Abstract
Seventeen isolates of Bartonella henselae from the region of Freiburg, Germany, obtained from blood cultures of domestic cats, were examined for their genetic heterogeneity. On the basis of different DNA fingerprinting methods, including pulsed-field gel electrophoresis (PFGE), enterobacterial repetitive intergenic consensus (ERIC)-PCR, repetitive extragenic palindromic (REP) PCR, and arbitrarily primed (AP)-PCR, three different variants were identified among the isolates (variants I to III). Variant I included 6 strains, variant II included 10 strains, and variant III included only one strain. By all methods used, the isolates could be clearly distinguished from the type strain, Houston-1, which was designated variant IV. A previously published type-specific amplification of 16S rDNA differentiated two types of the B. henselae isolates (16S rRNA types 1 and 2). The majority of the isolates (16 of 17), including all variants I and II, were 16S rRNA type 2. Only one isolate (variant III) and the Houston-1 strain (variant IV) comprised the 16S rRNA type 1. Comparison of the 16S rDNA sequences from one representative strain from each of the three variants (I to III) confirmed the results obtained by 16S rRNA type-specific PCR. The sequences from variant I and variant II were identical, whereas the sequence of variant III differed in three positions. All methods applied in this study allowed subtyping of the isolates. PFGE and ERIC-PCR provided the highest discriminatory potential for subtyping B. henselae strains, whereas AP-PCR with the M13 primer showed a very clear differentiation between the four variants. Our results suggest that the genetic heterogeneity of B. henselae strains is high. The methods applied were found useful for typing B. henselae isolates, providing tools for epidemiological and clinical follow-up studies.
Bartonella henselae was identified as a new species in 1990 (12) and is the causative agent of cat-scratch disease (CSD) (2). Furthermore, mostly in immunocompromised patients, B. henselae causes bacillary angiomatosis, bacillary peliosis hepatis, osteolytic lesions, relapsing fever with bacteremia, endocarditis, and encephalitis (1, 11, 16, 22). Epidemiological studies have implicated asymptomatic cats as a major reservoir of B. henselae (6, 7, 15), but the route of transmission to humans remains unknown. Cat scratches or bites are possible modes of transmission. However, B. henselae could not be detected in gingival swabs obtained from domestic cats (15). Several studies demonstrated the presence of B. henselae in the bloodstream of healthy cats, and these animals can remain bacteremic from several months to several years (6, 7, 15). Until now, three Bartonella species had been detected in the blood of cats: B. henselae (6, 7, 15), Bartonella quintana (10), and recently Bartonella clarridgeiae (5). In the last few years, the number of Bartonella species as well as the number of diseases recognized as being caused by Bartonella species in humans has increased.
Bartonella bacteria are fastidious, slow-growing microorganisms, and distinguishing between the species is quite difficult because they are phenotypically and genotypically very similar. Definitive identification of the species requires molecular techniques. Recently, discrimination between Bartonella species and B. henselae isolates was successfully performed by repetitive-element PCR (13, 15, 18). By combining results of repetitive extragenic palindromic (REP)-PCR and enterobacterial repetitive intergenic consensus (ERIC)-PCR, Rodriguez-Barradas et al. identified five different fingerprint profiles among 17 B. henselae isolates (13). Using a PCR-based restriction fragment length polymorphism (RFLP) analysis of the 16S-23S rRNA intergenic spacer region and digestion of the amplicons with AluI and HaeII, Matar et al. (8) could differentiate between Bartonella species and obtained six different restriction patterns among 10 B. henselae isolates. Recently, Bergmans et al. (3) described the prevalence of two different B. henselae types by partial 16S rRNA gene sequence analysis. These types differ from each other in three nucleotides located at positions 172 to 175 of the 16S rRNA gene.
In a recent study (15), we investigated blood cultures of 100 domestic cats, and B. henselae was isolated from 13% of these animals. Genomic fingerprinting of nine of these isolates by ERIC-PCR identified two variants of B. henselae which were clearly distinguishable by specific patterns of PCR products. In order to investigate their genetic heterogeneity, we applied different DNA fingerprinting techniques to a total of 18 isolates of B. henselae, including the type strain Houston-1 and the 12 strains published previously (15).
(This work was presented in part at the 13th Sesqui Annual Meeting, American Society for Rickettsiology, Seven Springs Mountain Resort, Pa., 21 to 24 September 1997 [17].)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A total of 17 strains of B. henselae were obtained from domestic cats in the region of Freiburg, Germany (Table 1). Five of these isolates were obtained from cats in households with children suffering from CSD. All strains were cultured on chocolate agar plates containing 10% defibrinated sheep blood. Columbia blood agar was used as base medium for preparing the chocolate agar plates. B. henselae 103737 (which is identical to B. henselae ATTC 49882 type strain Houston-1) was obtained from the Collection de L’Institut Pasteur, Paris, France, and was used as a control for further testing.
TABLE 1.
Characteristics of the B. henselae isolates studied
| Isolate | Position in the geld | Source | No. of CFU/ml of blood | Growth time (days) | Reference or source |
|---|---|---|---|---|---|
| Houston-1a | C | HIVb | 11 | ||
| FR96/BK3 | D | Cat | 200 | 10 | 15 |
| FR96/BK8 | E | Cat | 120 | 12 | 15 |
| FR96/BK26II | F | Cat | 30 | 6 | 15 |
| FR96/BK36 | G | Cat | 100 | 7 | 15 |
| FR96/BK36II | H | Cat | 100 | 8 | 15 |
| FR96/BK38 | I | Cat | 600 | 6 | 15 |
| FR96/BK74 | K | Cat | 200 | 6 | 15 |
| FR96/BK75 | L | Cat | 120 | 8 | 15 |
| FR96/BK75II | M | Cat | 100 | 8 | 15 |
| FR96/BK77 | N | Cat | 400 | 7 | 15 |
| FR96/BK78 | O | Cat | 240 | 7 | 15 |
| FR96/BK79 | P | Cat | 800 | 7 | 15 |
| FR96/K2 | Q | Cat (CSD)c | 400 | 10 | This study |
| FR96/K4 | R | Cat (CSD) | 1,200 | 6 | This study |
| FR96/K5 | S | Cat (CSD) | 2,000 | 6 | This study |
| FR96/K6 | T | Cat (CSD) | 4,000 | 7 | This study |
| FR97/K7 | U | Cat (CSD) | 10,000 | 10 | This study |
DNA purification.
