Abstract
To assess genetic variation in hepatitis C virus (HCV) sequences accurately, we optimized a method for identifying distinct viral clones without determining the nucleotide sequence of each clone. Twelve serum samples were obtained from seven individuals soon after they acquired HCV during a prospective study, and a 452-bp fragment from the E2 region was amplified by reverse transcriptase PCR and cloned. Thirty-three cloned cDNAs representing each specimen were assessed by a method that combined heteroduplex analysis (HDA) and a single-stranded conformational polymorphism (SSCP) method to determine the number of clonotypes (electrophoretically indistinguishable cloned cDNAs) as a measure of genetic complexity (this combined method is referred to herein as the HDA+SSCP method). We calculated Shannon entropy, incorporating the number and distribution of clonotypes into a single quantifier of complexity. These measures were evaluated for their correlation with nucleotide sequence diversity. Blinded analysis revealed that the sensitivity (ability to detect variants) and specificity (avoidance of false detection) of the HDA+SSCP method were very high. The genetic distance (mean ± standard deviation) between indistinguishable cloned cDNAs (intraclonotype diversity) was 0.6% ± 0.9%, and 98.7% of cDNAs differed by <2%, while the mean distance between cloned cDNAs with different patterns was 4.0% ± 3.2%. The sensitivity of the HDA+SSCP method compared favorably with either HDA or the SSCP method alone, which resulted in intraclonotype diversities of 1.6% ± 1.8% and 3.5% ± 3.4%, respectively. The number of clonotypes correlated strongly with genetic diversity (R2, 0.93), but this correlation fell off sharply when fewer clones were assessed. This HDA+SSCP method accurately reflected nucleotide sequence diversity among a large number of viral cDNA clones, which should enhance analyses to determine the effects of viral diversity on HCV-associated disease. If sequence diversity becomes recognized as an important parameter for staging or monitoring of HCV infection, this method should be practical enough for use in laboratories that perform nucleic acid testing.
Genetic variation of certain RNA viruses may explain their capability to cause persistent infections and evade traditional treatment and prevention efforts. Hepatitis C virus (HCV) frequently establishes chronic infection and has considerable sequence variation, especially in putative envelope proteins E1 and E2, for which <60% amino acid identity has been described worldwide (3, 15, 17, 18, 22, 37, 46).
Genetic variation may also refer to differences among the swarm of viral variants within a person, often called a quasispecies (10, 11). The variants in an HCV quasispecies generally have 94 to 99% nucleotide identity (2, 25, 35). Within a single specimen, such variation can be characterized in terms of diversity or complexity. Diversity is the mean genetic distance calculated for all pairs of sequences (26), where genetic distance is directly proportional to the number of nucleotide differences between two variants. Complexity refers to the population distribution of variants and has been calculated from sequence data (26) but has also been estimated more practically on the basis of either the number of distinct gel bands resulting from single-stranded conformational polymorphism (SSCP) analysis (24) or the number of indistinguishable cDNA clones (clonotypes) recognized by gel shift analysis (32).
Nucleic acid sequencing of cloned cDNAs remains the “gold standard” for the assessment of viral variation but is too cumbersome to be applied to large, population-based studies. Electrophoretic analysis of SSCP has been more expedient, but its sensitivity (ability to identify distinct clones) is limited and it does not provide an estimate of genetic distance (5, 6, 23). Heteroduplex analysis (HDA) is also convenient and provides information on both genetic complexity and distance (4, 8, 9, 13, 14, 21, 27, 32, 47). However, HDA alone may not be sufficiently sensitive (6). We sought to develop a method combining HDA and SSCP analysis (referred to herein as the HDA+SSCP method) that could assess genetic complexity with high sensitivity (ability to discriminate between distinct sequences) and specificity (chance that clones detected as distinct truly represent distinct sequences) and that could provide accurate estimates of genetic diversity in a large prospective investigation by sampling a sufficiently large number of cloned cDNAs. In addition, we tested the value of estimating quasispecies complexity by calculating the Shannon entropy of the clonotype distribution, thus incorporating information about both the number of clonotypes and their respective proportions.
MATERIALS AND METHODS
Study subjects.
As part of a prospective study of acute HCV infection, serial serum samples were obtained from individuals in the ALIVE cohort of injection drug users in Baltimore, Md. (45). These samples were tested for antibodies to HCV by using the second-generation HCV 2.0 enzyme immunoassay (Ortho Diagnostic Systems, Raritan, N.J.) as previously described (45). Individuals were identified as seroconverters when a sample tested positive following at least one negative result. Positive results were supplemented by a recombinant immunoblot assay (Chiron RIBA HCV 2.0 strip immunoblot assay; Chiron Corporation, Emeryville, Calif.) and confirmed by the detection of HCV RNA by a reverse transcriptase PCR (RT-PCR) assay (Amplicor HCV Monitor; Roche Diagnostic Systems, Branchburg, N.J.) as previously described (44). Twelve samples from seven subjects were arbitrarily selected for this investigation, without knowledge of risk factors or disease state (Table 1). Genotyping by analysis of Core-E1 HCV sequence, according to the nomenclature of Simmonds et al. (36), revealed that all subjects were infected with HCV genotype 1a except subject 11469 (sample E), who was infected with genotype 1b (44).
TABLE 1.
