Abstract
NET-1 is a key chemotropic ligand that signals commissural axon migration and change in direction. NET-1 and its receptor UNC-5B switch axon growth cones from attraction to repulsion. The biophysical properties of NET-1 + UNC-5B complex has been poorly characterized. Using multi-wavelength-AUC by adding a fluorophore to UNC-5B, we were able to separate the UNC-5B sedimentation from NET-1. Using both multi-wavelength- and single-wavelength AUC, we investigated NET-1 and UNC-5B hydrodynamic parameters and complex formation. The sedimentation velocity experiments show that NET-1 exists in a monomer–dimer equilibrium. A close study of the association shows that NET-1 forms a pH-sensitive dimer that interacts in an anti-parallel orientation. UNC-5B can form equimolar NET-1 + UNC-5B heterocomplexes with both monomeric and dimeric NET-1.
Keywords: Netrin-1, Uncoordinated-5, Hetero-interaction studies between proteins, Multi-wavelength analytical ultracentrifugation, UltraScan, UltraScan-SOMO, Hydrodynamic bead modeling, Axon guidance
Introduction
Protein–protein interactions are very common in biological processes and the formation of multi-protein assemblies is common during many cellular processes. However, the nature and driving forces for many of these interactions is poorly understood. To unravel the dynamics of these networks is an area of extensive research as suggested by many publications in recent years (Wang et al. 2020; Xu et al. 2022). Targeting protein–protein interactions for drug design is a challenging, but very attractive research field (Lu et al. 2020). A pivotal requirement to the understanding of protein assemblies is an understanding of the molecular and structural basis of the simplest system of two interacting proteins.
Netrin-1 (NET-1) is a chemotropic ligand responsible for the bi-functional regulation of commissural axon migration. NET-1 controls the attraction and repulsion of the growing end of axons called growth cones. It does so by binding to several dependence receptors, notably deleted in colorectal cancer (DCC) and uncoordinated-5 (UNC-5B). NET-1 and DCC complexes signal chemoattraction, whereas NET-1 binding to UNC-5B causes chemorepulsion (Hernaiz-Llorens et al. 2021; Shao et al. 2017). While direct NET-1 and DCC interactions are structurally and biophysically characterized, the molecular interactions between NET-1 and UNC-5B are poorly understood (Finci et al. 2014; Xu et al. 2014).
NET-1 belongs to the laminin superfamily; it has a laminin VI domain with a calcium binding loop to bind Ca2+, three laminin-type epidermal growth factor-like (LE-1, 2, 3) repeats, and a C-terminal domain. LE-2 is a highly positively charged domain that binds heparin sulfates (Serafini et al. 1994; Matsumoto et al. 2007). UNC-5B is a single-pass receptor containing the following extracellular domains: Tandem Ig-1 and Ig-2, and thrombospondin-1 and thrombospondin-2, and the following cytoplasmic domains: a ZU-5 domain, a DCC binding domain, and a death domain motif. Previous studies showed that NET-1 binds to UNC-5B via LE-2 to Ig-2 (Hong et al. 1999; Kruger et al. 2004; Grandin et al. 2016; Hernaiz-Llorens et al. 2021). In this study, we used multi-wavelength analytical ultracentrifugation (MW-AUC) to characterize the dynamic complex formed between NET-1 and soluble extracellular domains of UNC-5B. Through strategic addition of a fluorophore on UNC-5B we spectrally separated the UNC-5B signal from the NET-1 signal in the sedimentation profile (Henrickson et al. 2022). We discovered that NET-1 and UNC-5B form transient heterocomplexes independent from the oligomerization of NET-1.
Materials and methods
Protein expression and purification
Gallus gallus NET-1 with truncated C-terminal domain (Uniprot ID: Q90922, amino acid [aa] 26–458), and UNC-5B (Uniprot ID: Q8K1S3, aa 25–300) all containing a thrombin cleavable C-terminal double Strep II®-tag were expressed in human embryonic kidney 293 T cells using the Sleeping beauty transposon system (Kowarz et al. 2015). Confluent cells were induced with 0.5 μg/mL Doxycycline in Dulbecco’s Modified Eagle’s Media and 2.5% fetal bovine serum. Collected media was passed through a Strep-Tactin Superflow Plus cartridge (Qiagen Inc., Toronto, Canada) to affinity purify the proteins. Subsequently, the affinity tag was removed by thrombin digestion during dialysis. The receptor UNC-5B was dialyzed into 50 mM Tris pH 7.5, 200 mM NaCl and NET-1 was dialyzed into 50 mM Tris pH 7.5, 500 mM NaCl, 500 mM (NH4)2SO4 to remove bound heparins. Final purification was performed on a Superdex 200 with the appropriate dialysis buffer.
Alexa 488 labeling of UNC-5B
Purified UNC-5B was buffer exchanged into 50 mM HEPES pH 8, 200 mM NaCl using a pre-equilibrated PD-10 column. Then ten-fold molar excess Alexa Fluor™ 488 NHS ester was added to the protein and incubated in the dark for 60 min. The unbound dye was removed via desalting column PD-10, and buffer exchanged into the analysis buffers: 50 mM Tris pH 7.5, 200 mM NaCl (TRIS pH 7.5) or 50 mM MES pH 6.5, 154 mM NaCl (MES pH 6.5) using a PD-10 column. The molar extinction coefficients of labeled and unlabeled proteins were determined by measuring the spectrum at various concentrations from 200 to 550 nm with a Genesys 10S UV–Vis spectrophotometer.
Differential scanning fluorimetry
Measurements were performed using a Prometheus Panta (NanoTemper Technologies GmbH). Standard Prometheus capillaries (PR-C002) were filled with 10 μL of NET-1 (0.5 mg/mL) with either 1 mM CaCl2 or 1 mM EDTA. The temperature was increased from 25 to 65 °C at a ramp rate of 1 °C/min, intrinsic tryptophan fluorescence was measured at 330 nm and 350 nm. Data analysis was performed with the Panta Analysis software (Version 1.2).
