Abstract
With the recent introduction of combination therapy, human immunodeficiency virus type 1 (HIV-1) RNA levels in plasma have been dramatically reduced, frequently to below the limit of quantitation (400 copies/ml of plasma) of the AMPLICOR HIV-1 MONITOR Test (Roche Diagnostic Systems). To achieve enhanced sensitivity of the AMPLICOR HIV-1 MONITOR Test, a modified specimen preparation procedure that allows input of RNA from 10-fold more plasma per amplification reaction was developed. This “ultrasensitive” method allows the accurate quantitation of plasma HIV-1 RNA levels as low as 50 copies/ml. A precision study yielded average within-run and between-run coefficients of variation (CV) of 24.8 and 9.6%, respectively. A multicenter reproducibility study demonstrated that the laboratory-to-laboratory reproducibility of this assay is good, with an average CV of 32%. The linear range of this test is between 50 and 50,000 copies/ml of plasma. RNA concentrations measured by the ultrasensitive and standard HIV-1 MONITOR tests exhibited good agreement within the shared linear range of the two methods. The two measurements were within a factor of 2 for 91% of the specimens tested, with the concentration measured by the ultrasensitive method being only slightly lower (median, 22% lower). Preliminary studies suggest that this assay will prove to be useful for predicting the stability of viral suppression in patients whose RNA levels drop below 400 copies/ml in response to highly active antiretroviral therapy.
The measurement of plasma human immunodeficiency virus type 1 (HIV-1) RNA levels has become an important tool for identifying individuals likely to benefit from antiretroviral therapy (12, 15, 16, 21, 26, 29) as well as monitoring patients on therapy (5, 6, 9, 12, 18, 20, 23) and is now regarded as standard medical practice for managing the treatment of HIV-1-infected individuals (1–4, 22, 25, 28). Recently, the use of combination therapy resulted in rapid and potent antiretroviral and immunological effects which lead to sharp declines in the plasma HIV-1 RNA concentration, frequently to an undetectable level (6, 18, 23). A more sensitive method with a lower detection limit for plasma HIV-1 RNA is therefore required.
The AMPLICOR HIV-1 MONITOR Test, an in vitro nucleic acid amplification test for the quantitation of HIV-1 RNA in plasma, is intended to be used as an indicator of disease prognosis in conjunction with other laboratory markers and clinical presentation and as an aid in assessing the efficacy of antiretroviral therapy. The lower limit of quantitation of the AMPLICOR HIV-1 MONITOR Test is 400 RNA copies/ml of plasma (24). We introduce here a modified specimen preparation procedure (17, 27) that enhances the sensitivity of the standard MONITOR test. Increased sensitivity is obtained by increasing the input plasma volume by a factor of 2.5, performing high-speed centrifugation to concentrate the virus particles from the plasma, and reducing the final resuspension volume for the recovered nucleic acid by a factor of 4. If centrifugation yields 100% recovery of virus, this modified, ultrasensitive procedure should result in a 10-fold increase in the analytical sensitivity of the AMPLICOR HIV-1 MONITOR Test. We evaluated the sensitivity, specificity, linear range, reproducibility, and precision of the ultrasensitive test. We also analyzed the correlation between RNA concentrations measured by the ultrasensitive and the standard HIV-1 MONITOR Tests.
MATERIALS AND METHODS
Clinical specimens.
Clinical specimens for correlation and precision studies were obtained from Angela Caliendo at the Clinical Microbiology Laboratory, Massachusetts General Hospital; Anne Warford at the Diagnostic Virology Department, Stanford University Hospital; and Robert McPhee at the Pathology Reference Laboratory, University of Southern California. Clinical specimens for the reproducibility study were obtained from BioClinical Partners, Inc., Franklin, Mass. Whole blood was collected in sterile tubes with EDTA as the anticoagulant and was stored at 2 to 25°C for no more than 6 h. The plasma was then separated from whole blood by centrifugation at 800 to 1,600 × g for 20 min at room temperature, aliquoted, and stored at −20°C or lower.
Quantified viral stock.
An HIV stock concentrate was prepared by the Virology Quality Assurance (VQA) Laboratory of the AIDS Clinical Trial Group (ACTG) (30). The viral concentration was then determined by (i) electron microscopy (13), (ii) p24 antigen analysis (7), (iii) the AMPLICOR HIV-1 MONITOR Test, and (iv) branched chain DNA analysis (Chiron Corporation, Emeryville, Calif.). The viral stock was serially diluted in HIV-negative human plasma for linear range determination.
Ultra-low-level HIV RNA panel.
A randomized, blinded ultra-low-level HIV RNA proficiency panel designated PPUL01R was prepared by the VQA Laboratory of ACTG with viral stock concentrate diluted in defibrinated HIV-negative human plasma (Basematrix). This panel was used to determine the limit of detection of the ultrasensitive HIV-1 MONITOR Test.
Specimen preparation. (i) Standard method.
The standard specimen preparation procedure was performed as described in the package insert for the AMPLICOR HIV MONITOR Test. Briefly, HIV RNA was extracted from 200 μl of plasma with 600 μl of working HIV-1 MONITOR Lysis Buffer containing a known number of Quantitation Standard (QS) RNA molecules. The RNA was precipitated with isopropanol, recovered by microcentrifugation at maximum speed (at least 12,500 × g) for 15 min at room temperature, washed with 1 ml of 70% ethanol, and resuspended in 400 μl of HIV-1 MONITOR Specimen Diluent. Fifty microliters of the processed specimen was added to Working HIV-1 MONITOR Master Mix for the reverse transcription (RT)-PCR amplification reactions. The amplification reaction mixture contained the HIV RNA recovered from 25 μl of a plasma sample.