Characteristics of subjects and samples and results of the HDA+SSCP method and sequence analysis
| Subject | Sample | Visit date (day/mo/yr) | Log10 [HCV RNA]a | Clonotype distributionb | No. of clonotypesc | Entropyd | Dwe |
|---|---|---|---|---|---|---|---|
| 10388 | A1 | 1/30/91 | 4.5 | 27,3,1,1,1 | 5 | 0.20 | 0.52 |
| A2 | 1/11/96 | 5.4 | 9,6,2,2,2,1,1,1,1,1,1,1,1,1,1,1,1 | 17 | 0.70 | NDg | |
| A3 | 6/20/97 | NDf | 4,3,3,2,2,2,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1 | 22 | 0.85 | NDg | |
| 10718 | B1 | 12/6/93 | 4.4 | 6,6,3,3,3,2,1,1,1,1,1,1,1,1,1 | 15 | 0.70 | NDg |
| B2 | 6/21/95 | 5.4 | 6,5,5,2,2,1,1,1,1,1,1,1,1,1,1,1,1,1 | 18 | 0.74 | NDg | |
| 10960 | C1 | 5/20/88 | 3.3 | 30,1,1,1 | 4 | 0.12 | 0.09 |
| C2 | 6/14/95 | 5.1 | 25,2,2,1,1,1,1 | 7 | 0.28 | 0.66 | |
| C3 | 5/6/96 | 4.9 | 15,4,3,2,2,1,1,1,1,1,1,1 | 12 | 0.55 | 1.30 | |
| 10976 | D | 3/13/91 | 3.0 | 26,1,1,1,1,1,1,1 | 8 | 0.27 | 1.23 |
| 11469 | E | 3/5/93 | 5.9 | 12,7,4,2,2,1,1,1,1 | 9 | 0.51 | 0.94 |
| 12951 | F | 8/21/92 | 2.7 | 6,2,2,2,2,2,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1 | 23 | 0.85 | 6.17 |
| 40209 | G | 9/5/95 | 7.6 | 14,11,2,2,1,1,1,1 | 8 | 0.43 | 0.52 |
[HCV RNA], HCV RNA copies per milliliter of serum.
Each series shows the number of clones comprising each clonotype for the given samples.
Number of electrophoretically indistinguishable cloned cDNAs.
Normalized Shannon entropy, a calculated value that incorporates the number and distribution of clonotypes.
Calculated value that incorporates the distribution of clonotypes and their respective genetic diversities.
ND, not done. The most recent value was 5.7, from 6/10/96.
Insufficient sequence data were obtained for this calculation.
Reverse transcription and nested-PCR amplification.
Total RNA was extracted from 100 μl of plasma or serum by using 1 ml of Trizol LS Reagent (Life Technologies, Gaithersburg, Md.) at room temperature, followed by chloroform extraction and isopropanol precipitation in the presence of 20 μg of glycogen (Boehringer Mannheim, Indianapolis, Ind.). The RNA pellet was washed with 75% (vol/vol) ethanol and then air dried briefly and redissolved in 50 μl of diethyl pyrocarbonate-treated water with 10 mM dithiothreitol (Promega, Madison, Wis.) and 5 U of RNasin ribonuclease inhibitor (Promega). After incubation at 65°C for 5 min, 5 μl of purified RNA was used to generate cDNA in a 20-μl reaction mixture at 37°C for 1 h with 20 U of Moloney murine leukemia virus RT (Perkin-Elmer, Foster City, Calif.) and first-round PCR reverse primer (see below). The entire 20-μl cDNA synthesis reaction mixture was used for the first-round PCR in a 25-μl reaction mixture containing 0.625 U of Taq polymerase (Life Technologies), 1.5 mM MgCl2, 0.2 mM concentrations of deoxynucleoside triphosphates, and 400 μM concentrations of primers. The primers (and nucleotide positions in the HCV-H77 strain [12]) for the first round were 5′-GCCCACTGGGGTGTCCTAGCGGG-3′ (forward; positions 1380 to 1403) and 5′-GTGCAGGGGTAGTGCCAGAGCCT-3′ (reverse; positions 2191 to 2168). One microliter of the first-round reaction mixture was added to the second-round PCR, which had the same reagents as in the first round except for the primers. The second-round (nested) primers were 5′-TTCCATGGTGGGGAACTGGGC-3′ (forward; positions 1415 to 1436) and 5′-GGCTCGGAGTGAAGCAATAC-3′ (reverse; positions 1866 to 1846). Reverse transcription and PCR were performed in a Perkin-Elmer Cetus 9600 thermocycler. Each PCR round was 35 cycles of 94°C for 10 s, 65°C for 30 s, and 72°C for 30 s.
Plasmid cloning and amplification of cloned cDNA.
PCR products were purified with a QIAQuick Gel Extraction Kit (Qiagen, Chatsworth, Calif.) used according to the manufacturer’s protocol. Within 24 h of synthesis, 20 to 40 ng of PCR products were ligated to 50 ng of the pCR 2.1 vector (TA Cloning kit; Invitrogen, Carlsbad, Calif.), used according to the manufacturer’s protocol, and were used to transform Escherichia coli INVαF′ competent cells (Invitrogen). Transformants were grown for approximately 20 h at 37°C on ampicillin plates, and 33 colonies were randomly selected.
Cloned cDNA was amplified from bacterial colonies as follows. Each colony was subcultured in Luria-Bertani medium for 14 h at 37°C. Three microliters of this culture was diluted in 100 μl of distilled water, mixed with 10 μl of 1 M NaOH, incubated for 1 h at 37°C, and then neutralized with 10 μl of 1 M HCl. One microliter of this lysate was added to a 25-μl PCR mixture as described above for the second round. Taq polymerase was inactivated by adding EDTA to a final concentration of 5 mM. The PCR product was purified with a QIAQuick Gel Extraction Kit.
HDA+SSCP method.