Analytical ultracentrifugation (AUC)
NET-1 monomer–dimer sedimentation velocity experiments (SVEs) were performed using a ProteomeLab™ XL-I analytical ultracentrifuge at the University of Manitoba and an An50Ti 8-cell rotor (Beckman Coulter Inc., Mississauga, ON). Standard 12 mm double-sector cells were filled with 400 μL of samples and run at 30,000 rpm, 20 °C for 24 h measuring intensity mode at 278 or 295 nm. Multi-wavelength AUC experiments were performed in a Beckman Optima AUC instrument (Beckman Coulter, Indianapolis) at the University of Lethbridge. Standard 12 mm 2-channel centerpiece cells were filled with 450 μL of sample per channel, diluted to the desired concentration in TRIS pH 7.5 or MES pH 6.5 and spun at 60,000 rpm for 7.5 h in an An60Ti 4-cell rotor. SVEs were measured in intensity mode. The wavelength ranges 250–300 nm and 460–500 nm were selected, in increments of 3 and 2 based on the molar extinction spectrum of NET-1 and UNC-5B (Fig. 1D). Solutions of NET-1 and UNC-5B were diluted to absorb 0.1–1 OD in a 12 mm pathlength AUC cell within the selected wavelength range. A minimum of 50 scans per wavelength were measured.
Fig. 1.
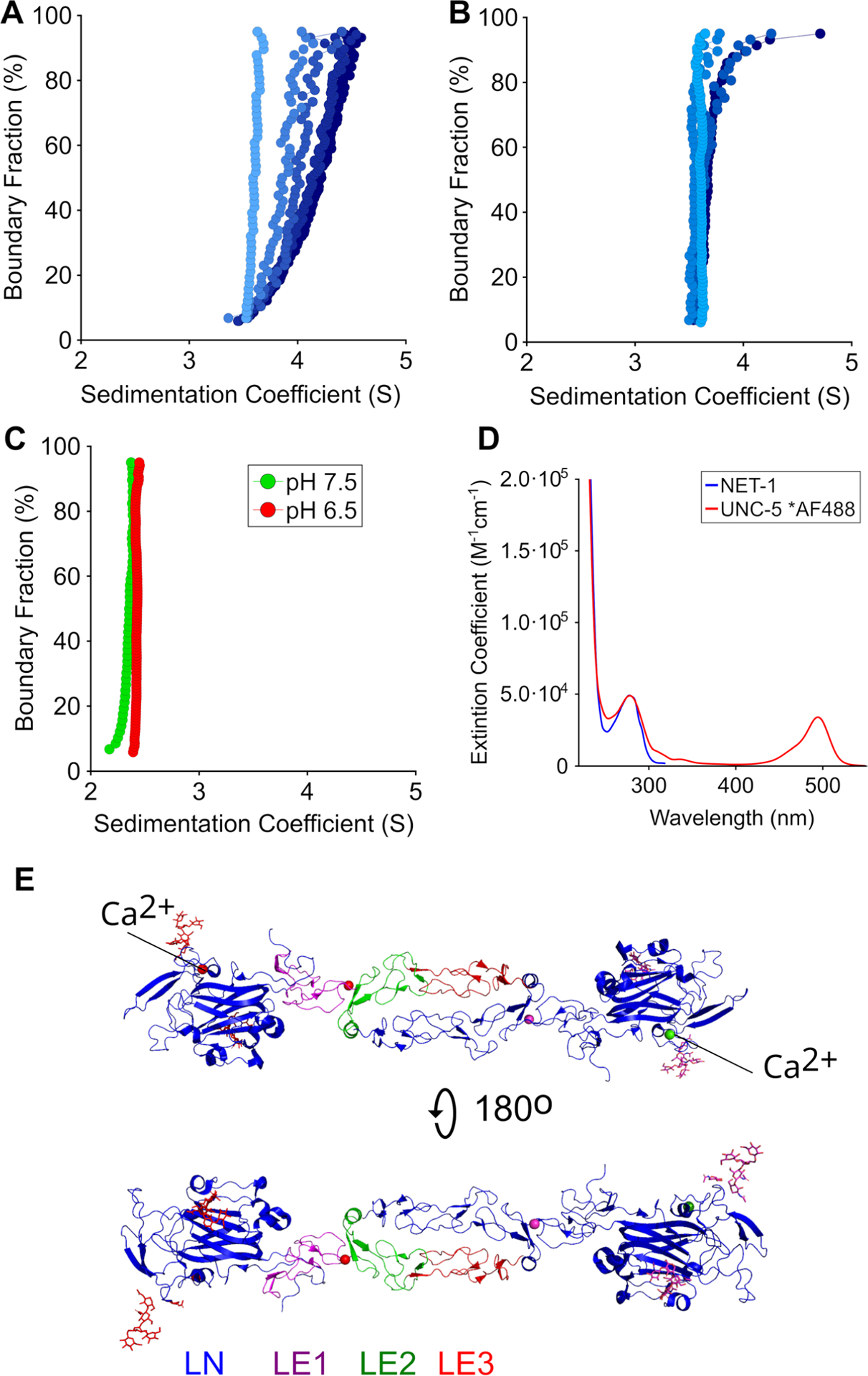
Diffusion-corrected integral sedimentation coefficient distributions of A 1, 8, 12, 33 and 40 μM NET-1 in 50 mM Tris pH 7.5, 200 mM NaCl, as indicated by the color gradient from light to dark blue; B 7, 10, 19, and 22 μM NET-1 in 50 mM MES pH 6.5, 154 mM NaCl, as indicated by the color gradient from light to dark blue; C UNC-5 in Tris pH 7.5 (green) and MES pH 6.5 (red); D Molar extinction spectrum of NET-1 (blue) and UNC-5 labeled with Alexa Fluor-488 (red); E Crystal structure of NET-1 dimer structure (PDB code: 4OVE)
AUC data analysis
Experimental data were processed with UltraScan-III (Demeler and Gorbet 2016) as described by Demeler et al. (2010) to remove time- and radially invariant noise and to determine boundary conditions. Multi-wavelength AUC (MW-AUC) data from the Optima AUC were processed as previously described (Henrickson et al. 2022). This results in iteratively refined two-dimensional spectrum analysis (2DSA) models (Brookes et al. 2010a, b) for each wavelength which are subsequently time-synchronized to account for serial data acquisition on the Optima AUC, and to allow spectral decomposition (Henrickson et al. 2022). The contribution of each analyte was spectrally deconvoluted using molar extinction profiles generated on a Genesys S10 spectrophotometer from a dilution series (Demeler and Gorbet 2016) and normalized to the molar extinction coefficient at 280 nm based on the analyte’s protein sequence. This process renders the MW-AUC data as separate datasets in molar concentration units for each spectrally unique analyte in the mixture, allowing each analyte’s contribution and stoichiometry in a complex to be studied separately. After deconvolution, and for single-wavelength experiments, diffusion-corrected sedimentation profiles were determined using the enhanced van Holde–Weischet (vHW) method (Demeler and van Holde 2004). Kd measurements were performed with the discrete model genetic algorithm analysis (DMGA) (Demeler et al. 2010) in combination with the Monte Carlo analysis (Demeler and Brookes 2008) to determine the confidence intervals (Fedorov et al. 2020).
