and NEF by calculating correlation coefficient matrix with threshold of p < 0.05 and correlation coefficient close to plus one or minus one. Topological Overlap Matrix (TOM) was used for cluster heatmap analysis, so as to reflect the correlation between protein expression levels among different co-expression modules. We used Gene Significance (GS) to calculate correlation between protein and sample/trait of each subgroup, Module Membership (MM) to represent correlation between module and sample/trait of each subgroup. Scatterplots to reflect correlation relationship between GS and MM was plotted to find proteins that their correlation with GS and correlation with MM had good concordance (p < 0.05). These proteins were defined as hub proteins in each module (MM threshold was set greater than 0.8). Pathway enrichment analysis on these hub proteins was carried out by using Metascape; protein-protein interaction networks (PPI) of these hub proteins were generated by String (https://string-db.org), and visualized by Cytoscape to dissect interaction network and core proteins.
Quantitative real-time PCR analysis
24 h after CLP or sham surgery, the cardiac tissues were isolated from mice. Total RNA was extracted from LV cardiac tissue using TransZol (TransGen Biotech, Beijing, China), and reverse-transcribed into cDNA using a cDNA Synthesis SuperMix with gDNA Eraser (Novoprotein, Suzhou, China) according to the manufacturer’s instructions. Quantitative PCR was performed using PerfectStart Green qPCR SuperMix (TransGen Biotech, Beijing, China) and specific primers (See Table S5) using a QuantStudio5 Real-Time PCR system (Applied Biosystems, USA).
Quantification and statistical analysis
The data in this study are shown as mean ± SEM as noted in individual figure legends. Statistical analyses were performed using GraphPad PRISM software. The unpaired Student’s 2-tailed t test were used to assess the difference between groups. Chi-square test was employed to analyze difference between three groups. The Kaplan-Meier survival curve was used to present cumulative incidence of percentage of death (POD) in period of 30 days for follow up, analyzed with log rank test, and p value was adjusted by Bonferroni method.
Acknowledgments
We thank all the ICU investigators and participant patients for the clinical data. This work was supported by the National Natural Science Foundation of China (#31872778, 82171506, 82172145); the Discipline Innovative Engineering Plan (111 Program) of China (#B13036); Key Research and Development Programs from Hunan Province (# 2021DK2001, 2022SK2039); innovative team program from Hunan Province (#2019RS1010); Science and Technology Major Project of Hunan Provincial Science and Technology Department (2021SK1010); key laboratory grant from Hunan province (2016TP1006); international science and technology innovative cooperation center from Hunan Province (022CB1004); Natural Science Foundation of Changsha City (grant numbers kq2202381); the innovation-driven team project from Central South University (#2020CX016); Central South University Research Programme of Advanced Interdisciplinary Studies (#2023QYJC022), Z.H. is supported by the Hunan Hundred Talents Program for Young Outstanding Scientists. The graphical abstract was drawn using Figdraw.
Author contributions
L.Z., Z.H., and Z.Q. conceived and supervised the study. D.Q. and L.Z. performed the primary experiments and analyses. M.P., B.M., X.W., and X.Z. contributed to the experiments and analyses. Z.Z. performed quantitative real-time PCR analysis. L.L. and Z.W. contributed to analyses and results interpretation. W.Z. performed TEM analysis. L.Z., D.Q., Z.H., and Z.Q. drafted the first version of the manuscript, and all authors contributed to the final version of the paper. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: September 7, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107825.
Contributor Information
Zhonghua Hu, Email: huzhonghua@csu.edu.cn.
Zhaoxin Qian, Email: xyqzx@csu.edu.cn.