The strains were grown on chocolate agar plates at 37°C in 5% CO2 for 4 to 5 days. Cultures from plates were harvested in 1 ml of 0.01 M phosphate-buffered saline. The DNA was extracted with a commercially available kit (Qiagen GmbH, Hilden, Germany) and used as template for the PCR.
PFGE.
For pulsed-field gel electrophoresis (PFGE), bacteria were harvested from chocolate agar plates after incubation at 37°C for 7 days. Bacteria were suspended in 0.9% NaCl, adjusted to an optical density at 600 nm (OD600) of 0.85, and washed three times with 0.9% NaCl. Agarose blocks were prepared by adding 500 μl of cell suspension to 700 μl of 2% PFGE agarose (Sigma, Deisenhofen, Germany). The solidified blocks were incubated in lysis buffer (0.5 mM EDTA, 1% lauroyl sarcosine, 1.8 mg of proteinase K per ml [pH 9.5]) at 56°C overnight. After a thorough washing with TE buffer (10 mM Tris, 10 mM EDTA [pH 7.5]), the blocks were incubated with 20 U of SmaI for 6 h under the conditions recommended by the enzyme manufacturer (New England BioLabs, Schwalbach, Germany). PFGE was done with a CHEF DRIII electrophoresis unit (BioRad, Munich, Germany). Agarose gel (2%) was prepared in 0.5× TBE running buffer (90 mM Tris, 90 mM boric acid, 2 mM EDTA [pH 8.5]). Electrophoresis was performed for 30 h at 5.9 V/cm with pulse times from 3 to 12 s at a constant temperature of 14°C. Agarose gels were stained with ethidium bromide and photographed under UV illumination with an EasyImage documentation system (Herolab, Wiesloch, Germany). Low-range PFG marker (New England BioLabs) was used as a molecular weight standard.
ERIC-PCR.
The primers ERIC1R (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC2 (5′-AAGTAAGTGACTGGGGTGAG-AGCG-3′) used in this study have been described previously by Versalovic et al. (21). The reaction mixture contained 8 ng of bovine serum albumin (Sigma) per ml, 200 μM (each) the four deoxynucleoside triphosphates (dNTPs), primers (117 nM each), 2 U of Taq polymerase (Pharmacia Biotech, Freiburg, Germany), and 100 ng of genomic DNA in 50.0 μl of TBE buffer. The mixture was overlaid with 2 drops of light mineral oil, and PCR amplification was performed with an automated thermal cycler (Robocycler 40; Stratagene) with initial denaturation (95°C, 7 min), followed by 30 cycles of denaturation (94°C, 1 min), annealing (40°C, 1 min), and extension (65°C, 8 min), with a single final extension (65°C, 16 min). The amplified products (20 μl) were electrophoretically separated in a 1% agarose gel at 120 V for 2 h in 0.5× TBE buffer, stained with ethidium bromide, visualized on a UV transilluminator, and photographed with Polaroid 665 film.
REP-PCR.
The primers Rep1R-Dt, 5′-(AGCT)CG(AGCT)CG(AGCT)CATC(AGCT)GGC-3′, and Rep2-D, 5′-(GA)CG(CT)CTTA-TC(CA)GGCCTAC-3′, used have been described previously by Versalovic et al. (21). Each 30-μl reaction mixture contained 2.5 μl of 10× reaction buffer (Pharmacia Biotech), 50 pmol of each of the primers, 100 ng of genomic DNA, 6.6 ng of bovine serum albumin (Sigma) per ml, 160 μM (each) dNTP, 2 U of Taq polymerase (Pharmacia Biotech), and 2.5 μl of dimethyl sulfoxide (Merck, Darmstadt, Germany). The amplification cycles were as follows: 1 cycle at 95°C for 7 min; 30 cycles at 90°C for 30 s, 43°C for 1 min, and 65°C for 8 min; and 1 cycle at 65°C for 16 min.
AP-PCR.
The core sequence of phage M13 (5′-GAGGGTGGCGGTTCT-3′) was used for arbitrarily primed (AP)-PCR as a single primer (4). Amplification reactions were carried out in a final volume of 50 μl containing 5 μl of 10× reaction buffer (Pharmacia Biotech), 10 ng of bovine serum albumin (Sigma) per ml, 250 pmol of the M13 primer, 100 ng of genomic DNA, 200 μM (each) dNTP, and 2 U of Taq polymerase (Pharmacia Biotech). The amplification cycles were as follows: 1 cycle at 94°C for 5 min, 4 cycles at 94°C for 30 s, 43°C for 2 min, and 72°C for 2 min; 30 cycles at 94°C for 20 s, 50°C for 1 min, and 72°C for 30 s; and 1 cycle at 72°C for 5 min.
16S rRNA type-specific amplification of B. henselae DNA.
The primers BH1, 5′-AATCCCTCTTTCTAAATAGCC-3′, and BH2, 5′-TAAACCTCTTTCTAAATAGCC-3′, in combination with the broad-host-range primer 16SF, 5′-AGAGTTTGATCCTGG(CT)TCAG-3′, were used for the type-specific amplification of B. henselae DNA as described by Bergmans et al. (3).
DNA amplification was carried out in 50-μl reaction volumes containing 5 μl of 10× reaction buffer (Pharmacia Biotech), 8 ng of bovine serum albumin (Sigma) per ml, 200 μM (each) dNTP, 20 pmol of each primer, 100 ng of genomic DNA, and 2 U of Taq polymerase (Pharmacia Biotech). PCR cycling consisted of 30 cycles of 20 s at 95°C, 30 s at 56°C, and 1 min at 72°C preceded by an initial denaturation of 3 min at 95°C and followed by a final extension of 5 min at 72°C. PCR products were separated on a 1.5% agarose gel and visualized by staining with ethidium bromide.
The DNA molecular weight marker VI (Boehringer GmbH, Mannheim, Germany) and a 1-kb ladder (ΦX174 replicative-form DNA HincII digest; Pharmacia Biotech) were used as standards for electrophoresis of DNA.