Hydrodynamic modeling
To compare the experimentally determined hydrodynamic parameters of NET-1 and UNC-5B to their respective crystal structures, the PDB structures were analyzed with ZENO implemented in UltraScan-SOMO (Brookes and Rocco 2018). Missing residues and side chains in the starting structure of NET-1 (PDB ID: 7LRF), and UNC-5 (PDB ID: 6OOL) were built into the models using Coot 0.9.8.5 to ensure that all residues in the expressed sequences (Emsley et al. 2010) were accounted and contributed to the hydrodynamic calculations. After the initial starting structures were confirmed, MD simulations were performed using the ff14SB force field parameters in GROMACS (Pronk et al. 2013). The system was solvated to a neutral charge using a TIP3P cubic periodic box with a minimal distance to the box border of 10 Å which included the addition of sodium and chloride ions at a concentration of 200 mM. Lastly, the system was energy minimized using a minimization method with 5000 steps, with steepest descent, and the Particle-mesh Ewald method (Darden et al. 1993). Final structure files (pdb format) for fully minimized UNC-5B, and NET-1 monomer and dimer structures are available for download from the Open Science Framework (Heide 2023).
Results
Macromolecular state of NET-1 and UNC-5B as a function of pH
To compare the oligomerization properties of NET-1 as a function of pH, we performed SVEs of NET-1 in 50 mM MES pH 6.5, 154 mM NaCl and in 50 mM Tris pH 7.5, 200 mM NaCl. Analysis by the enhanced van Holde– Weischet analysis (vHW) demonstrated oligomerization of NET-1 at pH 7.5, but not at pH 6.5, suggesting a monomer–dimer equilibrium at pH 7.5 (Fig. 1A and B). An increase in NET-1 concentration at pH 7.5 caused the sedimentation coefficient distributions to increase from 3.5S to 4.5S, indicating mass action. However, at pH 6.5, a change in the same concentration range only caused a negligible shift of the sedimentation coefficient distribution, with most of the sedimentation occurring at 3.5S, consistent with a decrease of the association constant compared to pH 7.5. To further quantify the mass action of NET-1 at pH 7.5 and pH 6.5, SVEs performed at 12.1 μM (pH 7.5) and 17.1 μM (pH 6.5) were analyzed with the DMGA analysis (Demeler et al. 2010). The hydrodynamic and thermodynamic results are shown in Table 1 and indicated a Kd of 17.6 μM for NET-1 at pH 7.5 and 137 μM at pH 6.5. Remarkably, the Zeno results were found to be in excellent agreement with the hydrodynamic parameters determined by the DMGA analysis (see Tables 1, 2). Since the Zeno predictions for the dimer complex present in the asymmetric unit of 7LRF and 4OVE NET-1 agree closely with the experimental AUC data, the dimer likely represents two anti-parallel NET-1 proteins interacting through the LE-2 and LE-3 domains as depicted in Fig. 1E.
| (1) |
Table 1.
Discrete model genetic algorithm fitting results of NET-1 monomer–dimer equilibrium (Eq. 1)
| Parameter | S (*10−13 s) | D (*10−7 cm/s) | f/f 0 | ν(cm3/g) | Kd (μM) | RMSD | |
|---|---|---|---|---|---|---|---|
| 12.1 μm | Monomer | 3.52 ± 0.04 | 5.90 ± 0.22 | 1.51 ± 0.06 | 0.71 ± 0.01 | 17.6 ± 3.6 | 0.00276 |
| Tris pH 7.5 | Dimer | 4.83 ± 0.17 | 4.04 ± 0.09 | 1.75 ± 0.04 | |||
| 17.1 μM | Monomer | 3.56 ± 0.03 | 5.09 ± 0.14 | 1.79 ± 0.06 | 0.66 ± 0.01 | 137 ± 26.1 | 0.00407 |
| MES pH 6.5 | Dimer | 5.13 ± 0.10 | 3.66 ± 0.09 | 1.97 ± 0.05 |
All parameters were floated, and 95% confidence intervals were determined by Monte Carlo analysis with 100 iterations
Table 2.
US-SOMO simulations of hydrodynamic parameters of NET-1 and UNC-5 using the Zeno method
| S (*10−13 s) | Translational D(*10−7 cm2/s) | f/f 0 | |
|---|---|---|---|
| NET-1 PDB: 7LRF | |||
| Monomer | 3.16 | 5.63 | 1.57 |
| Dimer | 4.64 | 4.13 | 1.70 |
| UNC-5 PDB: 6OOL | |||
| Monomer | 2.21 | 6.07 | 1.68 |
To characterize the hydrodynamic behavior of the receptor UNC-5B, we performed SVEs as a function of concentration and pH, 10 μM (pH 6.5) and 2.7 μM (pH 7.5) (Fig. 1C). Our data show that UNC-5B remains a homogeneous, monodisperse protein that sediments at 2.3S. In contrast to NET-1, UNC-5B does not change its sedimentation behavior as a function of either concentration nor pH (6.5 vs 7.5) and remains monomeric within the concentration range measured.
The dynamic nature of UNC-5B binding to monomeric and dimeric NET-1
To study the dynamics of the NET-1 + UNC-5B interaction, we first examined if UNC-5B interacts with monomeric NET-1. Therefore, we performed MW-AUC experiments at pH 6.5. UNC-5B was labeled with Alexa Fluor-488 to facilitate spectral separation. Both UNC-5B and NET-1 are homogeneous, monodisperse proteins that sediment at 2.3S and 3.5S, respectively (Fig. 2A). NET-1 was titrated (2–42 μM) into 5 μM UNC-5B with a multi-wavelength SVE obtained at each titration point (Fig. 2). As NET-1 concentration was increased, an increase in the sedimentation coefficient was observed, which clearly indicates NET-1 + UNC-5B complex formation (Fig. 2A and C). Spectral analysis of the MW-AUC data suggests a stoichiometric complex formation with an equimolar ratio of 1:1 (Fig. 2A and B), which is consistent with the hydrodynamic results. The small change in sedimentation coefficient of NET-1 and the gradual increase in UNC-5B sedimentation suggest the complex associates with relatively fast kinetics on the timescale of the velocity experiment. At lower NET-1 concentrations (Fig. 2A and B) a significant amount of unbound UNC-5B is evident, suggesting relatively weak binding. At higher molar excess concentrations of NET-1 (Fig. 2C and D) the slight shift in the sedimentation coefficient observed for the deconvoluted NET-1 signal suggests the presence of significant amounts of unbound monomeric NET-1.