(ii) Ultrasensitive method.
HIV particles were concentrated from 500 μl of plasma by centrifuging the samples at 17,000 rpm (24,000 × g) for 60 min at 4°C in a 17 RS Heraeus centrifuge equipped with rotor model HFA 22.1. The pelleted virus particles were lysed by treatment with 600 μl of working HIV-1 MONITOR Lysis Buffer, and the released RNA was precipitated with 600 μl of 100% isopropanol. The amount of QS added to the working HIV-1 MONITOR Lysis Buffer was adjusted so that the same number of QS molecules was added to each amplification reaction mixture in the standard and ultrasensitive tests. The precipitated RNA was recovered by centrifugation, washed with 1 ml of 70% ethanol, and resuspended in 100 μl of HIV-1 MONITOR Specimen Diluent. Fifty microliters of the processed specimen was added to 50 μl of Working HIV-1 MONITOR Master Mix for the RT-PCR amplification reactions. The amplification reaction mixture contained the HIV RNA recovered from 250 μl of a plasma sample.
RT-amplification.
The target sequence for the AMPLICOR HIV-1 MONITOR Test is a highly conserved region of the HIV-1 gag gene (11) that encodes the group-specific antigens (core structural proteins) of the virion. The working HIV-1 MONITOR Master Mix is a bicine-buffered solution containing glycerol, potassium acetate, deoxynucleoside triphosphates (including dATP, dCTP, dGTP, TTP, and dUTP), biotinylated primers (antisense primer SK431 and sense primer SK462), Thermus thermopilus DNA polymerase, manganese acetate, and sodium azide.
Specimens processed by both the standard and ultrasensitive methods were amplified on a GeneAmp PCR system 9600 thermal cycler (Perkin-Elmer Corporation, Norwalk, Conn.) as described in the AMPLICOR HIV-1 MONITOR package insert. The thermal cycling profile consisted of 2 min at 50°C; 30 min at 60°C (for RT); 4 PCR cycles of 10 s at 95°C, 10 s at 55°C, and 10 s at 72°C; 26 PCR cycles of 10 s at 90°C, 10 s at 60°C, and 10 s at 72°C; and 15 min at 72°C.
Hybridization and detection.
The two amplification products, 142-bp sequences generated from HIV-1 and QS target RNAs, were detected colorimetrically. Immediately upon completion of the amplification reaction, the amplification products were denatured by adding 100 μl of Denaturation Solution to each reaction mixture. HIV-1 amplification products were quantitatively detected by hybridizing six serial dilutions of each amplification reaction to microwell plate wells coated with an HIV-1-specific oligonucleotide probe. These dilutions were prepared by adding 25 μl of the denatured amplification product to 100 μl of Hybridization Buffer and performing five fivefold serial dilutions with Hybridization Buffer as the diluent. Similarly, QS amplification products were quantitatively detected by hybridizing two dilutions of each amplification reaction to microwell plate wells coated with a QS-specific oligonucleotide probe. These dilutions were prepared by adding 25 μl of the denatured amplification product to 100 μl of Hybridization Buffer and performing one fivefold dilution with Hybridization Buffer as the diluent. The HIV- and QS-specific wells were assembled onto the same microwell plate frame. The microwell plate was incubated at 37°C for 60 min to allow the amplification products to hybridize to the probe. The plate was washed, an avidin-horseradish peroxidase conjugate was added to each well, and the plate was incubated at 37°C for 15 min. The plate was washed again, and a substrate solution containing H2O2 and tetramethylbenzidine was added to each well. After a 10-min incubation at room temperature, the colorimetric reaction was terminated by adding Stop Reagent, and the optical density (OD) at 450 nm (single wavelength) was measured.
Calculation of RNA concentration.
For both the target and the QS, the dilution that yielded a signal within the linear range of the spectrophotometer was identified. The OD for this dilution was corrected by subtracting the background OD (A450, 0.07), and the corrected OD was multiplied by the dilution factor to determine the total signal generated. The input HIV-1 RNA concentration was calculated by comparing the total OD for HIV-1 in the sample to the total OD for the QS in the sample, as follows: number of RNA copies per milliliter of plasma = (total target OD/total QS OD) × QS copies per reaction ÷ plasma volume, where the plasma volume is 0.025 ml per reaction mixture for the standard test and 0.25 ml per reaction mixture for the ultrasensitive test.
RESULTS
Limit of detection.