PCR amplification and restriction analysis of the spacer region between the 16S and 23S DNAs.
Oligonucleotide primers RPC5 (5′-AAGTCGTAACAAGGTA-3′) and R23S2693 (5′-TACTGGTTCACTATCGGTCA-3′) described by Matar et al. (8) were used for the amplification of the spacer region separating the 16S and 23S regions. The amplification was performed in 100-μl reaction volumes containing 10 μl of 10× reaction buffer (Pharmacia Biotech), 8 ng of bovine serum albumin (Sigma) per ml, 200 μM (each) dATP, dCTP, dGTP, and dTTP, 20 pmol of each primer, 100 ng of DNA, and 2.5 U of Taq polymerase (Pharmacia Biotech). Amplifications were performed with an automated thermal cycler (Robocycler 40; Stratagene) with initial denaturation (3 cycles of 70 s at 94°C, 150 s at 55°C, and 3 min at 72°C), followed by 30 cycles of denaturation (94°C, 30 s), annealing (55°C, 150 s), and extension (72°C, 180 s), with a single final extension step (72°C, 10 min). Five microliters of each PCR product was digested with different restriction endonucleases (HindIII, TaqI, HaeIII, HinfI, PvuII, and AluI) according to the manufacturer’s instructions (New England BioLabs). The resulting DNA fragments were analyzed after electrophoresis on an ethidium-bromide-stained 2% agarose gel (Biozym).
Amplification and sequencing of 16S rRNA genes.
Universal prokaryotic primers TPU1 and RTU8 (Table 2) were used to amplify a 1,500-bp fragment of the 16S rRNA gene from the purified genomic DNA of the cultured isolates FR96/BK3, FR96/BK38, and FR97/K7. The PCR was carried out in a final volume of 100 μl containing 10 μl of 10× reaction buffer (Perkin-Elmer Cetus; Norwalk, Conn.), 200 μM (each) dNTP, 0.3 μM (each) primer, 2 U of Taq polymerase (AmpliTaq Gold; Perkin-Elmer Cetus), and 50 ng of DNA with an automated thermal cycler (model 2400; Perkin-Elmer Cetus) under the following conditions: an initial step at 94°C for 9 min; followed by 30 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 90 s; and a final elongation step at 72°C for 10 min. The purity of the amplified product was determined by electrophoresis in a 1.5% agarose gel (FMC Bioproducts; Rockland, Maine). After the PCR products were purified with the QIAquick PCR purification kit (Qiagen), both strands were sequenced with an automated thermal cycler (model 2400; Perkin-Elmer Cetus) with the Prism DyeDeoxy terminator cycle sequencing kit (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). Sequencing products were purified with MicroSpin S-200 HR columns (Amersham Pharmacia Biotech, Uppsala, Sweden) and analyzed with an ABI Prism 377 DNA sequencer (Perkin-Elmer, Applied Biosystems Division).
TABLE 2.
Oligonucleotide primers used for amplification and sequencing of 16S rDNA
| Primer | Sequence (5′ to 3′) | Positiona |
|---|---|---|
| TPU1 | AGA GTT TGA TCM TGG CTC AG | EC8–27 |
| TPU2 | CCA RAC TCC TAC GGG AGG CA | EC334–353 |
| TPU3 | CAG CMG CCG CGG TAA TWC | EC519–536 |
| TPU4 | GGA TTA GAT ACC CTG GTA GTC C | EC785–806 |
| TPU5 | AAA CTY AAA KGA ATT GAC GG | EC907–926 |
| TPU6 | GGG CKA CAC ACG TGC TAC AAT | EC1220–1240 |
| RTU2 | TGC CTC CCG TAG GAG TYT GG | EC334–353 |
| RTU3 | GWA TTA CCG CGG CKG CTG | EC519–536 |
| RTU4 | TAC CAG GGT ATC TAA TCC TGT T | EC781–802 |
| RTU5 | CCG TCA ATT CMT TTR AGT TT | EC907–926 |
| RTU6 | ATT GTA GCA CGT GTG TMG CCC | EC1220–1240 |
| RTU8 | AAG GAG GTG ATC CAK CCR CA | EC1525–1544 |
Numbering is based upon Escherichia coli sequence.
16S rRNA sequence analysis.
Comparison of 16S rDNA sequences with databases was done as previously described (19).
Visual analysis of band patterns.
The fingerprints obtained were compared for similarity by visual inspection of band patterns. Sizes of DNA fragments amplified by PCR were determined by direct comparison with the DNA marker. Fingerprints were considered highly similar when all visible bands obtained had the same migration distance for each isolate. Variations in intensity and shape of bands among isolates were not considered differences. The presence or absence of one distinct band was considered a difference.
Computer-assisted analysis of the DNA fingerprints.
All fingerprints were analyzed with the Windows version of GelCompar software version 4.0 (Applied Maths, Kortrijk, Belgium). The patterns produced by ERIC-, REP-, and AP-PCR were compared with the Pearson correlation coefficient, which considers both number of bands and band intensity. PFGE fingerprints were analyzed by applying the Dice coefficient to peaks. For clustering, the unweighted pair group method with arithmetic means (UPGMA method) was used. A tolerance in the band positions of 1.2% was applied for comparison of the fingerprint patterns. Fingerprint analysis and the methods and algorithms used in this study were performed according to the instructions of the manufacturer.
Nucleotide sequence accession numbers.
The 16S rDNA nucleotide sequence data reported in this study have been deposited in the EMBL, GenBank, and DDBJ nucleotide sequence databases. Accession numbers are as follows: FR96/BK3 to AJ223778, FR96/BK38 to AJ223779, and FR97/K7 to AJ223780.
RESULTS
PFGE.