Fig. 2.
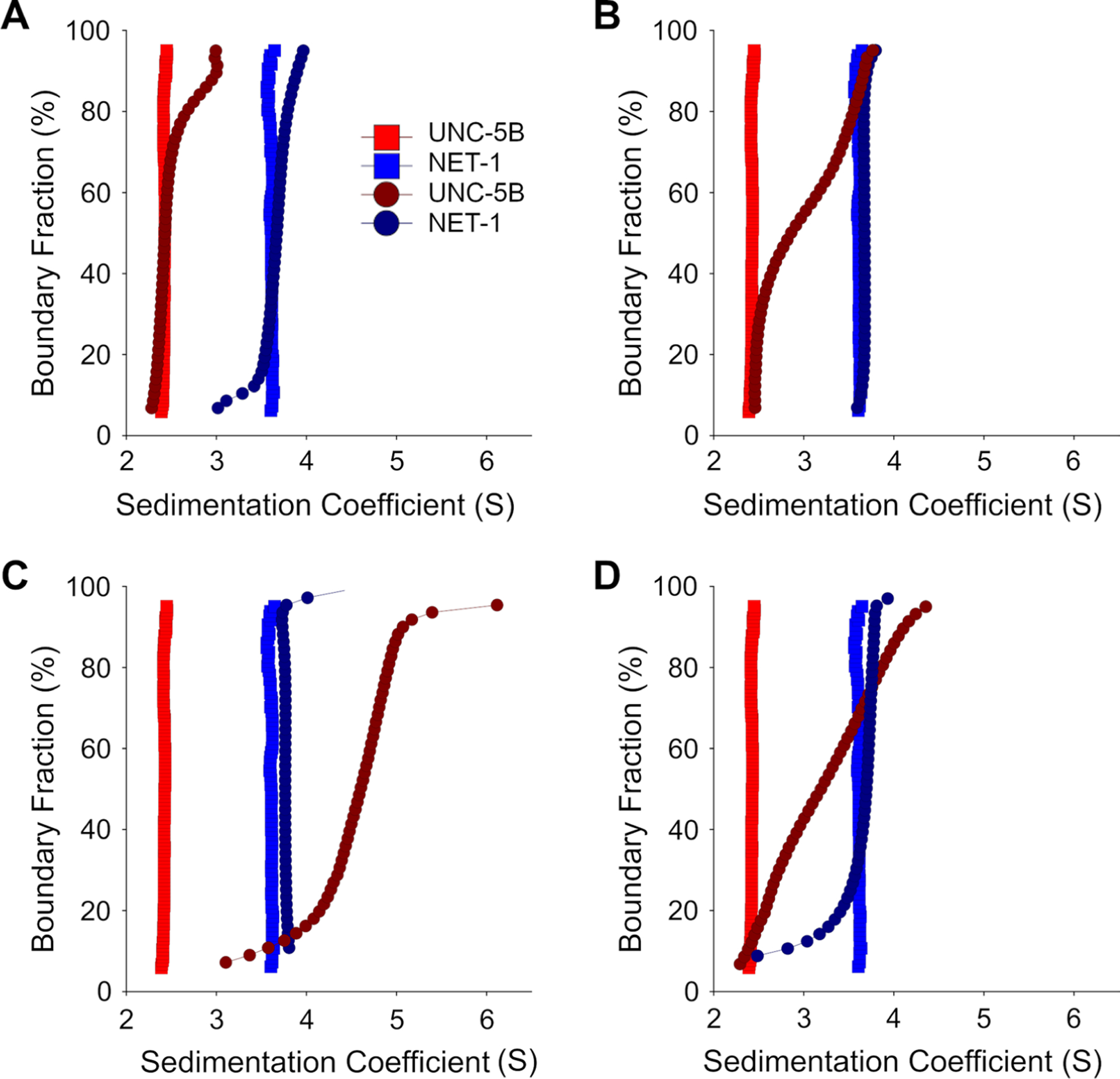
Diffusion-corrected integral sedimentation coefficient distributions of NET-1 and UNC-5 titration series in pH 6.5. Each plot shows 8 μM UNC-5 alone (red squares), 10 μM NET-1 alone (blue squares), deconvoluted UNC-5 (d-UNC-5, dark red circles) and deconvoluted NET-1 (d-NET-1, dark blue circles) at molar ratios of A 1:2, B 2:1 C 9:1 NET-1 to 5 μM UNC-5 and at D 2:1 molar ratios of NET-1 to 9 μM UNC-5
As described above, NET-1 exists in a monomer–dimer equilibrium at pH 7.5. To study whether UNC-5B can interact with dimeric NET-1, we repeated the NET-1 titration SVEs in TRIS buffer at pH 7.5 (Fig. 3). To account for the change in the sedimentation distribution as a function of NET-1 concentration, each NET-1 and UNC-5B solution is compared to NET-1 of similar concentration. As the concentration of NET-1 increased, the UNC-5B sedimentation distribution also increased (Fig. 3A and B). The sedimentation distribution of monomeric NET-1 also significantly increased, meaning most NET-1 is either self-associating or binding to UNC-5B. The wider UNC-5B sedimentation distribution in pH 7.5 compared to pH 6.5 further indicates UNC-5B interacts with NET-1 as a monomer and dimer (Fig. 3C and D).
Fig. 3.