Supplemental information
Document S1. Figures S1–S5, Tables S1, S2, and S5
Data and code availability
-
•
RNA-seq and proteomic data have been deposited at GEO and ProteomeXchange and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Hollenberg S.M., Singer M. Pathophysiology of sepsis-induced cardiomyopathy. Nat. Rev. Cardiol. 2021;18:424–434. doi: 10.1038/s41569-020-00492-2. [DOI] [PubMed] [Google Scholar]
- 2.Geri G., Vignon P., Aubry A., Fedou A.L., Charron C., Silva S., Repessé X., Vieillard-Baron A. Cardiovascular clusters in septic shock combining clinical and echocardiographic parameters: a post hoc analysis. Intensive Care Med. 2019;45:657–667. doi: 10.1007/s00134-019-05596-z. [DOI] [PubMed] [Google Scholar]
- 3.Habimana R., Choi I., Cho H.J., Kim D., Lee K., Jeong I. Sepsis-induced cardiac dysfunction: a review of pathophysiology. Acute Crit. Care. 2020;35:57–66. doi: 10.4266/acc.2020.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wehner G.J., Jing L., Haggerty C.M., Suever J.D., Leader J.B., Hartzel D.N., Kirchner H.L., Manus J.N.A., James N., Ayar Z., et al. Routinely reported ejection fraction and mortality in clinical practice: where does the nadir of risk lie? Eur. Heart J. 2020;41:1249–1257. doi: 10.1093/eurheartj/ehz550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvin J.E., Driedger A.A., Sibbald W.J. An assessment of myocardial function in human sepsis utilizing ECG gated cardiac scintigraphy. Chest. 1981;80:579–586. doi: 10.1378/chest.80.5.579. [DOI] [PubMed] [Google Scholar]
- 6.Parker M.M., Shelhamer J.H., Bacharach S.L., Green M.V., Natanson C., Frederick T.M., Damske B.A., Parrillo J.E. Profound but reversible myocardial depression in patients with septic shock. Ann. Intern. Med. 1984;100:483–490. doi: 10.7326/0003-4819-100-4-483. [DOI] [PubMed] [Google Scholar]
- 7.Parker M.M., Shelhamer J.H., Natanson C., Alling D.W., Parrillo J.E. Serial cardiovascular variables in survivors and nonsurvivors of human septic shock: heart rate as an early predictor of prognosis. Crit. Care Med. 1987;15:923–929. doi: 10.1097/00003246-198710000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Chotalia M., Ali M., Hebballi R., Singh H., Parekh D., Bangash M.N., Patel J.M. Hyperdynamic Left Ventricular Ejection Fraction in ICU Patients With Sepsis. Crit. Care Med. 2022;50:770–779. doi: 10.1097/CCM.0000000000005315. [DOI] [PubMed] [Google Scholar]
- 9.Sang Z., Zhang P., Wei Y., Dong S. miR-214-3p Attenuates Sepsis-Induced Myocardial Dysfunction in Mice by Inhibiting Autophagy through PTEN/AKT/mTOR Pathway. BioMed Res. Int. 2020;2020:1409038. doi: 10.1155/2020/1409038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han D., Li X., Li S., Su T., Fan L., Fan W.S., Qiao H.Y., Chen J.W., Fan M.M., Li X.J., et al. Reduced silent information regulator 1 signaling exacerbates sepsis-induced myocardial injury and mitigates the protective effect of a liver X receptor agonist. Free Radic. Biol. Med. 2017;113:291–303. doi: 10.1016/j.freeradbiomed.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Yang S., Chung C.S., Ayala A., Chaudry I.H., Wang P. Differential alterations in cardiovascular responses during the progression of polymicrobial sepsis in the mouse. Shock. 2002;17:55–60. doi: 10.1097/00024382-200201000-00010. [DOI] [PubMed] [Google Scholar]