The limit of detection was determined by using the ultra-low-level HIV-1 RNA proficiency panel prepared by the ACTG VQA Laboratory (30). Both the ultrasensitive and the standard HIV-1 MONITOR Tests were performed. The panel consisted of six different viral levels (1,000, 500, 100, 50, 20, and 0 copies/ml). In addition, we prepared viral concentrations of 30 and 40 copies/ml by diluting the sample with 1,000 copies/ml in the Basematrix provided by the VQA Laboratory. The standard HIV-1 MONITOR specimen preparation method detected HIV-1 in all of the samples tested with HIV-1 present at 1,000 and 500 copies/ml but failed to detect HIV-1 in most samples with HIV-1 present at 50 and 40 copies/ml and did not detect HIV-1 in any of the samples with HIV-1 present at 30 and 20 copies/ml (Table 1). In contrast, the ultrasensitive method detected HIV-1 in all of the samples tested with HIV-1 present at 1,000 to 50 copies/ml and also detected HIV-1 in most of the samples with HIV-1 present at 40, 30, and 20 copies/ml (Table 1). Thus, an approximately 10-fold increase in sensitivity was achieved by preparing specimens by the ultrasensitive method instead of the standard method. Because we define the detection limit of the test as the concentration required to yield positive results in at least 90% of replicate reactions, the detection limit for the ultrasensitive method is approximately 50 viral RNA copies/ml of plasma.
TABLE 1.
Limit of detection for the ultrasensitive and standard HIV-1 MONITOR Testsa
| Actual input concn (copies/ml) | No. of replicates | Standard test
|
Ultrasensitive test
|
||
|---|---|---|---|---|---|
| No. (%) positive | Avg no. of copies/mlb | No. (%) positive | Avg no. of copies/mlb | ||
| 1,000 | 3 | 3 (100) | 1,213 | 3 (100) | 1,190 |
| 500 | 3 | 3 (100) | 697 | 3 (100) | 612 |
| 100 | 6 | 2 (33) | 265 | 6 (100) | 134 |
| 50 | 7 | 1 (14) | 390 | 7 (100) | 67 |
| 40 | 7 | 2 (29) | 271 | 6 (86) | 90 |
| 30 | 7 | 0 (0) | Not detected | 7 (100) | 61 |
| 20 | 7 | 0 (0) | Not detected | 5 (71) | 55 |
| 0 | 6 | 0 (0) | Not detected | 0 (0) | Not detected |
The limit of detection was determined by using the ultra-low-level HIV RNA proficiency panel described in Materials and Methods.
The viral RNA concentration in each replicate reaction yielding a positive signal was calculated as described in Materials and Methods. The values presented here are the averages of the individual values for the positive replicates. Reactions yielding negative results were not included in the average.
Linear range.
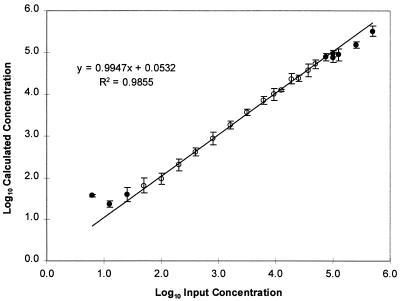
The linear range for the ultrasensitive method was determined by analysis of reconstructed HIV-positive plasma specimens prepared by serial dilution of quantified VQA viral stock in HIV-negative human plasma. This panel of reconstructed HIV-positive specimens contained HIV-1 RNA at concentrations from 500,000 to 6 copies/ml of plasma. Three individuals tested the panel in duplicate, for a total of six replicates for each virus concentration. The log10 calculated concentration for each replicate was compared to the log10 input concentration. Linear regression analysis showed that the linear range of the ultrasensitive method was 50 to 50,000 copies/ml, with a slope of 0.995, an intercept of 0.053, and an R2 value of 0.986 (Fig. 1). Within the linear range, the concentration measured by the ultrasensitive method is, on average, close to the actual concentration because the slope approaches 1.0 and the intercept approaches 0.0.
FIG. 1.
Linear range for the ultrasensitive AMPLICOR HIV-1 MONITOR Test. Six replicate tests were performed for each input viral RNA concentration. For input RNA concentrations below 50 copies/ml, some of the replicates gave negative results and were excluded from the data analysis. At each input RNA concentration, the mean of the log10 calculated RNA concentrations and the standard deviation were determined (circles). The open circles indicate the results for concentrations that were used for linear regression analysis. The closed circles indicate the results for concentrations that were not included in the regression analysis. Linear regression analysis was performed for input RNA concentrations of from 50 to 50,000 copies/ml (solid line and equation). The regression analysis was performed with the individual results for each replicate at each input RNA concentration.
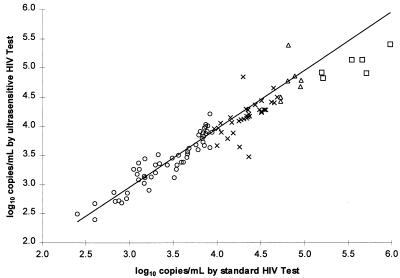
Correlation with the standard HIV-1 MONITOR Test.
The correlation between the standard and the ultrasensitive methods was determined by comparing the log10 calculated RNA concentrations for 100 positive clinical specimens (with titers of from 250 to 106 copies/ml) prepared by both methods (Fig. 2). The slope, intercept, and r values of the relationship between the two methods were determined through linear regression analysis. Separate regression analyses were performed for four intervals of virus concentration because the slope and intercept did not appear to be constant over the full concentration range.
FIG. 2.
Correlation between the ultrasensitive and standard AMPLICOR HIV-1 MONITOR Tests. The log10 calculated RNA concentration was determined by each method for a set of 100 specimens. Specimens are grouped by calculated RNA concentration (○, 250 to 9,999 copies/ml; ×, 10,000 to 49,999 copies/ml; ▵, 50,000 to 99,999 copies/ml; □, 100,000, to 499,999 copies/ml). Separate linear regressions were performed for each group of specimens (see Table 2). The regression line for specimens with <10,000 copies/ml is shown (solid line).
