Abstract
Background
Allergenic proteins can cause IgE-mediated adverse reactions in sensitized individuals. Although the sequences of many allergenic proteins have been identified, bioinformatics data analysis with advanced computational methods and modeling is needed to identify the basis for IgE binding and cross-reactivity.
Objective
We aim to present the features and use of the updated Structural Database of Allergenic Proteins 2.0 (SDAP 2.0) webserver, a unique, publicly available resource to compare allergens using specially designed computational tools and new high-quality 3-D models for most known allergens.
Methods
Previously developed and novel software tools for identifying cross-reactive allergens using sequence and structure similarity are implemented in SDAP 2.0. A comprehensive set of high-quality 3-D models of most allergens was generated with the state-of-the-art AlphaFold 2 software. A graphics tool enables the interactive visualization of IgE epitopes on experimentally determined and modeled 3-D structures.
Results
A user can search for allergens similar to a given input sequence with the FASTA algorithm or the window-based World Health Organization/International Union of Immunological Societies (WHO/IUIS) guidelines on safety concerns of novel food products. Peptides similar to known IgE epitopes can be identified with the property distance tool and conformational epitopes by the Cross-React method. The updated database contains 1657 manually curated sequences including all allergens from the IUIS database, 334 experimentally determined X-ray or NMR structures, and 1565 3-D models. Each allergen/isoallergen is classified according to its protein family.
Conclusions
SDAP provides access to the steadily increasing information on allergenic structures and epitopes with integrated bioinformatics tools to identify and analyze their similarities. In addition to serving the research and regulatory community, it provides clinicians with tools to identify potential coallergies in a sensitive patient and can help companies to design hypoallergenic foods and immunotherapies.
Key words: Protein evaluation for allergenicity, database of allergens, IgE binding, cross-reactivity, physical-chemical property distance (PD)
Proteins from different foods, plants, fungi, and animals can sensitize individuals and stimulate IgE-mediated allergic responses.1 After binding to IgE antibodies on mast cells,2 allergens can trigger an immune response ranging from rashes to difficulty breathing to potentially deadly anaphylaxis.3 It is known that allergens from many different sources can cause dangerous cross-reactions, which may be revealed by sequence and structural relationships.4,5 The Structural Database of Allergenic Proteins (SDAP), the first publicly available, cross-referenced database of allergenic proteins,6 allowed users to rapidly determine relationships between allergens or to any input sequence using specifically designed bioinformatic tools.7,8 Other websites (recently summarized9) with different goals complement this information.
SDAP 2.0 is a comprehensive update that includes sequences and structures for all proteins classified by the Allergen Nomenclature Sub-Committee of the World Health Organization and International Union of Immunological Societies (WHO/IUIS) according to general clinical and biochemical guidelines as allergens.10 The WHO/IUIS database lists proteins that conform to their clinical criteria for allergenicity with respect to the number of patients with documented IgE reactions. In addition to sequences from the WHO/IUIS database (which are marked as such), additional proteins described as having allergenic characteristics from the literature are included for comparison purposes. Bioinformatic tools developed by us and other groups, functioning “on the fly,” allow rapid comparison of allergens according to protein families (Pfams),11 physicochemical property motifs,12 physical relationships, and biological functions.4,6,13, 14, 15, 16, 17, 18, 19, 20 The tools now integrated into SDAP 2.0 and, most significantly, 3-dimensional (3D) structures of more than 1600 distinct allergens and their isoforms are a valuable resource for identifying cross-reactive allergens. Recently, a major advance in determining a conformational IgE epitope of the house dust mite allergen Der p 2 in a high-resolution X-ray crystal structure was achieved,21 which suggests that experimental data as input for our bioinformatics tools for predicting cross-reactivities will be available in increasing numbers.