Gel electrophoresis was carried out with the MDE Heteroduplex Kit (FMC Bioproducts, Rockland, Maine) at a 1× concentration according to the manufacturer’s protocol, with the addition of 15% (wt/vol) urea to increase the resolution. Driver and test-cloned cDNA (2.5 μl each) were mixed, denatured at 95°C for 3 min, and then immediately plunged into crushed ice. Five microliters of each reaction mixture plus 1 μl of Triple Dye loading buffer (FMC Bioproducts) was loaded on an MDE gel (19.0 by 16.0 by 0.1 cm) in a Protein II xi cell (Bio-Rad Laboratories, Hercules, Calif.), followed by electrophoresis at 140 V for 4,500 V · h (∼32 h). Gels were stained in a 1:10,000 dilution of SYBR Green II (FMC Bioproducts) for ≥30 min and documented with an Eagle Eye II Still Video System (Stratagene, Carlsbad, Calif.) with a SYBR Green Filter (Stratagene).
Selection of subject-specific driver.
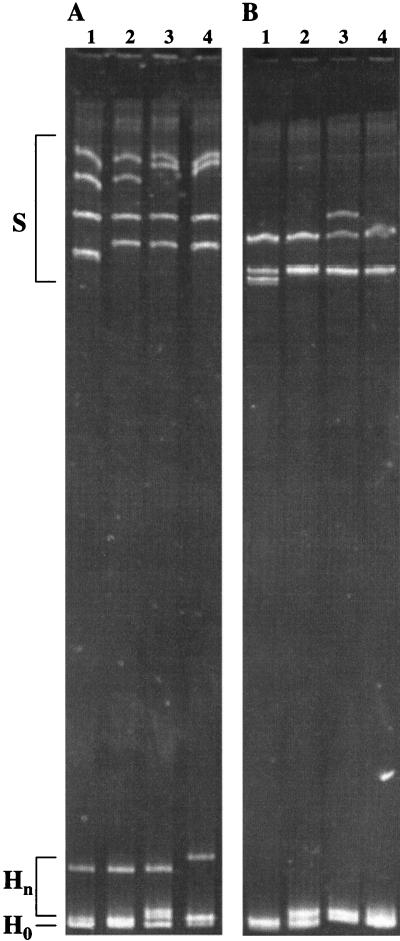
For each sample, a preliminary HDA+SSCP gel was performed with 20 cloned cDNAs, 1 of which was also randomly selected for use as a preliminary driver. Inspection of the preliminary gel always yielded a cloned cDNA with a relatively large gel shift as well as an SSCP pattern with four distinct bands that represented a minority of the SSCP patterns among the cloned cDNAs. This process resulted in the selection of a subject-specific driver that optimized resolution of all HDA and SSCP bands, in contrast to results obtained with a suboptimal driver (Fig. 1).
FIG. 1.
Example of combined HDA and SSCP analysis in one gel, illustrating the importance of driver selection. The E2 region of HCV was amplified from serum by RT-PCR and cloned, and the combined HDA+SSCP procedure was performed. Representative results are shown. (A) Lanes 1 to 4 contain DNA from four cloned cDNAs, with a driver sequence selected according to details given in Materials and Methods. (B) Lanes 1 to 4 contain DNAs from the same four clones, with a different driver sequence from the same sample. S, SSCP; Hn, heteroduplex bands; H0, homoduplex bands.
Complexity analysis.
For each of the three electrophoretic methods (HDA, SSCP analysis, and the HDA+SSCP method), a clonotype was defined as a group of indistinguishable cloned cDNAs based on inspection of the gel for the number and mobility of bands. The number of clonotypes and the number of clones comprising each clonotype were recorded.
To quantify complexity, the Shannon entropy (H) calculation (34, 40) was applied. Shannon entropy incorporates both the number of clonotypes and the number of cloned cDNAs in each clonotype. It is defined as H = −Σ (from i = 1 to N) P(i) Ln[P(i)], where N is the total number of clonotypes and P(i) is the number of clones represented in clonotype i. Because each sample in our study could have up to 33 clonotypes, which would yield a maximum value of Ln(33) for H, a normalized value of H, denoted H′, was defined as H/Ln(33). This resulted in a range of possible values for H′ from 0 to 1, representing 1 to 33 distinct clonotypes, respectively.
Nucleotide sequencing.
Sequences were unidirectionally determined from the M13 primer binding site of plasmid clones by using a PRISM automated sequencer (version 2.1.1; Applied Biosystems, Inc., Foster City, Calif.). Except as noted, sequences were obtained from up to three representatives of each clonotype. Sequences were assembled by using the ESEE3s program (E. Cabot, Madison, Wis.), and primer sequences were removed.
Calculation of diversity.
For a pair of sequences, distance was calculated as the Hamming distance, or number of nucleotide differences, per 100 bases. Intraclonotype diversity (di) was defined as the mean of pairwise distance values for cloned cDNAs from clonotype i. Interclonotype diversity (dij) was defined as the mean of distance values for cloned cDNAs from clonotypes i and j, where i ≠ j. The weighted diversity (Dw) for a specimen was defined as the mean of distance values for all pairs of cloned cDNAs from that specimen, with each pair’s contribution weighted according to the proportion of cloned cDNAs represented by its clonotype. This calculation was simplified by summing the intraclonotype and interclonotype diversities as separate terms and can be represented as follows:
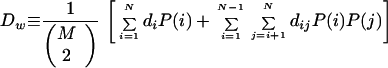 |
where M is the number of cloned cDNAs (generally 33), N is the number of clonotypes, and P(i) is the proportion of cDNAs represented by clonotype i. The denominator of the fractional expression is the binomial coefficient, yielding the number of possible paired comparisons for M sequences. If all cDNAs were sequenced, Dw would equal the mean of all pairwise distances.
A 32-bit Windows application called ClonoTyper was created by one of the authors (S.C.R.) to perform the complexity and diversity calculations and is available on request.