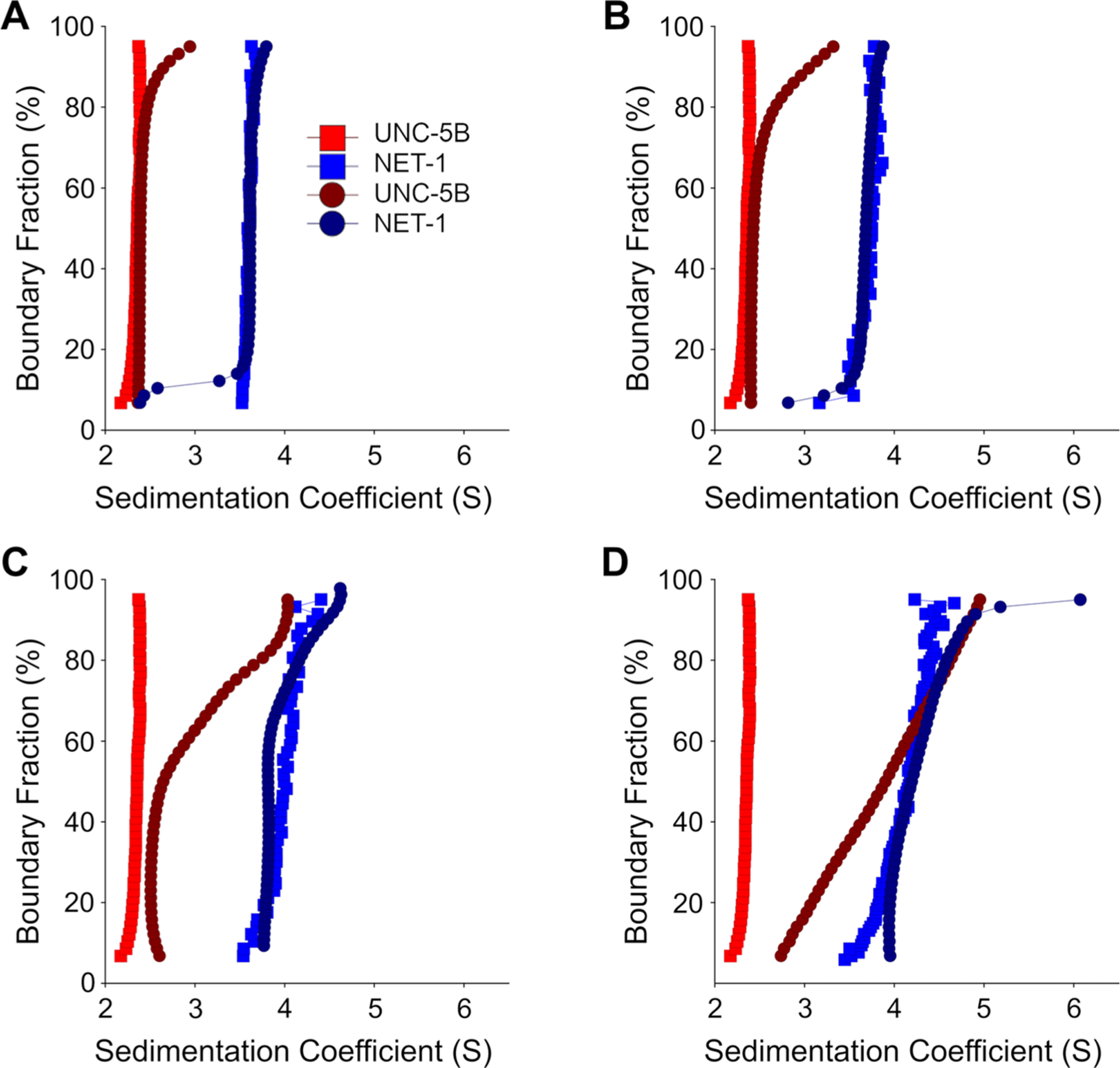
Diffusion-corrected integral sedimentation coefficient distributions of the deconvoluted NET-1 and UNC-5 titration series in pH 7.5. Each plot shows 8 μM UNC-5 alone (Red squares), 10 μM NET-1 alone (Blue squares), deconvoluted UNC-5 (Dark red circles) and NET-1 (Dark blue circles) at at A 1:2, B 1:1, C 2:1, and at D 6:1 molar ratios of NET-1 to 5 μM UNC-5
The role of Ca2+ and heparin in NET-1-mediated UNC-5B binding
In the literature is a controversial discussion about the potential role of extracellular short-chain heparins, and their role for NET-1 mediated receptor interactions (Bennett et al. 1997; Geisbrecht et al. 2003; Kruger et al. 2004). Also, the role of Ca2+ in the oligomerization of NET-1 and UNC-5B has not been established. Here, we performed multi-wavelength sedimentation velocity experiments in the presence and absence of Ca2+ and heparin and measured their impact on complex formation (Fig. 4). The addition of Ca2+ or short-chain heparin (dp8) to UNC-5B in the absence of NET-1 did not change the UNC-5B sedimentation coefficient (Fig. 4). In the absence of UNC-5B, addition of Ca2+ to NET-1 did not alter the vertical sedimentation distribution pattern in the van Holde–Weischet analysis, which indicates homogeneity, but induced a slight shift to a higher sedimentation coefficient.
Fig. 4.
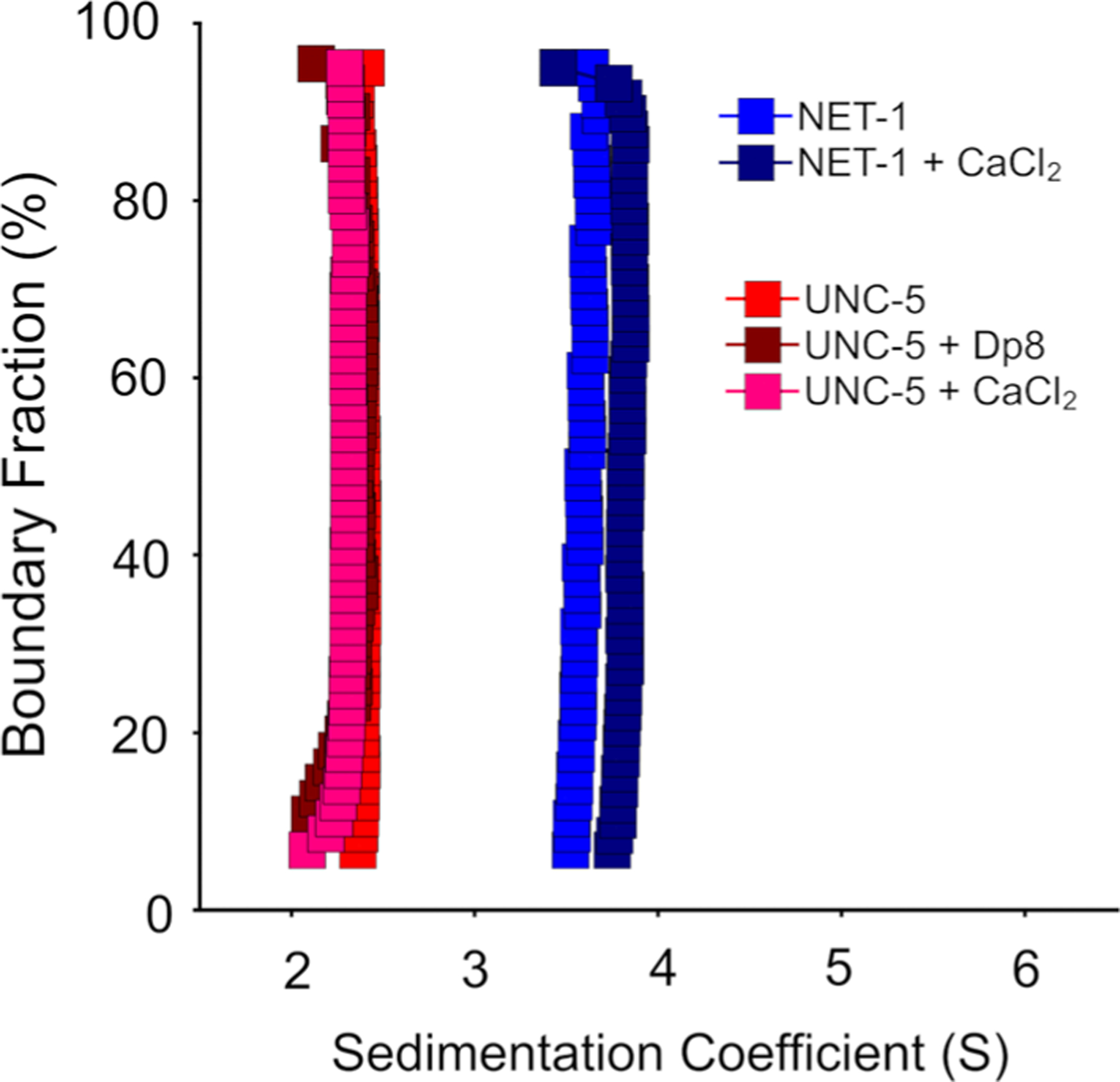
Diffusion-corrected integral sedimentation coefficient distributions of UNC-5B in 50 mM Tris pH 7.5, 200 mM NaCl (red), with the addition of 2 mM CaCl2 (pink), and 12 μM heparin oligosaccha-ride dp8 (brown), and NET-1 in the same buffer without (blue) and with 2 mM CaCl2 (dark blue)
Discussion
Biophysical techniques such as analytical ultracentrifugation, size-exclusion chromatography, and light scattering provide pivotal information required to understand protein assemblies under physiological conditions (Inagaki et al. 2013; Chouquet et al. 2022). A powerful addition to this arsenal is multi-wavelength analytical ultracentrifugation which adds a second characterization dimension to the hydrodynamic separation. It provides the ability to distinguish individual molecules with different chromophores based on their absorbance properties, offering spectral separation and assessment of the stoichiometry of hetero-associations and complex assemblies. Simultaneously, the hydrodynamic characterization identifies the effects of changes in ionic strength, pH, and the addition of small molecule ligands (Henrickson et al. 2022). By adding fluorescent tags to one of the molecules, a distinct chromophore is available to facilitate the effectiveness of MW-AUC, and extends the capabilities of protein–protein assembly studies. Here, we used MW-AUC to separate the contributions of NET-1 and UNC-5B and extracted critical details about their dynamic interactions in the solution phase.