SDAP was first publicly available in 2002,7,22 with a major update in 200823 that included 3-D models of allergens for which no experimental structure was available.24 New or updated features in SDAP 2.0 are the interactive graphics tool for the display of experimentally determined 3-D structures, an updated list of allergenic proteins classified by Pfam, a version of the previously validated Cross-React software14 integrated into the webserver, and a comprehensive set of highly reliable 3-D models of most allergen sequences generated with the highly precise and validated AlphaFold program.25 These models and the updated software tools now available in the webserver SDAP 2.0, can help in interpreting the experimental results and determining structural similarities in cross-reactive proteins even in the face of extremely variant sequences.5,26,27 We show the capabilities of the webserver with the identification of food allergens cross-reactive to the birch pollen allergen, Bet v 1, a linear peptide search in the database for an IgE epitope of the Juniper ashei pollen allergen, Jun a 1, with the PD tool, which identified other pectate lyases from Cupressaceae tree pollen, but not other pectate lyases from the Asterales plant family of mugwort and ragweed.28,29 New in SDAP 2.0, the integrated Cross-React software can be used to identify other potential conformational IgE epitopes, as illustrated by the conservation of a previously identified IgE epitope in other pathogenesis-related (PR)-5 thaumatin-like proteins.30,31 Finally, we document the reliability of the new 3-D models in SDAP 2.0 generated by AlphaFold.
Methods
Data collection
All allergen sequences available in SDAP were manually identified using existing databases. The curated information is stored in MySQL tables according to a unique allergen and sequence ID linked to major databases, SWISS-PROT and NCBI. Any sequence annotated by WHO/IUIS as an allergen is labeled as “1” in SDAP, otherwise labeled a “0.” In addition, existing scientific literature related to any allergen sequence is added and linked to the NCBI PubMed database. Epitope information collected from the literature search is also available in SDAP.
Sequence search
Internal search utilities in SDAP are implemented with MySQL. For example, users can search allergen name, species, and source. Basic sequence analysis within SDAP is performed using the FASTA sequence alignment program.32 In addition, allergen sequences from SDAP can be directly searched against protein sequences in the NR and PDB databases at the NCBI using a BLAST search33 implemented in SDAP.
Peptide search
Allergenic proteins are known to be cross-reactive if they have high sequence or structural similarities.34,35 Therefore, a peptide search utility is implemented in SDAP to find similar sequences in other allergenic proteins. Users can compare a given peptide sequence against all allergenic protein sequences in SDAP using either an exact sequence match or peptide similarity with a property distance (PD) search.7,36,37 The PD search method has been described earlier,8,37, 38, 39, 40 and is very useful to find small peptide sequences that share common amino acid physicochemical properties.38,41, 42, 43
3-D model structures
We used the AlphaFold 2 software,25,44 running locally on a Linux computer, to generate 3-D model structures of all allergenic proteins in SDAP 2.0. AlphaFold generates a high-quality 3-D structure of a protein, when the sequence similarity between the target sequence and template structures varies from moderate to very high. The quality of the modeled structures is measured in terms of predicted local distance difference test (pLDDT) score, which is a reliable indicator of the quality of the models.45,46 All model structures of the allergenic proteins are shown in the SDAP website interactively with the user-friendly 3DJmol molecule viewer plugin,47 and are freely available to download with proper citation of their SDAP-2 origin. In addition, epitope information, if available, can be visualized on the 3-D structure of a protein.
Pfam classification
Despite the rapid increase in the number of sequences, most allergenic proteins belong to a small set of Pfams.48,49 Using the Pfamscan method,50 all allergenic proteins in SDAP are classified on the basis of their Pfam nomenclature,51 and users can search allergens in SDAP based on their Pfam name.
Food and Allergy Organization/WHO guidelines
We have implemented the Food and Allergy Organization/WHO guidelines for allergenic proteins as a web server. Users can search any peptide or a protein sequence against all allergenic proteins in SDAP. According to the FAO/WHO guidelines, a query protein and a known allergen should be considered potentially cross-reactive if there is (a) more than 35% identity in the amino acid sequence of the expressed protein, using a window of 80 amino acids and a suitable gap penalty, or (b) a sequence identity of 6 contiguous amino acids. Because condition (b) is rather liberal and generates potentially many false positives, the user may choose another cutoff value for the length of the exact sequence match.