Nucleotide sequence accession number.
The nucleotide sequences presented in this article have been submitted to GenBank (accession no. AF073020 to AF073176).
RESULTS
Clonotypes detected by HDA, SSCP, and HDA+SSCP methods.
For each sample, 33 cloned cDNAs were examined by the HDA+SSCP method, and two investigators (Y.-M.W. and O.L.) independently assigned clonotypes based on the migration patterns revealed by HDA alone, the SSCP method alone, and the HDA+SSCP method. These assignments were >99% concordant, and rare discrepancies were resolved in a blinded fashion. By the HDA+SSCP method, the number of clonotypes varied from 4 to 23 per sample (Table 1). The nucleotide sequence was determined for up to three representatives of each clonotype indistinguishable by all gel shift assays, generating a total of 157 cDNA sequences from 12 samples from seven individuals.
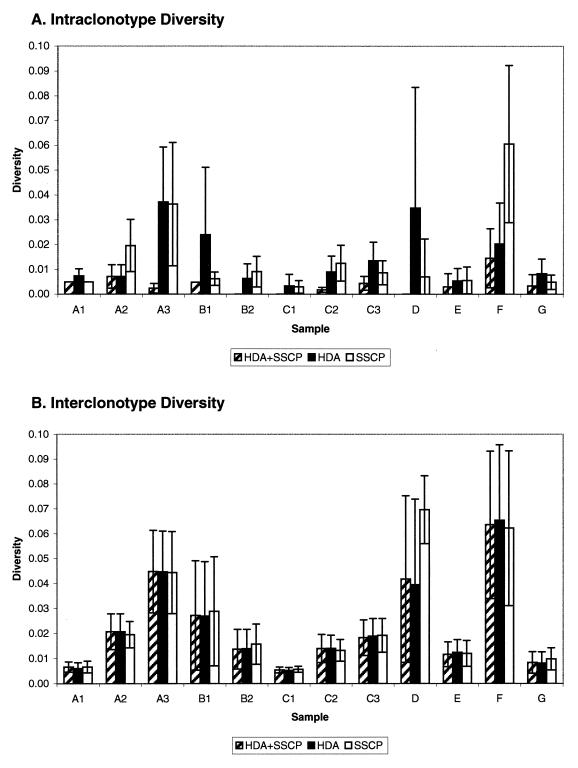
Assay sensitivity was determined from the sequence diversity within a clonotype (intraclonotype diversity): sensitivity (the ability of an electrophoretic method to distinguish nonidentical cloned cDNAs) varies inversely with the intraclonotype diversity. For 10 samples, intraclonotype diversity was lower for the combined method versus HDA or the SSCP method alone (Fig. 2A). Only samples A1 and A2 yielded similarly low intraclonotype diversity for one of the individual methods (HDA and the SSCP method, respectively). Intraclonotype diversities (mean ± standard deviation [SD]) for the SSCP method, HDA, and the HDA+SSCP method were 3.5% ± 3.4%, 1.6% ± 1.8%, and 0.6% ± 0.9%, respectively. Among cloned cDNAs indistinguishable by the combined method, the maximum distance was 3.5%; 98.7% of such sequences differed by <2%.
FIG. 2.
Intraclonotype versus interclonotype diversity, by specimen. For each specimen, intraclonotype (A) and interclonotype (B) nucleotide sequence comparisons were made and expressed as the mean pairwise genetic distance. Error bars indicate ±1 SD.
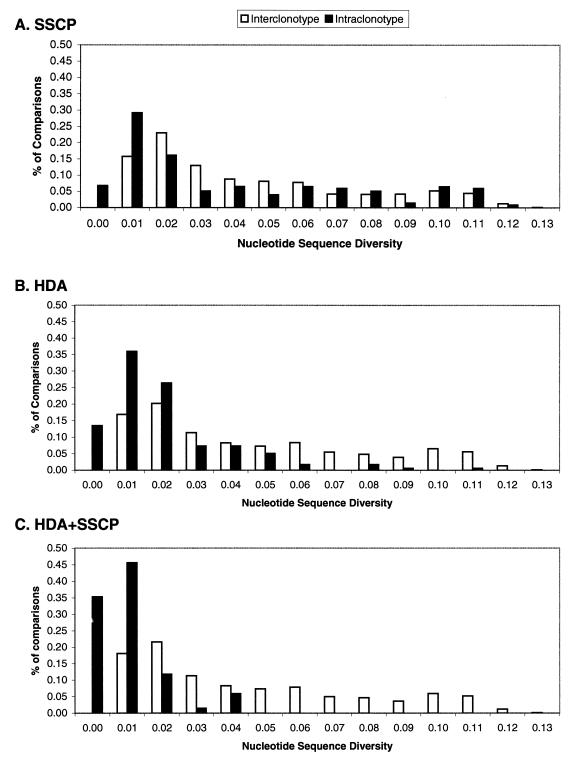
Assay specificity was determined from the sequence diversity among cloned cDNAs identified as electrophoretically different (interclonotype diversity). For all 12 samples, assay specificity for the combined method was as high as that for either HDA or SSCP alone (Fig. 2B). Interclonotype diversities for the SSCP method, HDA, and the HDA+SSCP method (mean ± SD) were 3.9% ± 3.1%, 4.2% ± 3.2%, and 4.0% ± 3.2%, respectively, and no two cloned cDNAs assigned to different clonotypes had identical sequences. That the high sensitivity of the combined method was achieved without loss of specificity was evident in the high proportion of clone pairs with low intraclonotype diversity by the combined method compared to those by the other methods (shift to the left in Fig. 3C compared with positions in Fig. 3A and B), while the distribution of interclonotype diversity remained the same.