For viral RNA concentrations below 10,000 copies/ml, the standard and ultrasensitive methods gave very similar results. The slope approached 1.0, the intercept approached 0.0, and the median of the ratio of the RNA concentration determined by the ultrasensitive method to the RNA concentration determined by the standard method approached 1.0 (Table 2). Furthermore, a twofold or greater difference between the RNA concentrations obtained by the two methods was found for only 2 of the 56 specimens.
TABLE 2.
Linear regression analysis of correlation between the standard and the ultrasensitive HIV-1 MONITOR Tests
| Viral load range (no. of copies/ml of plasma) | No. of specimens | Slope | Inter- cept | r | Ultrasensitive test RNA concn/standard test RNA concna |
|---|---|---|---|---|---|
| 250–9,999 | 56 | 0.999 | 0.044 | 0.936 | 0.896 |
| 10,000–49,999 | 32 | 0.927 | 0.135 | 0.630 | 0.650 |
| 50,000–99,999 | 7 | 0.935 | 0.248 | 0.287 | 0.664 |
| 100,000–499,999 | 6 | 0.555 | 1.976 | 0.805 | 0.347 |
For each specimen, the RNA concentration determined by the ultrasensitive method was divided by the RNA concentration determined by the standard method. The median of this ratio was determined for each group of specimens.
For intermediate viral RNA concentrations (between 10,000 and 100,000 copies/ml), the slope was somewhat lower, the intercept was somewhat higher, and the median ratio of the RNA concentrations determined by the two methods was approximately 0.7, indicating that the ultrasensitive method slightly underestimated the concentration compared to that estimated by the standard method (Table 2). Indeed, a twofold difference between the RNA concentrations obtained by the two methods was found for 7 of the 39 specimens. The standard method gave the higher result for five of the specimens and the ultrasensitive method gave the higher result for two of the specimens.
For high viral RNA concentrations (above 100,000 copies/ml), the slope was less than 0.9, the intercept was substantially greater than 0.0, and the median ratio of the RNA concentrations determined by the two methods was less than 0.4, indicating that the ultrasensitive method substantially underestimated the concentration (Table 2). Indeed, the standard method gave a more than twofold higher RNA concentration for five of the six specimens.
Although the ultrasensitive method was less accurate at higher concentrations, the two tests exhibited good agreement over their shared linear range (400 to 50,000 copies/ml). The median ratio of the RNA concentration determined by the ultrasensitive method to the RNA concentration determined by the standard method was 0.78. The concentrations determined by the two methods differed more than twofold for only 8 of 86 specimens; the standard method gave the higher result for six of these eight specimens.
Specificity.
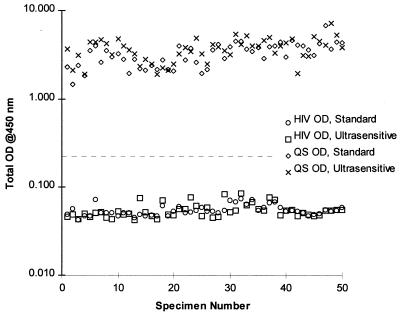
The analytical specificity of the standard AMPLICOR HIV-1 MONITOR Test was previously determined by analyzing 24 microbes and closely related viruses (24). None of the non-HIV organisms reacted in the standard test; three of the four HIV-2 isolates tested yielded positive results. Since the ultrasensitive and standard methods use identical primers, probe, and amplification conditions, the analytical specificity is expected to be the same and was not reevaluated. It was, however, necessary to demonstrate that the ultrasensitive method did not recover substances capable of causing false-positive results with clinical specimens. Therefore, the specificity of the ultrasensitive method was evaluated by testing 50 HIV-seronegative plasma specimens from healthy blood donors. All 50 specimens yielded negative results for HIV (Fig. 3). The average OD generated by the undiluted amplification products was 0.055 for both the standard and the ultrasensitive methods. The minimum OD observed was 0.043 for both methods. The maximum OD observed was 0.074 for the standard method and 0.085 for the ultrasensitive method. The two methods yielded similar total ODs for the QS.
FIG. 3.
Specificity of the ultrasensitive AMPLICOR HIV-1 MONITOR Test. A set of 50 HIV-1-negative specimens was tested by the ultrasensitive and standard methods. The HIV-1 and QS signals are shown. The dashed line indicates the test cutoff (A450, 0.2).
Precision study.
A precision study for the ultrasensitive method was performed by evaluating two dilutions of a viral stock (VQA Laboratory) containing HIV-1 RNA at 250 and 25,000 copies/ml in Basematrix, two dilutions of clinical specimen pools containing HIV-1 RNA at 500 and 25,000 copies/ml, and a negative control (Basematrix). The study was carried out over 10 consecutive working days by two laboratory operators. Five replicates for each HIV-containing sample and four replicates for the negative control (for a total of 24 samples) were tested by each operator on each day. All 80 negative controls yielded negative results. The test exhibited good total precision for all the HIV-1-positive samples, with coefficients of variation (CVs) ranging from 22 to 35% (Table 3). Most of the variability was due to variation between replicates within a run, with CVs ranging from 21 to 30% (Table 3). The between-day variation, with CVs ranging from 7 to 18%, was less than the within-run variation (Table 3). There was virtually no variation between operators (Table 3).