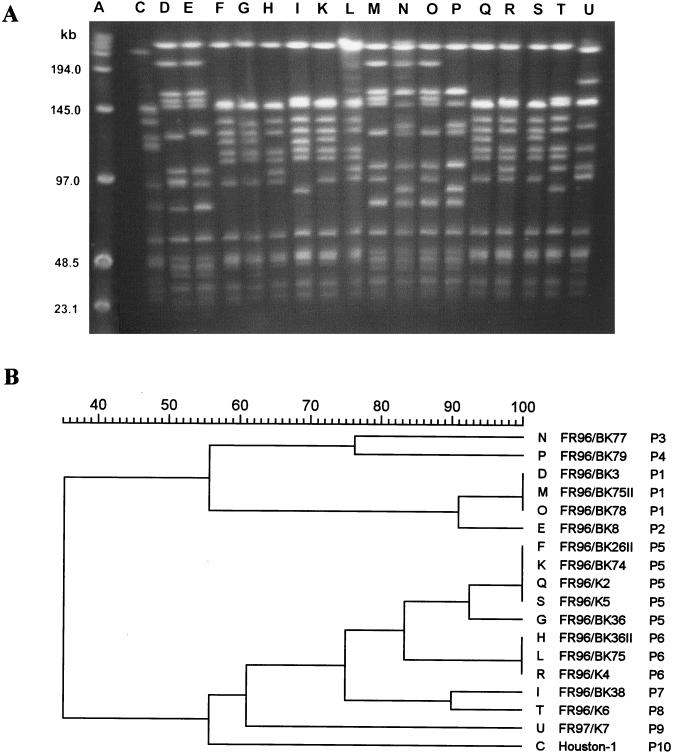
Digestion of DNA from the B. henselae strains with SmaI (CCCGGG) created between 10 and 15 chromosomal fragments for each isolate. The molecular sizes of the SmaI fragments ranged from 20 to more than 200 kb (Fig. 1A). Among the 18 strains, 10 different patterns were observed by visual inspection of band patterns (Table 3).
FIG. 1.
DNA fingerprint analysis of the 18 B. henselae strains by PFGE (A) and dendrogram of the fingerprints as determined by the Dice method (B). Lane A, molecular size markers; lane C, pattern P10 (variant IV); lanes D, E, M, N, O, and P, patterns P1, P2, P3, and P4 (variant I); lanes F, G, H, I, K, L, Q, R, S, and T, patterns P5, P6, P7, and P8 (variant II); lane U, pattern P9 (variant III).
TABLE 3.
Summary of results obtained by different typing methods for B. henselae variants
| Gel positionc | Isolate | Pattern or group obtained by indicated method
|
16S rRNA type-specific PCRa | Proposed variantb | ||||
|---|---|---|---|---|---|---|---|---|
| PFGE | ERIC-PCR | REP-PCR | AP-PCR | AluI RFLP | ||||
| C | Houston-1 | P10 | E4 | R4 | M4 | A1 | 1 | IV |
| D | FR96/BK3 | P1 | E1.1 | R1 | M1 | A2 | 2 | I |
| E | FR96/BK8 | P2 | E1 | R1 | M1 | A2 | 2 | I |
| M | FR96/BK75II | P1 | E1 | R1 | M1 | A2 | 2 | I |
| N | FR96/BK78 | P | E1 | R1 | M1 | A2 | 2 | I |
| O | FR96/BK78 | P1 | E1 | R1 | M1 | A2 | 2 | I |
| P | FR96/BK79 | P4 | E1 | R1 | M1 | A2 | 2 | I |
| F | FR96/BK26II | P5 | E2 | R2 | M2 | A2 | 2 | II |
| G | FR96/BK36 | P5 | E2 | R2 | M2 | A2 | 2 | II |
| H | FR96/BK36II | P6 | E2 | R2 | M2 | A2 | 2 | II |
| I | FR96/BK38 | P7 | E2 | R2 | M2 | A2 | 2 | II |
| K | FR96/BK74 | P5 | E2 | R2 | M2 | A2 | 2 | II |
| L | FR96/BK75 | P6 | E2 | R2 | M2 | A2 | 2 | II |
| Q | FR96/K2 | P5 | E2 | R2 | M2 | A2 | 2 | II |
| R | FR96/K4 | P6 | E2 | R2 | M2 | A2 | 2 | II |
| S | FR96/K5 | P5 | E2 | R2 | M2 | A2 | 2 | II |
| T | FR96/K6 | P8 | E2 | R2 | M2 | A2 | 2 | II |
| U | FR97/K7 | P9 | E3 | R3 | M3 | A1 | 1 | III |
With the Dice coefficient, these 10 types could be discriminated at a cutoff level of 92% similarity. The four variants (I to IV) could be differentiated at a similarity level of 74% in the dendrogram. Variant I could be subdivided into two groups (Fig. 1B). The average sum of fragment sizes was at least 1.5 Mbp for all B. henselae strains investigated.
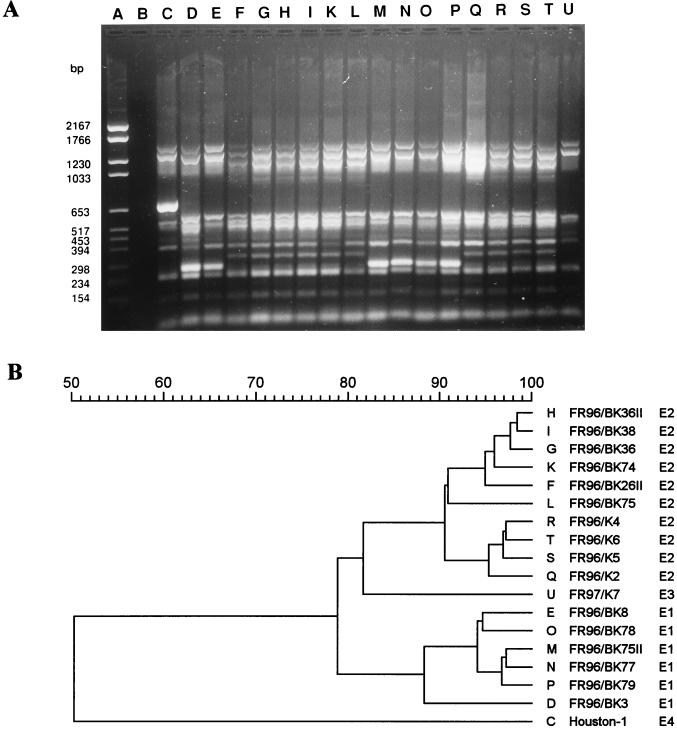
ERIC-PCR.