Results
Main functionalities in SDAP 2.0
An overview of the integrated bioinformatics tools for storing and accessing the allergenic proteins in SDAP 2.0 is shown in Fig 1. Buttons, clearly visible to the user, allow access to the allergen data entries and search tools according to keywords, sequence similarity searches, peptide similarities by PDs,37 allergenic risks factors according to the WHO/IUIS rules,1 and graphical display of experimentally determined and 3-D model structures. In addition, existing scientific literature related to any allergen sequence is added and linked to the NCBI PubMed database. Epitope information from periodically updated literature searches is available in the SDAP database. All sequences are annotated on the basis of their Pfam 35.0 database (November 2021 release) domains. We obtained 223 protein families of allergen proteins compared with 19,632 different families in the Pfam database.
Fig 1.
Main functions of SDAP and internal data storage of allergens: Left and middle columns contain the list of integrated search tools, display functions, and bioinformatics tool for epitope predictions; the right column lists the stored allergen information in the database.
Sequence and 3-D structural similarity searches in the database
FASTA search
Allergenic proteins are considered to be potentially cross-reactive if they have high sequence or structural similarities.34,35 Related allergens can be identified in SDAP 2.0 by a sequence search. For example, a FASTA search identified many food allergens in SDAP 2.0 with high sequence similarity to the birch pollen allergen Bet v 1 (Table I), many of which have been implicated in the pollen food syndrome.52,53 These sequence-related allergens are PR-10 proteins, activated in plants in response to stress. Their models also show 3-D structural similarity to Bet v 1, with a 7-stranded β-sheet wrapped around an α-helix (Fig 2). Most hazelnut-allergic patients in a Dutch study54 were sensitized to Bet v 1. Those with coallergy to peanut and high IgE levels to hazelnut Cor a 1 also had similar IgE levels to the peanut allergen Ara h 8 and pollen allergen Bet v 1. Cor a 1 and Ara h 8 sensitization was always accompanied by a Bet v 1 sensitization. The observed high correlation between IgE responses to Cor a 1 and Ara h 8 in the patient cohort, as well as the close correlation to IgE responses to Bet v 1, strongly suggest the cross-reactivity of these proteins despite their vastly different sources.
Table I.
List of food allergens obtained from a FASTA search in SDAP 2.0 starting with the pollen allergen Bet v 1
| Allergen name∗ | Source | Percentage of amino acid sequence identity with Bet v 1 |
|---|---|---|
| Cor a 1 | Hazelnut | 84 |
| Mal d 1 | Apple | 68 |
| Cas s 1 | Chestnut | 67 |
| Jug r 5 | English Walnut | 67 |
| Pru av 1 | Sweet Cherry | 61 |
| Pru p 1 | Peach | 61 |
| Pru ar 1 | Apricot | 61 |
| Pru du 1 | Almond | 57 |
| Pyr c 1 | Pear | 60 |
| Fra a 1 | Strawberry | 57 |
| Gly m 4 | Soybean | 51 |
| Ara h 8 | Peanut | 47 |
These allergens share high sequence and structural similarity with Bet v 1 but are from different sources known to induce pollen-food syndrome.
Fig 2.
3-D structures of cross-reacting food allergens to Bet v 1, Cor a 1 (hazelnut, PDB id: 6Y3I), Mal d 1 (apple, PDB id: 5MMU), and Ara h 8 (peanut, PDB id: 4M9B) were found by a FASTA search in SDAP-2 starting with the probable sensitizing allergen from birch pollen, Bet v 1 (PDB id: 1BV1). These allergens all group to the same Pfam, despite their diverse sources.