FIG. 3.
Frequency distributions of intraclonotype and interclonotype diversity for all samples. All possible pairwise sequence comparisons were performed and classified as interclonotype or intraclonotype based on the results of HDA, the SSCP method, or the HDA+SSCP method. A histogram of the resulting percent diversity is displayed.
Complexity versus sequence diversity.
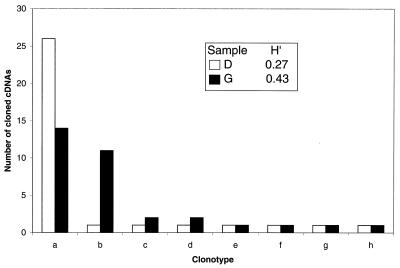
We compared Shannon entropy to the number of clonotypes as measures of quasispecies complexity. The two measures gave very different results for some samples, as illustrated in Fig. 4. The number of clonotypes correlated more strongly than the normalized Shannon entropy with the Dw, with correlation coefficients of 0.93 and 0.70, respectively. When the number of cloned cDNAs assessed was reduced from 33 by analyzing the leftmost lanes in each sample gel, the correlation between measures of complexity (number of clonotypes and entropy) and Dw was reduced in a nearly linear fashion (Table 2).
FIG. 4.
Measures of complexity. The clonotype distributions for samples D and G were plotted, and the normalized Shannon entropy values (H′) are indicated in the inset.
TABLE 2.
Measures of complexity: sensitivity to sampling
| No. of cloned cDNAs examinedb | Correlation coefficient (R2)a
|
|
|---|---|---|
| Clonotypesc vs Dw | Entropyc vs Dw | |
| 11 | 0.46 | 0.33 |
| 17 | 0.70 | 0.44 |
| 22 | 0.78 | 0.50 |
| 33 | 0.93 | 0.70 |
Least-squares linear regression R2 for the indicated relationship.
The leftmost n lanes were examined in rows for which n was <33.
The number of clonotypes and degree of entropy were calculated from the cloned cDNAs examined.
DISCUSSION
Our data indicate that the HDA+SSCP method identified distinct cloned cDNAs with high sensitivity and specificity. In addition, by screening 33 cloned cDNAs per sample (versus 2 to 10 as is customary), more precise measurements of viral complexity were obtained. Viral complexity measured by this method also correlated with diversity determined by nucleotide sequencing. When analyses were done with fewer cloned cDNAs, much less accurate estimates were obtained (Table 2), underscoring the merits of our approach (analysis of 33 cloned cDNAs by the HDA+SSCP method).
In developing this method we sought a highly sensitive means for detecting distinct sequences in a mixture of cloned cDNAs, so that we can screen a large number of samples for sequence variation in future investigations. The sensitivity we report for the HDA+SSCP method is comparable to that reported for more laborious and expensive methods which employ radiolabeling and purification of single-stranded drivers (14). Because we defined a clonotype according to any difference in electrophoretic migration, we expected that the HDA+SSCP method would be at least as sensitive as either method alone. This is illustrated by Fig. 1A, in which lanes 1 and 2 appear identical by HDA but are clearly different by SSCP analysis and lanes 3 and 4 appear identical by SSCP analysis but have different HDA patterns. Importantly, and not guaranteed by this design, the HDA+SSCP method maintained high specificity by not assigning identical cloned cDNAs to different clonotypes. Although methods combining HDA and SSCP for detection of genetic variation have been reported previously (1), either combined in one gel or performed separately, they used fewer samples and were not applied to sequences as variable as HCV-E2 (43).
We developed a subject-specific process for selecting a cloned cDNA driver because selection of an appropriate driver to which each variant is annealed is crucial for HDA. For other viruses, such as human immunodeficiency virus type 1, a single reference driver for cross-sectional or longitudinal assays allows comparison of gel shift both between individuals and over time (41). Like others, however, we were unable to form heteroduplexes when a driver representing this hypervariable region from another HCV quasispecies (i.e., another subject) was used (data not shown). Therefore, we could not use a single driver for all samples. We found that other approaches to driver selection, including random choice (14, 47) or selection of a majority-cloned cDNA (32), resulted in decreased sensitivity. Use of a driver which represented a majority clonotype for that specimen resulted in a large number of gel lanes with overlapping SSCP bands (rather than four distinct ones) and overlapping hetero- and homoduplex bands, both potentially obscuring small differences in gel shift (Fig. 1B). The approach described in Materials and Methods resulted in the selection of divergent minor variants for use as subject-specific drivers, maximizing clonotype identification while maintaining simplicity.
The combination of HDA and SSCP in one gel raises some unique methodological challenges. In order to obtain adequate resolution for both HDA and SSCP bands, which migrate with different rates, we found that at least a 15-cm migration distance was needed (data not shown). Prior studies of the HDA+SSCP method utilized either denaturing conditions which interfered with heteroduplex formation, producing indistinct bands (1, 28, 39, 42), or slow cooling, which decreased the yield of SSCP by favoring formation of homo- and heteroduplexes (33). We found that rapid cooling under nondenaturing conditions resulted in the best yield and resolution (data not shown). We also found that SYBR Green II or conventional silver staining gave equivalent results, whereas SYBR Green I or ethidium bromide yielded unacceptably faint staining of SSCP bands. In order to preserve the SSCP bands it was necessary to perform electrophoresis in a 4°C chamber or at a low voltage for 32 h as described in Materials and Methods.