- 12.Frangogiannis N.G. The extracellular matrix in myocardial injury, repair, and remodeling. J. Clin. Invest. 2017;127:1600–1612. doi: 10.1172/JCI87491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfeffer M.A., Shah A.M., Borlaug B.A. Heart Failure With Preserved Ejection Fraction In Perspective. Circ. Res. 2019;124:1598–1617. doi: 10.1161/CIRCRESAHA.119.313572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sverdlov A.L., Elezaby A., Qin F., Behring J.B., Luptak I., Calamaras T.D., Siwik D.A., Miller E.J., Liesa M., Shirihai O.S., et al. Mitochondrial Reactive Oxygen Species Mediate Cardiac Structural, Functional, and Mitochondrial Consequences of Diet-Induced Metabolic Heart Disease. J. Am. Heart Assoc. 2016;5:e002555. doi: 10.1161/JAHA.115.002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molina-Navarro M.M., Roselló-Lletí E., Tarazón E., Ortega A., Sánchez-Izquierdo D., Lago F., González-Juanatey J.R., García-Pavía P., Salvador A., Montero J.A., et al. Heart failure entails significant changes in human nucleocytoplasmic transport gene expression. Int. J. Cardiol. 2013;168:2837–2843. doi: 10.1016/j.ijcard.2013.03.192. [DOI] [PubMed] [Google Scholar]
- 16.Cortés R., Roselló-Lletí E., Rivera M., Martínez-Dolz L., Salvador A., Azorín I., Portolés M. Influence of heart failure on nucleocytoplasmic transport in human cardiomyocytes. Cardiovasc. Res. 2010;85:464–472. doi: 10.1093/cvr/cvp336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matkovich S.J., Al Khiami B., Efimov I.R., Evans S., Vader J., Jain A., Brownstein B.H., Hotchkiss R.S., Mann D.L. Widespread Down-Regulation of Cardiac Mitochondrial and Sarcomeric Genes in Patients With Sepsis. Crit. Care Med. 2017;45:407–414. doi: 10.1097/CCM.0000000000002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinkelbein J., Kalenka A., Schubert C., Peterka A., Feldmann R.E., Jr. Proteome and metabolome alterations in heart and liver indicate compromised energy production during sepsis. Protein Pept. Lett. 2010;17:18–31. doi: 10.2174/092986610789909520. [DOI] [PubMed] [Google Scholar]
- 19.Weckbach L.T., Schweizer L., Kraechan A., Bieber S., Ishikawa-Ankerhold H., Hausleiter J., Massberg S., Straub T., Klingel K., Grabmaier U., et al. Association of Complement and MAPK Activation With SARS-CoV-2-Associated Myocardial Inflammation. JAMA Cardiol. 2022;7:286–297. doi: 10.1001/jamacardio.2021.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houten S.M., Violante S., Ventura F.V., Wanders R.J.A. The Biochemistry and Physiology of Mitochondrial Fatty Acid beta-Oxidation and Its Genetic Disorders. Annu. Rev. Physiol. 2016;78:23–44. doi: 10.1146/annurev-physiol-021115-105045. [DOI] [PubMed] [Google Scholar]
- 21.Mann D.L., Barger P.M., Burkhoff D. Myocardial recovery and the failing heart: myth, magic, or molecular target? J. Am. Coll. Cardiol. 2012;60:2465–2472. doi: 10.1016/j.jacc.2012.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paonessa J.R., Brennan T., Pimentel M., Steinhaus D., Feng M., Celi L.A. Hyperdynamic left ventricular ejection fraction in the intensive care unit. Crit. Care. 2015;19:288. doi: 10.1186/s13054-015-1012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roche P.A., Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 2015;15:203–216. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu P., Boughner D.R., Sibbald W.J., keys J., Dunmore J., Martin C.M. Myocardial collagen changes and edema in rats with hyperdynamic sepsis. Crit. Care Med. 1997;25:657–662. doi: 10.1097/00003246-199704000-00017. [DOI] [PubMed] [Google Scholar]