PD tool
When the IgE epitopes of an allergen have been experimentally determined, potential cross-reactivity to other allergens can be found by a peptide similarity search, using the (implemented) property distance (PD) tool. The PD tool calculates how 2 peptides differ based on their quantitative value of 5 quantitative descriptors representing physical-chemical properties for each amino acid residue in each peptide. The PD value is 0 for identical sequences and assumes values up to 5-8, depending on length, for cross-reactive peptides. The PD search method has been validated,8,37, 38, 39, 40 where it was demonstrated to find peptide sequences of cross-reactive allergens that share common properties.38,41,42
In Fig 3 we give an example of a PD search in SDAP 2.0 for the pollen allergen Jun a 1. Using the Jun a 1 linear epitope 1,55,56 we found several pectate lyase allergens from other Cupressaceae species: common cypress (Cup s 1), Japanese cedar (Cry j 1), and Japanese cypress (Cha o 1). These allergens each have sequences with a low PD score (<3.0) to the input IgE-binding peptide of Jun a 1. Consistent with this result, pollen from all 3 of these Cupressaceae trees cross-react with IgE pollen-allergic patient sera in ELISA experiments.28 The PD scale clearly distinguished the pollen pectate lyases from Asteraceae species that have only limited cross-reactivity to Jun a 1, including Art v 6 from mugwort, Amb a 1 from ragweed, or Hel a 6 from sunflower that had PD values more than 7.0. In Table E1 (in the Online Repository available at www.jaci-global.org) we give a list of the top 20 allergens in the database that shows this drastic increase in PD value from Cupressaceae allergens to the Asteraceae pollen allergen Art v 6, illustrating how different the grass and tree pollen proteins are. Other allergens listed in Table E1 have high PD values, such as Chi t 6, Chi t 8, Bomb m 5, or Fag e 1, indicating their low degree of similarity to the query allergen. This indicates the power of the PD approach in distinguishing possible cross-reactive allergens using experimental IgE epitope data.
Fig 3.
Peptide search with the PD tool implemented in SDAP 2.0, starting with the experimentally determined, linear IgE epitope 1 (IFSQNMNIKLKMP) of Jun a 1. A, List of allergens with similar peptides as indicated by their low PD value. A more complete list of the top 20 allergens in SDAP 2.0 found by this search is given in Table E1. B-E, The common epitope areas mapped on the 3-D structures of the similar pectate lyase allergens from the Cupressaceae species Mountain cedar (Jun a 1), common cypress (Cup s 1), Japanese cedar (Cry j 1), and Japanese cypress (Cha o 1). IgE from both Texas and Japanese patients bind to the same regions of Jun a 1.
Cross-React
Our conformational epitope search method, Cross-React,14 has now been implemented in SDAP 2.0. This tool identifies exposed surface patches on the SDAP proteins that best map to known conformational epitopes. For existing linear epitopes in SDAP, we calculated the solvent-accessible surface areas of the amino acids in the epitope region of the 3D structure using GETAREA.57 The Cross-React program selects these surface-exposed residues and maps them on to the protein surfaces of all allergens in SDAP 2.0. Similar conformational epitopes are then found by a high correlation coefficient.14 A high Cross-React correlation score identified the allergens of apple (Mal d 1), cherry (Pru av 1), carrots (Dau c 1), and celery (Api g 1) as being cross-reactive with the birch pollen allergen Bet v 1, confirming previous data.14
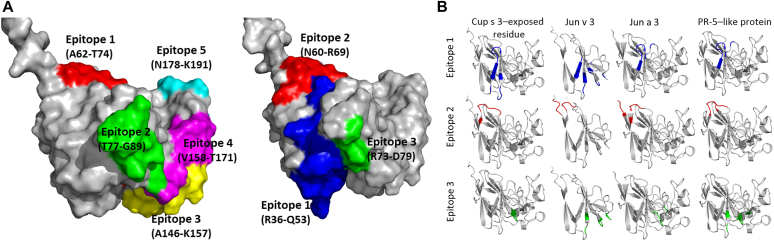