Compared to earlier evaluations of HCV quasispecies (19, 32), relatively little viral diversity was detected in our 12 serum samples. Because a large number of cloned cDNAs was sequenced, it is unlikely that diversity was substantially underestimated. Rather, the relatively low diversity probably resulted from selecting samples collected soon after HCV infection (16). The diversity of an HCV quasispecies increases with duration of infection, with estimates of 1.44 × 10−3 to 1.92 × 10−3 nucleotide substitutions per site per year (29, 30). In this study the five samples from later time points did show increases in entropy, Dw, and the number of clonotypes, compared with values from the earlier sample(s) from the same subject (Table 1). Viral loads also generally increased at later time points, but viral load did not correlate with diversity (e.g., Table 1, samples F and G).
A potential source of error in this type of investigation is inherent in amplification and sequencing with currently available polymerases. Even as higher-fidelity thermostable enzymes become available, investigators need to remain aware of polymerase error as a potential source of “unique clones” which differ by one or two bases from other variants (38). Gel-based methods are ideal for studying large cohorts because the same errors will affect both cases and controls, and their effects will be further reduced by assessing large numbers of samples. We propose that gel-based methods like ours can be used to assign a weight to each sequence in a diversity calculation, according to frequency (clonotype distribution), avoiding the bias of giving equal weight to an infrequent variant, regardless of its origin (polymerase artifact or divergent viral clone).
We calculated Shannon entropy (34) to evaluate its utility as a measure of complexity that reflects both the number and distribution of the clonotypes. In contrast, earlier reports have estimated complexity only from the number of clonotypes. The effect of neglecting clonotype distribution can be illustrated by samples D and G (Fig. 4). Both samples yielded eight clonotypes, but the distributions are clearly different, with sample D dominated by a major clonotype comprising nearly 80% of 33 cloned cDNAs while the major clonotype in sample G comprises less than a third of 33 clones. This difference, reflected in the higher entropy value for sample G, is ignored when only the number of clonotypes is used to estimate complexity. Unbalanced distribution of clones is the rule rather than the exception in HCV (Table 1), some of which may be due to artifacts incorporated during amplification and sequencing. Such errors give rise to “solitary clones” which have little effect on the Shannon entropy calculation but are given equal weight when the number of clonotypes is used as a measure of complexity. Shannon entropy has been used to describe the complexity of individual amino sequence positions in sequences representing human (40) and human immunodeficiency virus type 1 (20) genomes, as well as for automated gel analysis (7), and has recently been applied to HCV clone distributions (31).
Despite the theoretical power of entropy to model complexity, the number of clonotypes was more strongly correlated to sequence diversity in this study. While both methods of estimating complexity are very sensitive to the number of cloned cDNAs assessed (Table 2), our assessment of 33 clones per sample in this study favored the use of the number of clonotypes. By extrapolating from the data in Table 2, we predict that Shannon entropy would be more useful for assessing more than 50 cloned cDNAs per sample. These results are consistent with the findings of Pawlotsky et al., who used SSCP of a 185-bp fragment to identify clonotypes, examined 30 HCV clones amplified from each of 13 subjects, and sequenced up to three clones per clonotype. They found linear regression of the normalized Shannon entropy versus the (unweighted) diversity to have a correlation coefficient (R2) of 0.331 (31). Whether this lower correlation was due to the region amplified, the size of the amplicon, the gel-shift assay used to identify clonotypes, or weighting of the diversity calculation remains to be determined. When entropy of sequences from the 5 subjects who responded to alpha interferon therapy was compared to that of 8 subjects selected from the 40 subjects who did not respond, those who responded had lower entropy values (31). Further study with more extensive sampling is required to determine whether Shannon entropy is truly superior to the number of clonotypes as a predictor of diversity and whether it has biological significance.
We report a method for measuring HCV quasispecies complexity that combines HDA and SSCP in a single gel visualized with UV light. The method was sensitive and specific for detecting clonotypes, and the number of clonotypes detected correlated strongly with sequence diversity when 33 cloned cDNAs are assessed. We introduce the use of entropy as a measure of complexity, incorporating the distribution of variants as well as the number of clonotypes, but suggest that a larger number of cloned cDNAs needs to be assessed when this measure is to be used. Our approach is expected to facilitate accurate analysis of the large number of cross-sectional and longitudinal samples that are now becoming available and could lead to clinical-laboratory assays for diagnosis or monitoring of HCV patients.
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grants AI-40035, DA-04334, and DA-08004.
We are grateful to the subjects in the ALIVE cohort for their generosity in providing specimens and time. We also thank Stephen Villano for identifying appropriate specimens and Amy Weiner for assistance in the development of the HDA+SSCP method.