NET-1 is an extracellular matrix (ECM) protein and local changes of pH in the ECM were observed during brain injury, angiogenesis, immune reaction, bone loss or cancer cell invasion (Lardner 2001; Goerges and Nugent 2004; Arnett 2008; Tian and Bae 2012). We, therefore, investigated the NET-1 + UNC-5B interactions at both pH 7.5 and 6.5 because we noticed a difference in oligomerization and binding properties between the two pH values. The van Holde–Weischet integral sedimentation coefficient distributions provide information about the composition of a sample. A vertical distribution indicates homogeneity, while a half-parabola shaped distribution indicates monomer–dimer formation. UNC-5B alone remained homogeneous under all conditions tested, indicating the absence of oligomerization with a sedimentation coefficient of 2.3S, which is consistent with a monomer. Similarly, NET-1 at low concentration, and also at pH 6.5, sedimented primarily as a homogeneous monomer at 3.5S. At pH 7.5, a reversibly self-associating monomer–dimer system was observed, with increasing dimer formation as a function of increasing concentration. The same observation at pH 7.5 was made earlier in SVE studies of UNC-6, a NET-1 ortholog in C. elegans (Krahn et al. 2019). To quantify the thermodynamics of binding, we measured the Kd by SVE for the self-association of NET-1 at pH 7.5 (17.6 μM) and at pH 6.5 (137 μM), confirming significantly reduced dimer formation at lower pH. This suggests that concentration and pH both control the oligomeric conformation of NET-1 in the ECM, and may function differently based on these parameters.
Our interaction studies clearly demonstrate that UNC-5B binds to NET-1 at either pH. We also show that UNC-5B binding to NET-1 occurs regardless of the oligomerization state of NET-1. MW-AUC confirms a 1:1 molar interactions between NET-1 and UNC-5B. Because of the higher Kd at lower pH, much higher ratios of NET-1:UNC-5B are required to see the interaction of UNC-5B with dimeric NET-1 at low pH, but the interaction stoichiometry between NET-1 and UNC-5B seems not to be affected by the pH. The physiological function of NET-1 oligomerization is not clear, but it is conceivable that NET-1 oligomerization is a key regulatory function. Taylor et al. (2015) observed a reversal of the migration direction from attraction to repulsion when the NET-1 concentration increased. As a single-pass receptor, UNC-5 requires a second receptor to signal. This role could be filled by a different NET-1 receptor in the monomeric case, such as DCC, or a second UNC-5 molecule that could bind to the dimeric form of NET-1 at high concentration of NET-1. Either combination could have a different signaling pathway (Merz et al. 2001; Lim and Wadsworth. 2002; Kruger et al. 2004). Merz et al. (2001) also show UNC-5 alone signals repulsion. DCC binds at two sites (LN and LE-3 domains) to NET-1, and potentially via the heparin binding motif on the LE-2 domain (Finci et al. 2014), the same site where UNC-5 binds, but this is controversial. LN and LE-3 DCC binding sites identified in previous studies are compatible with our results and allow for a NET-1 + DCC + UNC-5 heterocomplex. Keleman and Dickson (2001) showed that a complex of NET-1 + DCC signaled attractive migration axon growth, but was reversed from attractive to repulsive when UNC-5B was present. Therefore, repulsive signaling can be achieved by modulating NET-1 concentration through NET-1 oligomerization and mass action, by forming a NET-1 + UNC-5 signaling complex with UNC-5 alone.