In another example of how structure can identify cross-reactive epitopes, our group previously identified a novel allergen in Juniper pollen, Jun a 3, and characterized a major area on its surface for IgE binding.31 Five IgE-reactive tryptic fragments, separated by HPLC, were mapped on a model based on the nearest structural template in the PDB, for the sweet-tasting protein thaumatin. Subsequently, similar PR-5 homologs of Jun a 3 were identified as allergens in peach, cherry, bell pepper, and apple and could be related to observed pollen-related food sensitivities (oral allergy syndrome).58 In Fig 4, A, we show the location of the IgE-reactive tryptic fragments on Jun a 3 and compare them, structurally, to recently detected epitopes in another PR-5 allergen, Cup s 3 from common cypress (Cupressus sempervirens).30 Cup s 3 and Jun a 3 are known to be cross-reactive from earlier studies by ELISA inhibition assays.38 The epitope 1 from Jun a 3 and epitope 2 of Cup s 3, and epitope 2 from Jun a3 and epitope 3 from Cup s 3, overlap despite the different methods used to identify them. The Cross-React program identified other structurally related proteins in SDAP with common conformational epitopes. Cross-React found 3 structurally similar proteins related to Cup s 3: Jun v 3, Jun a 3, and a thaumatin-like protein homolog from Cupressus arizonica (a non-WHO/IUIS allergen protein in SDAP).59,60 All WHO/IUIS allergens are labeled as 1 in SDAP or 0 if they are not included in this list. These allergens share a high sequence identity with Cup s 3 and have a high correlation coefficient calculated by Cross-React (Table II) of 0.84 or higher. We suggest that the exposed surface area overlapping with epitope 1 from Jun a 3 and epitope1/epitope 2 from Cup s 3 is a conserved conformational epitope among the PR-5 thaumatin-like allergens. The banana allergen Mus a 4, also a PR-5-TLP, also has a high correlation coefficient, but a lower sequence identity. Earlier structural studies of Mus a 461 detected a similar IgE-binding groove of the banana allergen and the pollen allergen Jun a 3, and suggested that this similarity could be the molecular basis for the cross-reactivity between aeroallergens and fruit allergens from the TLP family.
Fig 4.
A, Linear epitopes of Jun a 3 (left) and Cup s 3 (right) mapped on the 3-D structures of the PR-5 allergens. B, Cross-React search in SDAP 2.0 with the linear epitopes of Cup s 3. We found similar surface-exposed patches on the pollen allergens Jun v 3, Jun a 3, and a thaumatin-like protein homolog from Cupressus arizonica with a high Pearson correlation value. N-terminals of proteins are not shown (from M1-L19 in Jun a 1, and M1-A16 in Cup s 3) because they are part of the signal peptides and not part of the mature protein. Epitope location is labeled on the basis of the original publication related to Jun a 1 and Cup s 3.
Table II.
Mapping of linear epitopes using the Cross-React∗ search
| Allergen name | Predicted residues in the patch (epitope 1) | PCC | Sequence identity (%) |
|---|---|---|---|
| Jun v 3 | T18V19W20G28K29R30G52T54G55C66L75 | 0.84 | 95.90 |
| Jun a 3 | L44P45G46G47G48K49S96T98 | 0.81 | 95.10 |
| PR-5–like protein | L18P19G20G21G22K23S70T72 | 0.81 | 98.00 |
| Mus a 4 | W14G20G21G22R23W31L69S70 | 0.82 | 55.70 |
| Allergen name | Predicted residues in the patch (epitope 2) | PCC | Sequence identity (%) |
|---|---|---|---|
| Jun v 3 | G3A4G5A6A42A43G44T45 | 0.94 | 95.90 |
| Jun a 3 | A24G25V26N60A62A63G64T65 | 0.93 | 95.10 |
| PR-5–like protein | L35A36A37G38T39A40Q86S87 | 0.88 | 98.00 |
| Mus a 4 | A1T2N34A37G190G191N193 | 0.91 | 55.70 |
| Allergen name | Predicted residues in the patch (epitope 3) | PCC | Sequence identity (%) |
|---|---|---|---|
| Jun v 3 | T18W20T54G55T57C66Q67T68 | 0.87 | 95.90 |
| Jun a 3 | G75C76T77F78D79G84S85 | 0.89 | 95.10 |
| PR-5–like protein | G49C50T51F52G58S59 | 0.92 | 98.00 |
| Mus a 4 | T12W14T48G49C50S51Q61G137 | 0.84 | 55.70 |
The Cross-React method uses all surface-exposed residues in the linear peptide and locates surface patches in all 3-D structures of allergens in the SDAP 2.0 database with a similar amino acid composition. Using the 3 linear epitopes of Cup s 3 (RYTVWAAGLPGGGKRLDQ, NLAAGTASAR, and RTGCTFD) we found Jun v 3, Jun a 3, and a thaumatin-like protein homolog from Cupressus arizonica (labeled as PR-5–like protein in the table) and Mus a 4 allergens as potential cross-reactive allergens. All amino acid labels are based on the sequence information in the database.