Investigation of the 18 strains of B. henselae by ERIC-PCR resulted in banding patterns, which allowed the definition of subtypes. The patterns consisted of approximately 7 to 11 bands per isolate (Fig. 2A). The molecular sizes of the fragments ranged from 200 to 1,500 bp. The patterns obtained allowed a visual differentiation of four major fingerprint patterns (E1, E2, E3, and E4). Comparison of the banding patterns within strains was done by the method of Pearson (Fig. 2B). The four variants (E1 to E4) could be identified at 88% similarity of PCR fingerprints in the dendrogram. The first variant (E1) contained 6 isolates, the second (E2) contained 10 isolates, and the third (E3) contained only 1 isolate (Table 3). The type strain of B. henselae (Houston-1) was different from all of our isolates, indicating an additional variant, E4. The isolate FR96/BK3 (lane D) had an additional band of approximately 500 bp by visual inspection (E1.1) compared with the other strains of variant I. This was also recognized in the computer-assisted analysis, in which this isolate was clearly discriminated from all other strains within variant E1. The fingerprint patterns of the four isolates from cats to which patients with CSD were exposed (FR96/K2, FR96/K4, FR96/K5, and FR96/K6) formed a single closely related group within the cluster E2 and showed a similarity of 95% between the isolates.
FIG. 2.
DNA fingerprint analysis of the 18 B. henselae strains by ERIC-PCR (A) and dendrogram of the fingerprints as determined by the Pearson method (B). Lane A, molecular size marker; lane B, negative control; lane C, pattern E4 (variant IV); lanes D, E, M, N, O, and P, pattern E1 (variant I); lanes F, G, H, I, K, L, Q, R, S, and T, pattern E2 (variant II); lane U, pattern E3 (variant III).
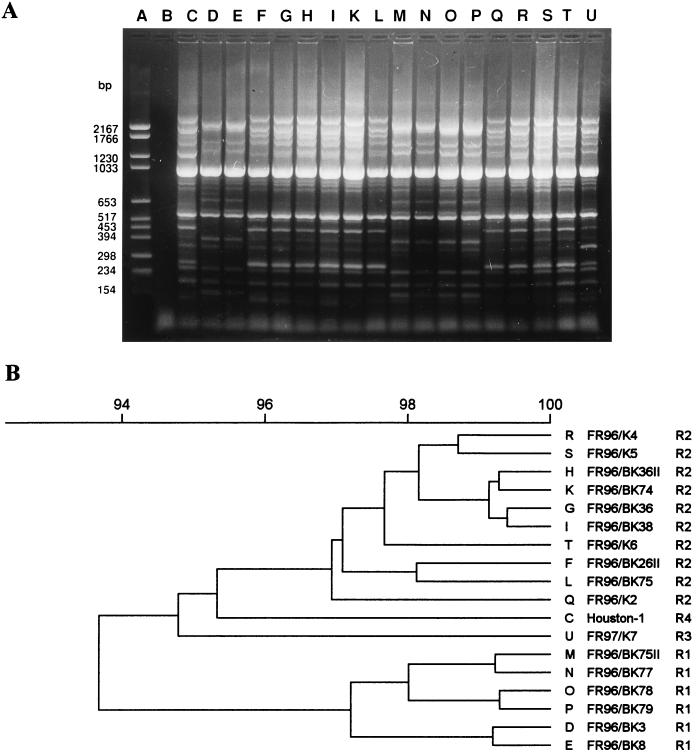
REP-PCR.
Fingerprints created by REP-PCR were more complex than the patterns generated by ERIC- or AP-PCR but less complex than those obtained by PFGE. They consisted of 17 to 19 bands per isolate, showing a molecular size of up to 2,200 bp (Fig. 3A). Visual inspection of the band patterns and computer-assisted analysis of the DNA fingerprints determined by the Pearson method (Fig. 3B), with a cutoff level of 97% correlation on the dendrogram, revealed four major fingerprint profiles (R1 to R4). The reference strain (pattern R4) and the isolate FR97/K7 (pattern R3) were different from all other strains. The first and second clusters (patterns R1 and R2, respectively) contained the same isolates as those seen by ERIC-PCR (Table 3). By REP-PCR, the distinction between the isolate FR96/BK3 (lane D) and the other strains within group E1 as shown by ERIC-PCR could not be detected. Within the groups R1 and R2 no differences were observed between the patterns by visual inspection.
FIG. 3.
DNA fingerprint analysis of the 18 B. henselae strains by REP-PCR (A) and dendrogram of the fingerprints as determined by the Pearson method (B). Lane A, molecular size marker; lane B, negative control; lane C, pattern R4 (variant IV); lanes D, E, M, N, O, and P, pattern R1 (variant I); lanes F, G, H, I, K, L, Q, R, S, and T, pattern R2 (variant II); lane U, pattern R3 (variant III).
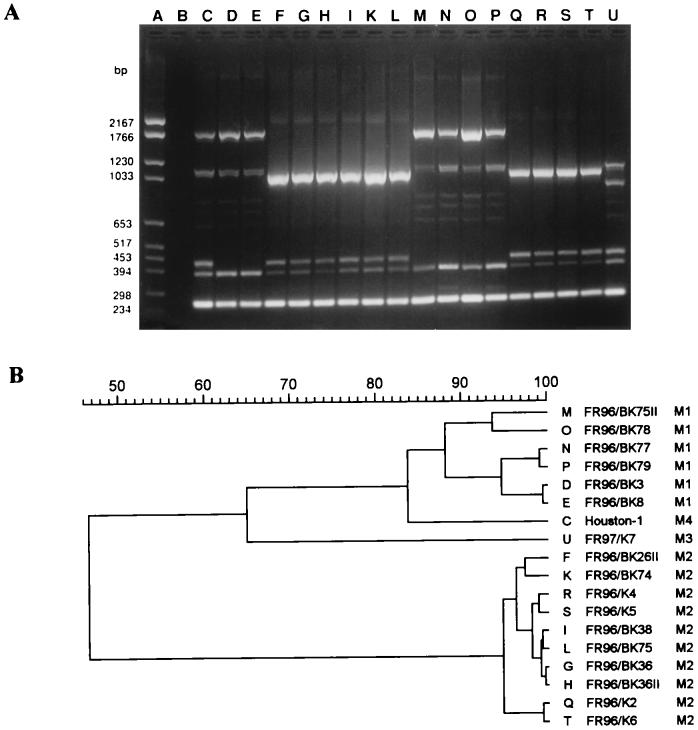
AP-PCR.