TABLE 3.
Precision of ultrasensitive HIV-1 MONITOR Testa
| Specimen and no. of input RNA copies/mlb | Mean calculated RNA concn (no. of copies/ml) | RNA concn (no. of copies/ml)
|
Within-run variation
|
Between-day variation
|
Between-operator variation
|
Total
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Minimum | Maximum | SD (no. of RNA copies/ml) | CV (%) | SD (no. of RNA copies/ml) | CV (%) | SD (no. of RNA copies/ml) | CV (%) | SD (no. of RNA copies/ml) | CV (%) | ||
| VQA | |||||||||||
| 250 | 375 | 165 | 1,287 | 111 | 29.5 | 57 | 15.2 | 38 | 10.1 | 130 | 34.7 |
| 25,000 | 27,844 | 13,695 | 90,526 | 8,348 | 30.0 | 5,023 | 18.0 | 0c | 0c | 9,742 | 35.0 |
| Clinical | |||||||||||
| 500 | 710 | 355 | 1,202 | 132 | 18.7 | 90 | 12.7 | 0c | 0c | 160 | 22.5 |
| 25,000 | 29,444 | 17,380 | 55,540 | 6,111 | 20.8 | 1,988 | 6.8 | 0c | 0c | 6,426 | 21.8 |
These calculations were performed as described in the EP5T guideline of the National Committee for Clinical Laboratory Standards (19) for within-run, between-day, and between-operator variations and total precision. Between-day variation includes variation between runs; between-run variation within 1 day could not be calculated because each operator performed only one run per day. The total number of replicates for each specimen is 100.
VQA are dilutions of the viral stock from ACTG. Clinical specimens were obtained from HIV-positive patients.
The estimate of variance was negative and was set to zero in accordance with the guideline of the National Committee for Clinical Laboratory Standards (19).
Interlaboratory variation.
To determine the variability of the ultrasensitive method among laboratories, a panel was constructed by aliquoting one HIV-negative and four HIV-positive clinical specimens (obtained from Bioclinical Partners) with different viral RNA concentrations. The panel consisted of 12 coded, single-use aliquots; the negative specimen was represented three times, and each of the HIV-positive specimens was represented two or three times. At each of three sites, the University of New Mexico (UNM), Johns Hopkins University (JHU) and Roche Molecular System (RMS), one operator tested one panel each day for 2 days. The median CVs for the duplicate determinations in the RMS, UNM, and JHU laboratories were 24, 8, and 30%, respectively, and the median CV for the combined results from all three sites was 35% (Table 4).
TABLE 4.
Results of multicenter reproducibility study
| Sample no. | RMS
|
UNM
|
JHU
|
Combined resultsa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calculated RNA concn (no. of copies/ml)
|
CV (%) | Calculated RNA concn (no. of copies/ml)
|
CV (%) | Calculated RNA concn (no. of copies/ml)
|
CV (%) | Calculated RNA concn (no. of copies/ml)
|
CV (%) | |||||
| Day 1 | Day 2 | Day 1 | Day 2 | Day 1 | Day 2 | Mean | SD | |||||
| 1 | 66 | 28 | 57 | 55 | 50 | 7 | 79 | 14 | 99 | 49 | 31 | 64 |
| 2 | 9,182 | 4,924 | 43 | 5,052 | 5,243 | 3 | 6,034 | 3,994 | 29 | 5,738 | 1,929 | 34 |
| 3 | 111 | 47 | 57 | 67 | 126 | 43 | 87 | 67 | 18 | 84 | 36 | 43 |
| 4 | 12,010 | 7,647 | 31 | 9,785 | 9,000 | 6 | 10,097 | 6,535 | 30 | 9,179 | 2,322 | 25 |
| 5 | 588 | 823 | 24 | 686 | 608 | 9 | 1,114 | 882 | 16 | 784 | 212 | 27 |
| 6 | 18,522 | 14,584 | 17 | 8,190 | 8,886 | 6 | 12,237 | 9,921 | 15 | 12,057 | 4,308 | 36 |
| 7 | 4,492 | 3,384 | 20 | 3,241 | 3,652 | 8 | 4,792 | 2,484 | 45 | 3,674 | 1,059 | 29 |
| 8 | 15,189 | 10,763 | 24 | 9,513 | 12,600 | 20 | 8,637 | 5,295 | 34 | 10,333 | 3,575 | 35 |
| 9 | 923 | 651 | 24 | 654 | 525 | 15 | 1,059 | 451 | 57 | 711 | 277 | 39 |
| Median CV | 24 | 8 | 30 | 35 | ||||||||
The mean was calculated by averaging the six individual determinations for each test sample. The standard deviations and CVs among laboratories were determined by performing a nested analysis of variance similar to that described in guideline EP5T of the National Committee for Clinical Laboratory Standards (19).
Detection of HIV-1 RNA in low-titer clinical specimens.