- 25.Rosca M.G., Hoppel C.L. Mitochondria in heart failure. Cardiovasc. Res. 2010;88:40–50. doi: 10.1093/cvr/cvq240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller K.A.L., Heinzmann D., Klingel K., Fallier-Becker P., Kandolf R., Kilias A., Walker-Allgaier B., Borst O., Kumbrink J., Kirchner T., et al. Histopathological and Immunological Characteristics of Tachycardia-Induced Cardiomyopathy. J. Am. Coll. Cardiol. 2017;69:2160–2172. doi: 10.1016/j.jacc.2017.02.049. [DOI] [PubMed] [Google Scholar]
- 27.Santulli G., Xie W., Reiken S.R., Marks A.R. Mitochondrial calcium overload is a key determinant in heart failure. Proc. Natl. Acad. Sci. USA. 2015;112:11389–11394. doi: 10.1073/pnas.1513047112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durand A., Duburcq T., Dekeyser T., Neviere R., Howsam M., Favory R., Preau S. Involvement of Mitochondrial Disorders in Septic Cardiomyopathy. Oxid. Med. Cell. Longev. 2017;2017:4076348. doi: 10.1155/2017/4076348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin L., Derwall M., Al Zoubi S., Zechendorf E., Reuter D.A., Thiemermann C., Schuerholz T. The Septic Heart: Current Understanding of Molecular Mechanisms and Clinical Implications. Chest. 2019;155:427–437. doi: 10.1016/j.chest.2018.08.1037. [DOI] [PubMed] [Google Scholar]
- 30.van der Velden J., Stienen G.J.M. Cardiac Disorders and Pathophysiology of Sarcomeric Proteins. Physiol. Rev. 2019;99:381–426. doi: 10.1152/physrev.00040.2017. [DOI] [PubMed] [Google Scholar]
- 31.Frank D., Kuhn C., Katus H.A., Frey N. Role of the sarcomeric Z-disc in the pathogenesis of cardiomyopathy. Future Cardiol. 2007;3:611–622. doi: 10.2217/14796678.3.6.611. [DOI] [PubMed] [Google Scholar]
- 32.Frank D., Kuhn C., Katus H.A., Frey N. The sarcomeric Z-disc: a nodal point in signalling and disease. J. Mol. Med. 2006;84:446–468. doi: 10.1007/s00109-005-0033-1. [DOI] [PubMed] [Google Scholar]
- 33.Rutai A., Zsikai B., Tallósy S.P., Érces D., Bizánc L., Juhász L., Poles M.Z., Sóki J., Baaity Z., Fejes R., et al. A Porcine Sepsis Model With Numerical Scoring for Early Prediction of Severity. Front. Med. 2022;9:867796. doi: 10.3389/fmed.2022.867796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chalkias A., Laou E., Papagiannakis N., Spyropoulos V., Kouskouni E., Theodoraki K., Xanthos T. Assessment of Dynamic Changes in Stressed Volume and Venous Return during Hyperdynamic Septic Shock. J. Person. Med. 2022;12:724. doi: 10.3390/jpm12050724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merico D., Isserlin R., Stueker O., Emili A., Bader G.D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rittirsch D., Huber-Lang M.S., Flierl M.A., Ward P.A. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat. Protoc. 2009;4:31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.D., Coopersmith C.M., et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ponikowski P., Voors A.A., Anker S.D., Bueno H., Cleland J.G.F., Coats A.J.S., Falk V., González-Juanatey J.R., Harjola V.P., Jankowska E.A., et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. J. Heart Fail. 2016;18:891–975. doi: 10.1002/ejhf.592. [DOI] [PubMed] [Google Scholar]
- 40.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reimand J., Isserlin R., Voisin V., Kucera M., Tannus-Lopes C., Rostamianfar A., Wadi L., Meyer M., Wong J., Xu C., et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019;14:482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bader G.D., Hogue C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Figures S1–S5, Tables S1, S2, and S5
Data Availability Statement
-
•
RNA-seq and proteomic data have been deposited at GEO and ProteomeXchange and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.