The M13 core sequence primer (Fig. 4) generated the lowest number of amplification products. Only three to seven fragments between 0.2 and 1.7 kbp in size could be amplified. However, the patterns obtained with the M13 primer showed a very clear differentiation between the four fingerprint patterns seen by ERIC- and REP-PCR at a cutoff level of 88% correlation on the dendrogram. The patterns were sufficiently distinct to allow division of the strains into four subgroups, M1, M2, M3, and M4 (Table 3).
FIG. 4.
DNA fingerprint analysis of the 18 B. henselae strains by AP-PCR with the M13 core sequence primer (A) and dendrogram of the fingerprints as determined by the Pearson method (B). Lane A, molecular size marker; lane B, negative control; lane C, pattern M4 (variant IV); lanes D, E, M, N, O, and P, pattern M1 (variant I); lanes F, G, H, I, K, L, Q, R, S, and T, pattern M2 (variant II); lane U, pattern M3 (variant III).
16S rRNA type-specific amplification of B. henselae DNA.
With the primers BH1 and BH2 and the broad-host-range primer 16SF, the two types described by Bergmans et al. (3) could be determined for all strains (Table 3). Interestingly, Bergmans type 1 was detected in only one isolate (FR97/K7) and in the reference strain (Fig. 5A) corresponding to variants III and IV, respectively, observed in this study. The 16 isolates of variants I and II were Bergmans type 2 (Fig. 5B).
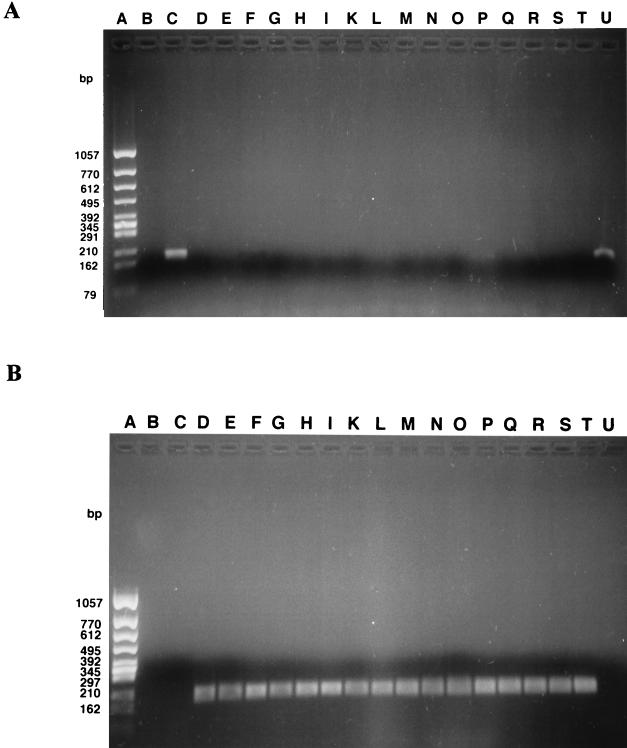
FIG. 5.
Type-specific PCR products of the 18 B. henselae strains. PCR products were obtained with primer BH1 (A) and primer BH2 (B), which detect B. henselae Bergmans type 1 and Bergmans type 2, respectively. Lanes A, molecular size markers; lanes B, negative controls; lanes C and U, Bergmans type 1; lanes D, E, F, G, H, I, K, L, M, N, O, P, Q, R, S, and T, Bergmans type 2. Bergmans type 1 includes variants III and IV. Bergmans type 2 includes variants I and II.
PCR amplification and restriction analysis of the spacer region between the 16S and 23S DNAs.
PCR with primers described earlier by Matar et al. (8) generated DNA fragments of approximately 1,600 bp in size from all strains investigated. From the restriction endonucleases evaluated, only AluI created a fragment pattern which allowed differentiation between the strains (Fig. 6). The reference strain and isolate FR97/K7 (resembling Bergmans type 1) showed six bands (pattern A1, Table 3), whereas all other B. henselae strains showed seven bands (pattern A2, Table 3) corresponding to Bergmans type 2.
FIG. 6.
AluI RFLP patterns of PCR-amplified 16S-23S spacer regions from the 18 B. henselae strains. Lane A, molecular size marker; lane B, negative control; lanes C and U, pattern A1; lanes D, E, F, G, H, I, K, L, M, N, O, P, Q, R, S, and T, pattern A2. Pattern A1 corresponds to Bergmans type 1 or to variants IV and III. Pattern A2 corresponds to Bergmans type 2 or to variants I and II.
16S rRNA sequences.
The 16S rRNA sequences were determined for one representative strain from each of the three variant groups of the German isolates (FR96/BK3, corresponding to variant I; FR96/BK38, corresponding to variant II; and FR97/K7, corresponding to variant III). Sequencing of 16S rRNA genes from isolates FR96/BK3, FR96/BK38, and FR97/K7 revealed that FR96/BK3 and FR96/BK38 were identical and contained the type 2 sequence pattern described by Bergmans et al. (3). The sequence from isolate FR97/K7 differed in 3 bp located at positions 172 to 175 of the 16S rRNA gene, showing the Bergmans type 1 sequence pattern (data not shown). Comparison of isolate sequences with EMBL and GenBank databases with the FASTA-opt algorithm of the Heidelberg Unix Sequence Analysis Resources program package (version 4.0; Deutsches Krebsforschungszentrum, Heidelberg, Germany) showed that the isolate sequences showed highest similarity (>99%) to the reference sequences M73229 (Bergmans type 1) and Z11684 from B. henselae (Bergmans type 2).