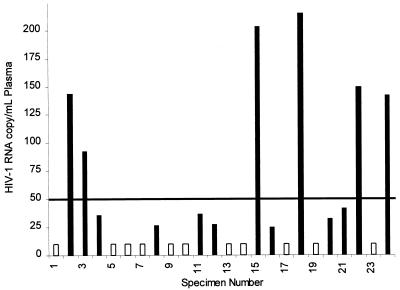
To demonstrate the improved sensitivity of the ultrasensitive method, we evaluated 24 clinical specimens that tested negative by the standard method and that thus had titers below 400 copies/ml (the quantitation limit of the standard method). When processed by the ultrasensitive method, 13 of the 24 specimens gave positive results (Fig. 4). Six of the 13 positive specimens had viral RNA titers within the linear range of the assay, and 7 had titers below the linear range of the assay. The highest titer observed was 215 copies/ml. We also further increased the sensitivity of the ultrasensitive method by increasing the input plasma volume from 0.5 to 1.0 ml. This twofold increase in plasma input produced an approximately twofold additional increase in analytical sensitivity, for a total increase in analytical sensitivity of 20-fold compared to that of the standard method (data not shown). When the ultrasensitive method was used with a 1.0-ml plasma input, 3 additional low-titer specimens were detected, for a total of 16 of 24 low-titer specimens in which HIV-1 RNA could be detected (data not shown). For routine use, the convenience afforded by the 0.5-ml sample size outweighs the small increase in sensitivity achieved with a 1.0-ml sample.
FIG. 4.
Detection of HIV-1 RNA in clinical specimens with low titers of HIV-1 RNA. RNA titers were measured by the ultrasensitive AMPLICOR HIV-1 MONITOR Test in a set of 24 specimens that gave negative results by the standard AMPLICOR HIV-1 MONITOR Test. The solid line indicates the lower limit of the linear range. Filled bars indicate the RNA titer in specimens that gave positive results, and open bars indicate specimens that gave negative results.
DISCUSSION
The analytical sensitivity of the AMPLICOR HIV-1 MONITOR Test was increased approximately 10-fold through a relatively simple modification of the specimen processing procedure. An ultracentrifugation step was performed prior to extracting RNA to concentrate the HIV particles. By eliminating soluble plasma components that potentially interfere with RT-PCR, we were able to increase the volume of specimen tested 10-fold, from the equivalent of 25 μl of plasma by the standard method to the equivalent of 250 μl of plasma by the ultrasensitive method.
The linear range for the ultrasensitive method is 50 to 50,000 copies of viral RNA/ml of plasma. This represents an approximate 10-fold shift compared to the linear range for the standard method (400 to 750,000 copies of viral RNA/ml of plasma). This is expected because the linearity of the test depends on the number of target molecules amplified. The number of viral RNA molecules added to the amplification reaction mixture will be the same for an extract prepared by the ultrasensitive method from plasma containing 50,000 copies/ml and an extract prepared by the standard method from plasma containing 500,000 copies/ml.
The RNA concentrations measured by the standard and ultrasensitive methods differed by less than a factor of 2 for 91% (78 of 86) of the specimens that had RNA titers within the linear range of both methods. The RNA concentrations calculated by the ultrasensitive method were slightly lower (median, 22% lower) than those calculated by the standard method. A laboratory that developed its own, similar ultrasensitive version of the AMPLICOR test reported results comparable to those reported here and suggested that they could be explained by the observation that 10 to 16% of the HIV-1 RNA in plasma is not recovered during high-speed centrifugation (27). Nevertheless, the relatively good agreement implies that laboratories can switch between the two methods to obtain accurate measurements of viral burden at all stages of HIV infection. For example, the standard method could be used to obtain titers at the baseline and the ultrasensitive method could be used after the initiation of therapy or after the viral titer exhibits a substantial decrease. Should the viral titer rise due to therapy failure, the standard method could be used again.
The limit of detection for the ultrasensitive method was 50 viral RNA copies/ml of plasma. While the method was also able to detect samples with lower titers, a single measurement cannot accurately assess viral burden when the titer is below 50 viral RNA copies/ml of plasma. By performing replicate tests, we demonstrated that such specimens will not always yield positive results. Furthermore, when positive results were obtained, the viral titer was overestimated. Statistical fluctuation probably contributes to this apparent irreproducibility. For a titer in plasma of 20 RNA copies/ml, an average of at most 5 copies (assuming 100% recovery of RNA) is introduced into the amplification reaction. The actual number of RNA molecules delivered to a reaction will be distributed according to the Poisson statistics. Thus, the standard deviation of the number of molecules per reaction will be 2.2 (√5), and any individual test could receive as few as 1 or as many as 9 target molecules. The actual fluctuation is likely to be somewhat larger because this simplified analysis does not take into account sampling variation that also occurs when drawing the initial 0.5-ml aliquot of plasma.
The ultrasensitive method exhibited good reproducibility. The overall coefficient of variation was approximately 30%, which implies that a single test result will provide an estimate of the actual titer within a factor of 2. Within-run differences between replicates was the largest source of variation. When different laboratories tested the same sample, the individual viral titer determinations generally differed from each other by a factor of 3 at most, which indicates that the results obtained in different laboratories can be compared.