REFERENCES
- 1.Axton R A, Hanson I M, Love J, Seawright A, Prosser J, van Heyningen V. Combined SSCP/heteroduplex analysis in the screening for PAX6 mutations. Mol Cell Probes. 1997;11:287–292. doi: 10.1006/mcpr.1997.0117. [DOI] [PubMed] [Google Scholar]
- 2.Bukh J, Miller R H, Purcell R H. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liver Dis. 1995;15:41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 3.Bukh J, Purcell R H, Miller R H. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc Natl Acad Sci USA. 1993;90:8234–8238. doi: 10.1073/pnas.90.17.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvo P L, Kansopon J, Sra K, Quan S, Dinello R, Guaschino R, Calabrese G, Danielle F, Brunetto M R, Bonino F, Massaro L, Polito A, Houghton M, Weiner A J. Hepatitis C virus heteroduplex tracking assay for genotype determination reveals diverging genotype 2 isolates in Italian hemodialysis patients. J Clin Microbiol. 1998;36:227–233. doi: 10.1128/jcm.36.1.227-233.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrington M, Millter T, White M, Gerrard B, Stewart C, Dean M, Mann D. Typing of HLA-DQA1 and DQB1 using DNA single-stranded conformational polymorphism. Hum Immun. 1992;33:208–212. doi: 10.1016/0198-8859(92)90073-v. [DOI] [PubMed] [Google Scholar]
- 6.Cotton R G. Current methods of mutation detection. Mutat Res. 1993;285:125–144. doi: 10.1016/0027-5107(93)90060-s. [DOI] [PubMed] [Google Scholar]
- 7.Delwart E L, Pan H, Sheppard H W, Wolpert D, Neumann A U, Korber B, Mullins J I. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J Virol. 1997;71:7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen W H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 9.Delwart E L, Shpaer E G, Mullins J I. Heteroduplex mobility assays for phylogenetic analysis. In: Innis M A, Gelfand D H, Sninsky J J, editors. PCR strategies. San Diego, Calif: Academic Press; 1995. pp. 154–160. [Google Scholar]
- 10.Domingo E, Martinez-Salas E, Sobrino F, de la Torre J C, Portela A, Ortin J, Lopez-Galindez C, Perez-Brena P, Villanueva N, Najera R. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance—a review. Gene. 1985;40:1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- 11.Eigen M. Self organization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 12.Feinstone S M, Alter H J, Dienes H P, Shimizu Y, Popper H, Blackmore D, Sly D, London W T, Purcell R H. Non-A, non-B hepatitis in chimpanzees and marmosets. J Infect Dis. 1981;144:588–598. doi: 10.1093/infdis/144.6.588. [DOI] [PubMed] [Google Scholar]
- 13.Gavier B, Martínez-González M A, Riezu-Boj J I, Lasarte J J, Garcia N, Civeira M P, Prieto J. Viremia after one month of interferon therapy predicts treatment outcome in patients with chronic hepatitis C. Gastroenterology. 1997;113:1647–1653. doi: 10.1053/gast.1997.v113.pm9352868. [DOI] [PubMed] [Google Scholar]
- 14.Gretch D R, Polyak S J, Wilson J J, Carithers R L, Jr, Perkins J D, Corey L. Tracking hepatitis C virus quasispecies major and minor variants in symptomatic and asymptomatic liver transplant recipients. J Virol. 1996;70:7622–7631. doi: 10.1128/jvi.70.11.7622-7631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higashi Y, Kakumu S, Yoshioka K, Wakita T, Mizokami M, Ohba K, Ito Y, Ishikawa T, Takayanagi M, Nagai Y. Dynamics of genome change in the E2/NS1 region of hepatitis C virus in vivo. Virology. 1993;197:659–668. doi: 10.1006/viro.1993.1641. [DOI] [PubMed] [Google Scholar]
- 16.Honda M, Kaneko S, Sakai A, Unoura M, Murakami S, Kobayashi K. Degree of diversity of hepatitis C virus quasispecies and progression of liver disease. Hepatology. 1994;20:1144–1151. [PubMed] [Google Scholar]
- 17.Kao J-H, Chen P-J, Lai M-Y, Wang T-H, Chen D-S. Quasispecies of hepatitis C virus and genetic drift of the hypervariable region in chronic type C hepatitis. J Infect Dis. 1995;172:261–264. doi: 10.1093/infdis/172.1.261. [DOI] [PubMed] [Google Scholar]
- 18.Kato N, Ootsuyama Y, Ohkoshi S, Nakazawa T, Sekiya H, Hijikata M, Shimotohno K. Characterization of hypervariable regions in the putative envelope protein of hepatitis C virus. Biochem Biophys Res Commun. 1992;189:119–127. doi: 10.1016/0006-291x(92)91533-v. [DOI] [PubMed] [Google Scholar]
- 19.Kato N, Ootsuyama Y, Tanaka T, Nakagawa M, Nakazawa T, Muraiso K, Ohkoshi S, Hijikata M, Shimotohno K. Marked sequence diversity in the putative envelope proteins of hepatitis C viruses. Virus Res. 1992;22:107–123. doi: 10.1016/0168-1702(92)90038-b. [DOI] [PubMed] [Google Scholar]
- 20.Korber B T, Kunstman K J, Patterson B K, Furtado M, McEvilly M M, Levy R, Wolinsky S M. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J Virol. 1994;68:7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreis S, Whistler T. Rapid identification of measles virus strains by the heteroduplex mobility assay. Virus Res. 1997;47:197–203. doi: 10.1016/s0168-1702(96)01413-x. [DOI] [PubMed] [Google Scholar]
- 22.Kurosaki M, Enomoto N, Marumo F, Sato C. Rapid sequence variation in the hypervariable region of hepatitis C virus during the course of chronic infection. Hepatology. 1993;18:1293–1299. [PubMed] [Google Scholar]
- 23.Lee J H, Stripf T, Roth W K, Zeuzem S. Non-isotopic detection of hepatitis C virus quasispecies by single strand conformation polymorphism. J Med Virol. 1997;53:245–251. doi: 10.1002/(sici)1096-9071(199711)53:3<245::aid-jmv11>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 24.Le Guen B, Squadrito G, Nalpas B, Berthelot P, Pol S, Brechot C. Hepatitis C virus genome complexity correlates with response to interferon therapy: a study in French patients with chronic hepatitis C. Hepatology. 1997;25:1250–1254. doi: 10.1002/hep.510250531. [DOI] [PubMed] [Google Scholar]