DISCUSSION
The aim of this study was to compare the usefulness of various molecular methods for typing B. henselae isolates. Our results should allow further analysis of strain relatedness and may provide a useful basis for further studies on the epidemiology and the pathogenicity of this bacterium. Six different subtyping techniques, including PFGE, ERIC-PCR, REP-PCR, AP-PCR, 16S rRNA type-specific PCR, and PCR amplification and restriction analysis of the spacer region between the 16S and the 23S rDNAs, were established to investigate the genetic relatedness of 17 B. henselae strains isolated from blood cultures of domestic cats in the region of Freiburg, Germany, and the Houston-1 reference strain. By all methods, four different major variants of the B. henselae isolates could be distinguished. The cat isolates showed three variants (I to III) which differed from the Houston-1 reference strain (variant IV).
PFGE has been previously shown to be a suitable method for differentiation of Bartonella strains at the species level (9, 14) and for isolate identification (14). Genomic DNA analysis by PFGE with SmaI revealed 10 different subgroups (P1 to P10). Analysis of PFGE fingerprints was performed initially with the Pearson correlation coefficient according to the other typing techniques. This method considers not only bands but also the densitogram of a whole pattern, thus being advantageous in the analysis of fingerprints containing bands of different brightness (e.g., AP-PCR). On the other hand, this method is strongly affected by differences in the amount of DNA from lane to lane or by artifacts, such as incomplete restriction. Lane L of the pulsed-field gel (strain FR96/BK75) exhibits such an artifact, disarranging the correspondent dendrogram as calculated with the Pearson coefficient (dendrogram not shown). Therefore, PFGE fingerprints were analyzed by the UPGMA method, with the Dice coefficient applied to peak positions.
The molecular weight sums of the DNA fragments revealed a genome size of approximately >1.5 Mbp in all strains investigated. This result is in good agreement with the genome size of 2.0 Mbp determined earlier by Roux and Raoult (14). The fact that the sum of the fragments was not exactly 2.0 Mbp can be explained by the eventual presence of double bands, especially in the high-molecular-weight range above 194 kbp.
Analysis by REP- and ERIC-PCR generates species- and strain-specific DNA fingerprints of gram-negative enteric bacteria (21). Repetitive sequences of the bacterial genome are useful targets for DNA-based typing due to their restricted length and their widespread occurrence, although little is known about their function (20, 21). However, it is not known whether such sequences exist in Bartonella spp. Arbitrary binding of primers cannot be excluded. Both methods have been applied earlier to differentiate Bartonella organisms on the species level and to subtype various B. henselae isolates (13). The 17 isolates of B. henselae (7 cultured from tissue and blood of human immunodeficiency virus-infected patients, 2 from patients suffering from CSD, 8 from the blood of cats) investigated by Rodriguez-Barradas et al. (13) comprised five different fingerprint profiles with the combined results of REP-PCR and ERIC-PCR. We could confirm the applicability of ERIC-PCR for distinguishing between B. henselae and B. quintana isolates in a recent study (15). Concordant with the results of Rodriguez-Barradas et al. (13), ERIC-PCR banding patterns (7 to 11 bands per isolate) were less complex than those obtained by REP-PCR (17 to 19 bands per isolate). Variants I to IV were detected by both methods, although the divergence within the clusters was higher by ERIC-PCR (pattern E1.1 within cluster E1).
In addition to DNA repeat-based genetic variation, DNA polymorphisms can be detected by the selective amplification of sequences with arbitrarily chosen primers. The core sequence of phage M13 was used successfully for molecular typing of other bacteria, such as Acinetobacter baumanii (4). When this primer was used with AP-PCR, only four to seven fragments could be amplified, but the patterns were highly discriminative for the Bartonella strains.
RFLP analysis of the 16S-23S rDNA spacer region with AluI allowed us to differentiate only two groups (A1 and A2). In contrast, Matar et al. (8) found six distinct AluI RFLP patterns in 11 culture-grown B. henselae strains. The correlation in our isolates between the AluI RFLP pattern and the 16S rRNA type was 100%. By the 16S rRNA type-specific PCR described by Bergmans et al. (3), the 18 B. henselae strains yielded two different types: 16 were Bergmans type 2, whereas strain FR97/K7 and the reference strain were Bergmans type 1. The majority (82%; 32 of 41 samples) of the lymph nodes from patients with CSD investigated by Bergmans et al. (3) contained type 1 B. henselae. In contrast, 16 of 17 of our isolates from cats contained type 2 B. henselae. In contrast, 16 of 17 of our isolates from cats contained type 2 B. henselae. These results suggest that B. henselae type 1 might be more pathogenic than B. henselae type 2 in humans. The predominance of different types in various geographic regions (The Netherlands and Germany) may also explain the discrepancies between our results and those of Bergmans et al. (3). Whereas B. henselae type 1 seems to be predominant in CSD patients in The Netherlands, B. henselae type 2 is predominant in bacteremic German domestic cats.
On the basis of the typing results obtained by different methods, the cat isolates were divided into three variant groups. Comparison of the 16S rDNA sequences/ of one member of each group revealed that isolates FR96/BK3 (variant I) and FR96/BK38 (variant II) showed no differences in their 16S rRNA gene sequences, while the sequence of FR97/K7 (variant III) differed at three positions. These intraspecies sequence variations were previously reported by Bergmanns et al. (3). The findings that FR96/BK3 and FR96/BK38 contained the B. henselae type 2 16S rRNA, whereas FR97/K7 contained the B. henselae type 1, confirmed the results of the 16S rRNA type-specific PCR with the respective isolates.
In conclusion, all fingerprinting methods applied were found to be useful for subtyping B. henselae. All methods allowed differentiation of four major variants (I to IV). However, PFGE showed the highest discriminatory power, followed by ERIC-PCR. Lower banding patterns, such as that obtained by AP-PCR with the M13 primer, showed the clearest differentiation of the four variants. Sequence analysis of the 16S rRNA demonstrated that the differences between isolates of variants I and II are not localized within the 16S rRNA sequence. Therefore, PFGE and the PCR-based fingerprint methods (ERIC-PCR, REP-PCR, and AP-PCR) have more discriminatory potency than the 16S rRNA type-specific PCR and the PCR-based AluI RFLP analysis of the 16S rRNA. Major advantages of the PCR-based fingerprint methods (ERIC-PCR, REP-PCR, and AP-PCR) are technical simplicity, wide availability of equipment and reagents, and rapid feasibility. However, macrorestriction endonuclease analysis of genomic DNA by PFGE is more discriminatory but also more expensive and time-consuming.