Several recent studies indicate that the ultrasensitive assay will provide clinically useful information. The assay can measure HIV-1 RNA titers in patients whose titers fall below the detection limit of the standard AMPLICOR HIV-1 MONITOR Test. For example, 11 of 15 patients treated with a combination of nevirapine, indinavir, and lamivudine had viral titers below the limit of detection for the standard test; an ultrasensitive version of the test revealed that 5 of these 11 patients had RNA titers of between 20 and 200 copies/ml and that 6 had RNA titers below 20 copies/ml (8). Patients whose viral loads drop below 50 RNA copies/ml in response to therapy may be more likely to sustain suppression of viral replication. In one study, only 12% (4 of 32) of patients who achieved undetectable viral RNA loads ultimately had relapses (exhibited >1,000 RNA copies/ml) during follow-up, whereas 33% (26 of 83) of patients who achieved viral loads of between 50 and 400 RNA copies/ml had relapses (10). In a second study, 9 of 10 patients who achieved undetectable viral RNA loads did not exhibit an increase during follow-up (14). In contrast, 7 of 10 patients who achieved viral loads of between 50 and 200 copies/ml did exhibit increases during follow-up; the 3 patients who did not exhibit increased viral RNA titers were switched to a more potent therapy during follow-up (14).
The ultrasensitive processing method has been incorporated into the latest version of the AMPLICOR HIV-1 MONITOR Test. The test is designed to offer the option of using the ultrasensitive or standard processing method, which should enable it to be used throughout the course of HIV-1 infection. With a detection limit of 50 viral RNA copies/ml of plasma, it should prove to be useful for monitoring the response to the new, highly effective combination therapies. Improvements in therapeutic regimens may ultimately result in viral titers that are below the limit of detection of the ultrasensitive method. Additional modifications to the ultrasensitive method, such as further increases in the plasma input volume, can be used to enhance its sensitivity.
ACKNOWLEDGMENTS
We thank Kelly Mohan from JHU and Ray Mills from UNM for performing the reproducibility study, Brian Staes from the VQA Laboratory for providing us with the concentrated quantified VQA HIV-1 stock and ultra-low-level HIV-1 RNA panel, and Alex Wesolowski from the Regulatory Department, Roche Molecular Systems, for helpful discussions.
REFERENCES
- 1.BHIVA Guidelines Co-ordinating Committee. British HIV Association guidelines for antiretroviral treatment of HIV seropositive individuals. Lancet. 1997;349:1086–1092. [PubMed] [Google Scholar]
- 2.Carpenter C C, Fischl M A, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S, Richman D D, Saag M S, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A for the International AIDS Society-USA. Antiretroviral therapy for HIV infection in 1996. Recommendations of an international panel. JAMA. 1996;276:146–154. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Public Health Service task force recommendations for the use of antiretroviral drugs in pregnant women infected with HIV-1 for maternal health and for reducing perinatal HIV-1 transmission in the United States. Morbid Mortal Weekly Rep. 1998;47(No. RR-2):1–39. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Guidelines for the use of antiretroviral agents in pediatric HIV infection. Morbid Mortal Weekly Rep. 1998;47(No. RR-4):1–51. [PubMed] [Google Scholar]
- 5.Eron J J, Benoit S L, Jemsek J, MacArthur R D, Santana J, Quinn J B, Kuritzkes D R, Fallon M A, Rubin and the North American HIV Working Party M. Treatment with lamivudine, zidovudine, or both in HIV-positive patients with 200 to 500 CD4+ cells per cubic millimeter. N Engl J Med. 1995;333:1662–1669. doi: 10.1056/NEJM199512213332502. [DOI] [PubMed] [Google Scholar]
- 6.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 7.Hammer S, Crumpacker C, D’Aquila R, Jackson B, Lathey J, Livnat D, Reichelderfer P. Use of virologic assays for detection of human immunodeficiency virus in clinical trials: recommendations of the AIDS Clinical Trials Group Virology Committee. J Clin Microbiol. 1993;31:2557–2564. doi: 10.1128/jcm.31.10.2557-2564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris M, Durakovic C, Rae S, Raboud J, Fransen S, Shillington A, Conway B, Monaner J S G. A pilot study of nevirapine, indinavir, and lamivudine among patients with advanced human immunodeficiency virus disease who have had failure of combination nucleoside therapy. J Infect Dis. 1998;177:1514–1520. doi: 10.1086/515317. [DOI] [PubMed] [Google Scholar]
- 9.Hughes M D, Johnson V A, Hirsch M S, Bremer J W, Elbeik T, Erice A, Kuritzkes D R, Scott W A, Spector S A, Basgoz N, Fischl M A, D’Aquila R T for the ACTG 241 Protocol Virology Substudy Team. Monitoring plasma HIV-1 RNA levels in addition to CD4+ lymphocyte count improves assessment of antiretroviral therapeutic response. Ann Intern Med. 1997;126:929–938. doi: 10.7326/0003-4819-126-12-199706150-00001. [DOI] [PubMed] [Google Scholar]
- 10.Izopet I, Salama G, Pasquier C, Sandres K, Kuhlein E, Blancher A, Marchou B, Massip P, Puel J. Abstracts of the 5th Conference on Retroviruses and Opportunistic Infections. Alexandria, Va: Foundation for Retroviruses and Human Health; 1998. Ultra sensitive detection of plasma HIV-1 RNA for predicting the durability of 3-drug antiretroviral therapy, abstr. 326; p. 140. [Google Scholar]