- 25.Martell M, Esteban J I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navas S, Martín J, Quiroga J A, Castillo I, Carreno V. Genetic diversity and tissue compartmentalization of the hepatitis C virus genome in blood mononuclear cells, liver, and serum from chronic hepatitis C patients. J Virol. 1998;72:1640–1646. doi: 10.1128/jvi.72.2.1640-1646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson J A E, Fiscus S A, Swanstrom R. Evolutionary variants of the human immunodeficiency virus type 1 V3 region characterized by using a heteroduplex tracking assay. J Virol. 1997;71:8750–8758. doi: 10.1128/jvi.71.11.8750-8758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Offermans M T, Struyk L, de Geus B, Breedveld F C, van den Elsen P J, Rozing J. Direct assessment of junctional diversity in rearranged T cell receptor beta chain encoding genes by combined heteroduplex and single strand conformation polymorphism (SSCP) analysis. J Immunol Methods. 1996;191:21–31. doi: 10.1016/0022-1759(95)00283-9. [DOI] [PubMed] [Google Scholar]
- 29.Ogata N, Alter H J, Miller R H, Purcell R H. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:3392–3396. doi: 10.1073/pnas.88.8.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamoto H, Kojima M, Okada S, Yoshizawa H, Iizuka H, Tanaka T, Muchmore E E, Peterson D A, Ito Y, Mishiro S. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology. 1992;190:894–899. doi: 10.1016/0042-6822(92)90933-g. [DOI] [PubMed] [Google Scholar]
- 31.Pawlotsky J M, Germanidis G, Neumann A U, Pellerin M, Frainais P O, Dhumeaux D. Interferon resistance of hepatitis C virus genotype 1b: relationship to nonstructural 5A gene quasispecies mutations. J Virol. 1998;72:2795–2805. doi: 10.1128/jvi.72.4.2795-2805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polyak S J, Faulkner G, Carithers R L, Jr, Corey L, Gretch D R. Assessment of hepatitis C virus quasispecies heterogeneity by gel shift analysis: correlation with response to interferon therapy. J Infect Dis. 1997;175:1101–1107. doi: 10.1086/516448. [DOI] [PubMed] [Google Scholar]
- 33.Pursall M C, Clay T M, Bidwell J L. Combined PCR-heteroduplex and PCR-SSCP analysis for matching of HLA-A, -B and -C allotypes in marrow transplantation. Eur J Immunogenet. 1996;23:41–53. doi: 10.1111/j.1744-313x.1996.tb00263.x. [DOI] [PubMed] [Google Scholar]
- 34.Shannon C E. A mathematical theory of communication. Bell Syst Technol J. 1948;27:379–423. [Google Scholar]
- 35.Simmonds P. Variability of hepatitis C virus. Hepatology. 1995;21:570–583. doi: 10.1002/hep.1840210243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simmonds P, Holmes E C, Cha T-A, Chan S-W, McOmish F, Irvine B, Beall E, Yap P L, Kolberg J, Urdea M S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 37.Simmonds P, Smith D B, McOmish F, Yap P L, Kolberg J, Urdea M S, Holmes E C. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J Gen Virol. 1994;75:1053–1061. doi: 10.1099/0022-1317-75-5-1053. [DOI] [PubMed] [Google Scholar]
- 38.Smith D B, McAllister J, Casino C, Simmonds P. Virus ’quasispecies’: making a mountain out of a molehill? J Gen Virol. 1997;78:1511–1519. doi: 10.1099/0022-1317-78-7-1511. [DOI] [PubMed] [Google Scholar]
- 39.Spritz R A, Holmes S A, Ramesar R, Greenberg J, Curtis D, Beighton P. Mutations of the KIT (mast/stem cell growth factor receptor) proto-oncogene account for a continuous range of phenotypes in human piebaldism. Am J Hum Genet. 1992;51:1058–1065. . (Erratum, 52:654, 1993.) [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart J J, Lee C Y, Ibrahim S, Watts P, Shlomchik M, Weigert M, Litwin S. A Shannon entropy analysis of immunoglobulin and T cell receptor. Mol Immunol. 1997;34:1067–1082. doi: 10.1016/s0161-5890(97)00130-2. [DOI] [PubMed] [Google Scholar]
- 41.Strunnikova N, Ray S C, Livingston R A, Rubalcaba E, Viscidi R P. Convergent evolution within the V3 loop domain of human immunodeficiency virus type 1 in association with disease progression. J Virol. 1995;69:7548–7558. doi: 10.1128/jvi.69.12.7548-7558.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sud R, Talbot I C, Delhanty J D. Infrequent alterations of the APC and MCC genes in gastric cancers from British patients. Br J Cancer. 1996;74:1104–1108. doi: 10.1038/bjc.1996.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas A W, Morgan R, Sweeney M, Rees A, Alcolado J. The detection of mitochondrial DNA mutations using single stranded conformation polymorphism (SSCP) analysis and heteroduplex analysis. Hum Genet. 1994;94:621–623. doi: 10.1007/BF00206954. [DOI] [PubMed] [Google Scholar]
- 44.Villano, S. A., D. Vlahov, K. E. Nelson, S. Cohn, and D. L. Thomas. Persistence of viremia and the importance of long-term follow-up after acute hepatitis C infection. Unpublished data. [DOI] [PubMed]
- 45.Villano S A, Vlahov D, Nelson K E, Lyles C M, Cohn S, Thomas D L. Incidence and risk factors for hepatitis C among injection drug users in Baltimore, Maryland. J Clin Microbiol. 1997;35:3274–3277. doi: 10.1128/jcm.35.12.3274-3277.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiner A J, Brauer M J, Rosenblatt J, Richman K H, Tung J, Crawford K, Bonino F, Saracco G, Choo Q L, Houghton M, et al. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 47.Wilson J J, Polyak S J, Day T D, Gretch D R. Characterization of simple and complex hepatitis C virus quasispecies by heteroduplex gel shift analysis: correlation with nucleotide sequencing. J Gen Virol. 1995;76:1763–1771. doi: 10.1099/0022-1317-76-7-1763. [DOI] [PubMed] [Google Scholar]