NET-1 is a well-known heparin binding protein (Serafini et al. 1994; Bennet et al. 1997; Finci et al. 2014). Heparin is also known to mediate the assembly of other proteins in the ECM. One possibility is that UNC-5B binding to NET-1 is only possible by heparin-mediated assembly. To examine any potential effects of heparin binding to UNC-5B which may cause UNC-5B assembly, we measured the sedimentation of UNC-5B in the presence and absence of dp8, a short-chain heparin molecule. These experiments conclusively show that heparin does not cause UNC-5B assembly. This result suggests that UNC-5B interacts directly with NET-1, and not through heparin-mediated binding.
To check the effect of Ca2+ on the oligomerization properties of NET-1 and UNC-5B, we performed control experiments on UNC-5B and NET-1 in the presence and absence of Ca2+. We detected no effect on oligomerization. In the case of UNC-5B, the sedimentation coefficient remained unchanged in the presence of Ca2+. However, in the case NET-1, we detected a slight increase in the sedimentation NET-1 when Ca2+ was added. We explain the shift by the fact that NET-1, which has a Ca2+ binding site, may change conformation to a slightly more compact state when Ca2+ is bound. Since compaction reduces friction, the sedimentation coefficient increases slightly. The compaction is also supported by increased thermal stability observed in tryptophan fluorescence differential scanning fluorimetry assays when Ca2+ is bound to NET-1 (Supplemental Fig. 1).
NET-1 signaling is complicated, in addition to the dynamic monomer–dimer equilibrium of NET-1 alone, NET-1 also binds heparins, forms large assemblies with DCC as shown by Finci et al. (2014) and Xu et al. (2014) and forms a transient heterocomplex with UNC-5B. Here, we demonstrate that MW-AUC is a potent analytical tool to assist with further characterizing the complex assemblies that abound in NET-1 signaling systems. With this MWAUC investigation, we demonstrate that MW-AUC is an excellent approach not only for investigating complicated interactions involved in NET-1 signaling complexes, but suggest its use for other systems involving two or more interacting molecules.
Supplementary Material
Acknowledgements
This work was supported by the Canada 150 Research Chairs program (C150-2017-00015), the Canada Foundation for Innovation (CFI-37589), the National Institutes of Health (1R01GM120600) and the Canadian Natural Science and Engineering Research Council (DG-RGPIN-2019-05637). UltraScan supercomputer calculations were supported through NSF/XSEDE grant TGMCB070039N, and University of Texas grant TG457201. (All grants to B.D.). JS is a Tier-1 Canada Research Chair in Structural Biology and Biophysics. This research is funded by the Canadian Institutes of Health Research (CIHR)—Grant 201610PJT-152935 (JS).
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00249-023-01644-1.
Data availability
All AUC data (primary sedimentation velocity data, fitted model results, and reports) are stored in the UltraScan LIMS database at the Canadian Center for Hydrodynamics.
References
- Ahmed I, Hahn J, Henrickson A et al. (2022) Structure-function studies reveal ComEA contains an oligomerization domain essential for transformation in gram-positive bacteria. Nat Commun 13:7724. 10.1038/s41467-022-35129-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett TR (2008) Extracellular pH regulates bone cell function 1,2,3. J Nutr 138:S415–S418. 10.1093/jn/138.2.415S [DOI] [PubMed] [Google Scholar]
- Bennett KL, Bradshaw J, Youngman T et al. (1997) Deleted in colorectal carcinoma (DCC) binds heparin via its fifth fibronectin type III domain *. J Biol Chem 272:26940–26946. 10.1074/jbc.272.43.26940 [DOI] [PubMed] [Google Scholar]
- Brookes E, Rocco M (2018) Recent advances in the UltraScan SOlution MOdeller (US-SOMO) hydrodynamic and small-angle scattering data analysis and simulation suite. Eur Biophys J 47:855–864. 10.1007/s00249-018-1296-0 [DOI] [PubMed] [Google Scholar]
- Brookes E, Demeler B, Rocco M (2010a) Developments in the USSOMO bead modeling suite: new features in the direct residue-to-bead method, improved grid routines, and influence of accessible surface area screening. Macromol Biosci 10:746–753. 10.1002/mabi.200900474 [DOI] [PubMed] [Google Scholar]
- Brookes E, Cao W, Demeler B (2010b) A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur Biophys J : EBJ 39(3):405–414. 10.1007/s00249-009-0413-5 [DOI] [PubMed] [Google Scholar]
- Chouquet A, Pinto AJ, Hennicke J et al. (2022) Biophysical characterization of the oligomeric states of recombinant immunoglobulins type-M and their C1q-binding kinetics by biolayer interferometry. Front Bioeng Biotechnol 10:816275. 10.3389/fbioe.2022.816275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- Demeler B, Brookes E (2008) Monte Carlo analysis of sedimentation experiments. Colloid Polym Sci 286:129–137. 10.1007/s00396-007-1699-4 [DOI] [Google Scholar]
- Demeler B, Gorbet GE (2016) Analytical ultracentrifugation data analysis with UltraScan-III. In: Uchiyama S, Arisaka F, Stafford WF, Laue T (eds) Analytical ultracentrifugation instrumentation, software, and applications. Springer; Japan, Tokyo, pp 119–143. 10.1007/978-4-431-55985-6_8 [DOI] [Google Scholar]
- Demeler B, van Holde KE (2004) Sedimentation velocity analysis of highly heterogeneous systems. Anal Biochem 335:279–288. 10.1016/j.ab.2004.08.039 [DOI] [PubMed] [Google Scholar]
- Demeler B, Brookes E, Wang R et al. (2010) Characterization of reversible associations by sedimentation velocity with UltraScan. Macromol Biosci 10:775–782. 10.1002/mabi.200900481 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov D, Batys P, Hayes DB et al. (2020) Analyzing the weak dimerization of a cellulose binding module by analytical ultracentrifugation. Int J Biol Macromol 163:1995–2004. 10.1016/j.ijbiomac.2020.09.054 [DOI] [PubMed] [Google Scholar]
- Finci LI, Krüger N, Sun X et al. (2014) The crystal structure of netrin-1 in complex with DCC reveals the bi-functionality of netrin-1 as a guidance cue. Neuron 83:839–849. 10.1016/j.neuron.2014.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht BV, Dowd KA, Barfield RW et al. (2003) Netrin binds discrete subdomains of DCC and UNC5 and mediates interactions between DCC and heparin. J Biol Chem 278:32561–32568. 10.1074/jbc.M302943200 [DOI] [PubMed] [Google Scholar]