- 11.Kwok S, Sninsky J J. PCR detection of human immunodeficiency virus type 1 proviral DNA sequences. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 309–315. [Google Scholar]
- 12.Lathey J L, Hughes M D, Fiscus S A, Pi T, Jackson J B, Rasheed S, Elbeik T, Reichman R, Japour A, D’Aquila R T, Scott W, Griffith B P, Hammer S M, Katzenstein D A for the AIDS Clinical Trials Group Protocol 175 Team. Variability and prognostic values of virologic and CD4 cell measures in human immunodeficiency virus type 1-infected patients with 200–500 CD4 cells/mm3. J Infect Dis. 1998;177:617–624. doi: 10.1086/514250. [DOI] [PubMed] [Google Scholar]
- 13.Layne S P, Merges M J, Dembo M, Spouge J L, Conley S R, Moore J P, Raina J L, Renz H, Gelderbloom H R, Nara P. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology. 1992;189:695–714. doi: 10.1016/0042-6822(92)90593-e. [DOI] [PubMed] [Google Scholar]
- 14.Maeland A, Holberg M, Bruun J N, Løvgàrden G. Abstracts of the 5th Conference on Retroviruses and Opportunistic Infections. Alexandria, Va: Foundation for Retroviruses and Human Health; 1998. Subtle differences in low-level HIV viral load on treatment is important for outcome, abstr. 327; p. 140. [Google Scholar]
- 15.Mellors J W, Kingsley L A, Rinaldo C R, Jr, Todd J A, Hoo B S, Kokka R P, Gupta P. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 16.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 17.Mulder J, Resnick R, Saget B, Scheibel S, Herman S, Payne H, Harrigan R, Kwok S. A rapid and simple method for extracting human immunodeficiency virus type 1 RNA from plasma: enhanced sensitivity. J Clin Microbiol. 1997;35:1278–1280. doi: 10.1128/jcm.35.5.1278-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers M W, Montaner J S G the INCAS Study Group. Program and abstracts of the XI International Conference on AIDS. XI International Conference on AIDS Society, Vancouver, British Columbia, Canada. 1996. A randomized, double-blinded comparative trial of the effects of zidovudine, didanosine and nevirapine combinations in antiviral naïve, AIDS-free HIV-infected patients with CD4 counts 200-600/mm3, abstr. Mo B294; p. 1. [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. Evaluation of precision performance of clinical chemistry devices. 2nd ed. Tentative guideline. NCCLS document EP5-T2. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1992. [Google Scholar]
- 20.O’Brien W A, Hartigan P M, Daar E S, Simberkoff M S, Hamilton J D for the VA Cooperative Study Group on AIDS. Changes in plasma HIV RNA levels and CD4+ lymphocyte counts predict both response to antiretroviral therapy and therapeutic failure. Ann Intern Med. 1997;126:939–945. doi: 10.7326/0003-4819-126-12-199706150-00002. [DOI] [PubMed] [Google Scholar]
- 21.O’Brien W A, Hartigan P M, Martin D, Esinhart J, Hill A, Benoit S, Rubin M, Simberkoff M S, Hamilton J D for the VA Cooperative Study Group on AIDS. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 22.Office of Public Health and Science, U.S. Department of Health and Human Services. Guidelines for the use of antiretroviral agents in HIV-infected adults. Fed Regist. 1997;62:33417–33418. [Google Scholar]
- 23.Perelson A S, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D D. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 24.Roche Diagnostic Systems. AMPLICOR HIV-1-MONITOR test package insert. Branchburg, N.J: Roche Diagnostic Systems; 1996. pp. 8–9. [Google Scholar]
- 25.Saag M S, Holodniy M, Kuritzkes D R, O’Brien W A, Coombs R, Poscher M E, Jacobsen D M, Shaw G M, Richman D D, Volberding P A. HIV viral load markers in clinical practice. Nat Med. 1996;2:625–629. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 26.Saksela K, Stevens C E, Rubinstein P, Taylor P E, Baltimore D. HIV-1 messenger RNA in peripheral blood mononuclear cells as an early marker of risk for progression to AIDS. Ann Intern Med. 1995;123:641–648. doi: 10.7326/0003-4819-123-9-199511010-00001. [DOI] [PubMed] [Google Scholar]
- 27.Schockmel G A, Yerly S, Perrin L. Detection of low HIV-1 RNA levels in plasma. J Acquired Immune Defic Syndr Hum Retrovirol. 1997;14:179–183. doi: 10.1097/00042560-199702010-00013. [DOI] [PubMed] [Google Scholar]
- 28.Volberding P A. HIV quantification: clinical applications. Lancet. 1996;347:71–73. doi: 10.1016/s0140-6736(96)90205-6. [DOI] [PubMed] [Google Scholar]
- 29.Wong M T, Dolan M J, Kozlow E, Doe R, Melcher G P, Burke D S, Boswell R N, Vahey M. Patterns of virus burden and T cell phenotype are established early and are correlated with the rate of disease progression in human immunodeficiency virus type 1-infected persons. J Infect Dis. 1996;173:877–887. doi: 10.1093/infdis/173.4.877. [DOI] [PubMed] [Google Scholar]
- 30.Yen-Lieberman B, Brambilla D, Jackson B, Bremer J, Coombs R, Cronin M, Herman S, Katzenstein D, Leung S, Lin H J, Palumbo P, Rasheed S, Todd J, Vahey M, Reichelder P. Evaluation of a quality assurance program for quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trials Group Virology Laboratories. J Clin Microbiol. 1996;34:2695–2701. doi: 10.1128/jcm.34.11.2695-2701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]