Our results suggest that the genetic heterogeneity of B. henselae strains is high, providing tools for epidemiological and clinical follow-up studies.
REFERENCES
- 1.Anderson B E, Neuman M A. Bartonella spp. as emerging human pathogens. Clin Microbiol Rev. 1997;10:203–219. doi: 10.1128/cmr.10.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson B, Sims K, Regnery R, Robinson L, Schmidt M J, Goral S, Hager C, Edwards K. Detection of Rochalimaea henselae DNA in specimens from cat scratch disease patients by PCR. J Clin Microbiol. 1994;32:942–948. doi: 10.1128/jcm.32.4.942-948.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmans A M C, Schellekens J F P, van Embden J D A, Schouls L M. Predominance of two Bartonella henselae variants among cat-scratch disease patients in The Netherlands. J Clin Microbiol. 1996;34:254–260. doi: 10.1128/jcm.34.2.254-260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gräser Y, Klare I, Halle E, Gantenberg R, Buchholz P, Jacobi H D, Presber W, Schönian G. Epidemiological study of an Acinetobacter baumanii outbreak by using polymerase chain reaction fingerprinting. J Clin Microbiol. 1993;31:2417–2420. doi: 10.1128/jcm.31.9.2417-2420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurfield A N, Boulouis H J, Chomel B B, Heller R, Kasten R W, Yamamoto K, Piemont Y. Coinfection with Bartonella clarridgeiae and Bartonella henselae and with different Bartonella henselae strains in domestic cats. J Clin Microbiol. 1997;35:2120–2123. doi: 10.1128/jcm.35.8.2120-2123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koehler J E, Glaser A, Tappero J W. Rochalimaea henselae infection. A new zoonosis with domestic cat as reservoir. JAMA. 1994;271:531–535. doi: 10.1001/jama.271.7.531. [DOI] [PubMed] [Google Scholar]
- 7.Kordick D L, Breitschwerdt E B. Intraerythrocytic presence of Bartonella henselae. J Clin Microbiol. 1995;33:1655–1656. doi: 10.1128/jcm.33.6.1655-1656.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matar G M, Swaminathan B, Hunter S B, Slater L N, Welch D. Polymerase chain reaction-based restriction fragment length polymorphism analysis of a fragment of the ribosomal operon from Rochalimaea species for subtyping. J Clin Microbiol. 1993;31:1730–1734. doi: 10.1128/jcm.31.7.1730-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maurin M, Roux V, Stein A, Ferrier F, Viraben R, Raoult D. Isolation and characterization by immunofluorescence, sodium dodecyl sulfate-polycrylamide gel electrophoresis, Western blot, restriction fragment length polymorphism-PCR, 16S rRNA gene sequencing, and pulsed-field gel electrophoresis of Rochalimaea quintana from a patient with bacillary angiomatosis. J Clin Microbiol. 1994;32:1166–1171. doi: 10.1128/jcm.32.5.1166-1171.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raoult D, Drancourt M, Carta A, Gastaut J A. Bartonella (Rochalimaea) quintana isolation in a patient with chronic adenopathy, lymphopenia and a cat. Lancet. 1994;343:977. doi: 10.1016/s0140-6736(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 11.Regnery R L, Anderson B E, Clarridge III J E, Rodriguez-Barradas M C, Jones D C, Carr J H. Characterization of a novel species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J Clin Microbiol. 1992;30:265–274. doi: 10.1128/jcm.30.2.265-274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez-Barradas M C, Hamill R J, Houston E D, Georghiou P R, Clarridge J E, Regnery R L, Koehler J E. Genomic fingerprinting of Bartonella species by repetitive element PCR for distinguishing species and isolates. J Clin Microbiol. 1995;33:1089–1093. doi: 10.1128/jcm.33.5.1089-1093.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roux V, Raoult D. Inter- and intraspecies identification of Bartonella (Rochalimaea) species. J Clin Microbiol. 1995;33:1573–1579. doi: 10.1128/jcm.33.6.1573-1579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sander A, Bühler C, Pelz K, von Cramm E, Bredt W. Detection and isolation of two Bartonella henselae variants in domestic cats in Germany. J Clin Microbiol. 1997;35:584–587. doi: 10.1128/jcm.35.3.584-587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sander A, Kaliebe T, Bredt W. Bartonella (Rochalimaea)-Infektionen: Katzenkratzkrankheit und bazilläre Angiomatose. Dtsch Med Wochenschr. 1996;121:65–69. doi: 10.1055/s-2008-1042973. [DOI] [PubMed] [Google Scholar]
- 17.Sander A, Ruess M, Steinbrueckner B, Bredt W. Presented at the 13th Sesqui Annual Meeting, American Society for Rickettsiology, 21 to 24 September 1997, Seven Springs Mountain Resort, Pa. 1997. DNA-fingerprinting of Bartonella henselae by PFGE, ERIC-PCR, REP-PCR and AP-PCR. [Google Scholar]
- 18.Schmidt H U, Kaliebe T, Poppinger J, Bühler C, Sander A. Isolation of Bartonella quintana from an HIV-positive patient with bacillary angiomatosis. Eur J Clin Microbiol Infect Dis. 1996;15:736–741. doi: 10.1007/BF01691961. [DOI] [PubMed] [Google Scholar]
- 19.Schuppler M, Mertens F, Schön G, Göbel U B. Molecular characterization of nocardioform actinomycetes in activated sludge by 16S rRNA analysis. Microbiology. 1995;141:513–521. doi: 10.1099/13500872-141-2-513. [DOI] [PubMed] [Google Scholar]
- 20.van Belkum A. DNA fingerprinting of medically important microorganisms by use of PCR. Clin Microbiol Rev. 1994;7:174–184. doi: 10.1128/cmr.7.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch D F, Pickett D A, Slater L N, Steigerwalt A G, Brenner D J. Rochalimaea henselae sp. nov., a cause of septicemia, bacillary angiomatosis, and parenchymal bacillary peliosis. J Clin Microbiol. 1992;30:275–280. doi: 10.1128/jcm.30.2.275-280.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]