- Goerges AL, Nugent MA (2004) pH regulates vascular endothelial growth factor binding to fibronectin: a mechanisms for control of extracellular matrix storage and release. J Biol Chem 279:2307–2315. 10.1074/jbc.M308482200 [DOI] [PubMed] [Google Scholar]
- Gorbet GE, Pearson JZ, Demeler AK et al. (2015) Next-generation AUC: analysis of multiwavelength analytical ultracentrifugation data. Methods Enzymol 562:27–47. 10.1016/bs.mie.2015.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin M, Meier M, Delcros JG (2016) Structural decoding of the netrin-1/UNC5 interaction and its therapeutical implications in cancers. Cancer Cell 29(2):173–85. 10.1016/j.ccell.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Heide F (2023) Molecular dynamics structure models for NET-1 and Unc5B. 10.17605/OSF.IO/2QCZE. https://osf.io/2qcze/ [DOI] [Google Scholar]
- Henrickson A, Gorbet GE, Savelyev A, Kim M, Hargreaves J, Schultz SK, Kothe U, Demeler B (2022) Multi-wavelength analytical ultracentrifugation of biopolymer mixtures and interactions. Anal Biochem 652:114728. 10.1016/j.ab.2022.114728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernaiz-Llorens M, Roselló-Busquets C, Durisic N et al. (2021) Growth cone repulsion to Netrin-1 depends on lipid raft microdomains enriched in UNC5 receptors. Cell Mol Life Sci 78:2797–2820. 10.1007/s00018-020-03663-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Hinck L, Nishiyama M et al. (1999) A ligand-gated association between cytoplasmic domains of UNC5 and DCC family receptors converts netrin-induced growth cone attraction to repulsion. Cell 97:927–941. 10.1016/S0092-8674(00)80804-1 [DOI] [PubMed] [Google Scholar]
- Inagaki S, Ghirlando R, Grisshammer R (2013) Biophysical characterization of membrane proteins in nanodiscs. Methods 59:287–300. 10.1016/j.ymeth.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang E-H, Mansfield ML, Douglas JF (2004) Numerical path integration technique for the calculation of transport properties of proteins. Phys Rev E Stat Nonlin Soft Matter Phys 69:031918. 10.1103/PhysRevE.69.031918 [DOI] [PubMed] [Google Scholar]
- Keleman K, Dickson BJ (2001) Short- and long-range repulsion by the Drosophila Unc5 netrin receptor. Neuron 32:605–617. 10.1016/S0896-6273(01)00505-0 [DOI] [PubMed] [Google Scholar]
- Kowarz E, Löscher D, Marschalek R (2015) Optimized sleeping beauty transposons rapidly generate stable transgenic cell lines. Biotechnol J 10:647–653. 10.1002/biot.201400821 [DOI] [PubMed] [Google Scholar]
- Krahn N, Meier M, Reuten R et al. (2019) Solution structure of C. elegans UNC-6: a nematode paralogue of the axon guidance protein netrin-1. Biophys J 116:2121–2130. 10.1016/j.bpj.2019.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger RP, Lee J, Li W, Guan K-L (2004) Mapping netrin receptor binding reveals domains of Unc5 regulating its tyrosine phosphorylation. J Neurosci 24:10826–10834. 10.1523/JNEUROSCI.3715-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardner A (2001) The effects of extracellular pH on immune function. J Leukoc Biol 69:522–530. 10.1189/jlb.69.4.522 [DOI] [PubMed] [Google Scholar]
- Lim Y, Wadsworth WG (2002) Identification of domains of netrin UNC-6 that mediate attractive and repulsive guidance and responses from cells and growth cones. J Neurosci 22:7080–7087. 10.1523/JNEUROSCI.22-16-07080.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zhou Q, He J et al. (2020) Recent advances in the development of protein–protein interactions modulators: mechanisms and clinical trials. Sig Transduct Target Ther 5:1–23. 10.1038/s41392-020-00315-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Irie F, Inatani M et al. (2007) Netrin-1/DCC signaling in commissural axon guidance requires cell-autonomous expression of heparan sulfate. J Neurosci 27:4342–4350. 10.1523/JNEUROSCI.0700-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz DC, Zheng H, Killeen MT et al. (2001) Multiple signaling mechanisms of the UNC-6/netrin receptors UNC-5 and UNC-40/DCC in vivo. Genetics 158:1071–1080. 10.1093/genetics/158.3.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk S, Páll S, Schulz R et al. (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29:845–854. 10.1093/bioinformatics/btt055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J, Plotnikov AN, Ibrahimi OA et al. (2000) Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell 6:743–750. 10.1016/S1097-2765(00)00073-3 [DOI] [PubMed] [Google Scholar]
- Serafini T, Kennedy TE, Galko MJ et al. (1994) The netrins define a family of axon outgrowth-promoting proteins homologous to C. elegans UNC-6. Cell 78:409–424. 10.1016/0092-8674(94)90420-0 [DOI] [PubMed] [Google Scholar]
- Shao Q, Yang T, Huang H et al. (2017) Uncoupling of UNC5C with polymerized TUBB3 in microtubules mediates netrin-1 repulsion. J Neurosci 37:5620–5633. 10.1523/JNEUROSCI.2617-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Menon S, Gupton SL (2015) Passive microfluidic chamber for long-term imaging of axon guidance in response to soluble gradients. Lab Chip 15:2781–2789. 10.1039/c5lc00503e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Bae YH (2012) Cancer nanomedicines targeting tumor extra-cellular pH. Colloids Surf B 99:116–126. 10.1016/j.colsurfb.2011.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Chen W, Zhu L et al. (2020) Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation. Protein Cell 11:366–373. 10.1007/s13238-020-00711-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Wu Z, Renier N et al. (2014) Neural migration. Structures of netrin-1 bound to two receptors provide insight into its axon guidance mechanism. Science 344:1275–1279. 10.1126/science.1255149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Ericson CF, Lien Y-W et al. (2022) Identification and structure of an extracellular contractile injection system from the marine bacterium Algoriphagus machipongonensis. Nat Microbiol 7:397–410. 10.1038/s41564-022-01059-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All AUC data (primary sedimentation velocity data, fitted model results, and reports) are stored in the UltraScan LIMS database at the Canadian Center for Hydrodynamics.