Accuracy of our 3-D models of allergenic proteins in SDAP 2.0
We used the AlphaFold 2 software25,44 running locally on a Linux computer to generate 3-D model structures of all allergenic proteins in SDAP. The qualities of the modeled structures are measured in terms of pLDDT scores.45 Based on the CASP experiment,46 an AlphaFold model is considered reliable if the pLDDT score is above 70. Fig 5, A, shows the average pLDDT scores compared with the sequence identities of the top template structures obtained by a BLAST sequence search against the protein data bank. This indicates that AlphaFold can generate high-quality structures even for difficult targets with low sequence identities to potential template structures. Most of our 3-D model structures have very high average pLDDT scores (Fig 5, B), with 1283 modeled structures above 80%, 1404 structures above 70%, and only 120 allergen structures with average pLDDT score values less than 70%.
Fig 5.
A, Average values of pLDDT scores of 3-D model structures obtained with the AlphaFold program vs the sequence identities of the top template structures in the protein data bank using a BLAST search. B, Histogram of the pLDDT scores. Most of the 3-D models in SDAP 2.0 have average pLDDT scores greater than 70, indicating they are highly reliable 3-D models.
Fig 6 shows an example of a model structure of the major peanut allergen Ara h 2 obtained from the AlphaFold method. Although multiple IgE epitopes have been identified on the basis of peptide mapping,62, 63, 64, 65 Ara h 2 resisted experimental structure determination for years, due to high mobility especially in the flexible loop region, which is one of its most important IgE epitopes. The partial X-ray crystal structure of Ara h 2 was solved in 2011, but major disordered loop regions are missing from the coordinate list in the PDB.66 The loop region with correct stereochemistry is included in the SDAP-2 model, permitting visualization of this area and rational interpretation of clinical data. The model can be used as a starting structure in molecular dynamics calculations, allowing interpretation of mutations affecting IgE binding that are consistent with experimental data (unpublished data). In the current version of SDAP 2.0, we have also provided snapshots of the sequence alignment used to generate each 3-D model structure and its pLDDT score profile plot, allowing easily identification of the reliability of individual segments.
Fig 6.
Quality profile and experimental and 3-D model of the peanut allergen Ara h 2. A, Sequence coverage of Ara h 2 obtained from a database search. Sequence coverage plot (or head-map representation of the multiple sequence alignment) shows the number of homologous sequences identified. These are colored according to sequence identity with Ara h 2. Red color shows low sequence identity, while yellow to blue color shows moderate to high sequence identity. The gap (white space) in the alignment shows regions not covered, and bold black line shows the relative coverage of the query sequence with respect to the total number of aligned sequences. B, X-ray-crystal structure of Ara h 2 in the Protein Data Bank. C, 3-D model of Ara h 2 obtained from AlphaFold 2. Although the X-ray crystal structure has low sequence coverage in the disordered loop region (residue number ∼45-85), AlphaFold builds a good quality complete model of this area, allowing visualization of IgE epitopes, 59-64 RDPYSP and 65-72 SQDPYSPD. Quality of the predicted model is measured by the pLDDT score. A model structure with pLDDT score greater than 70 is modeled well, whereas those with less than 70 need more attention.
Discussion
Allergenic diseases including asthma, allergic rhinitis (also known as hay fever), eczema, hives, and food allergies affect more than 100 million people worldwide. The societal and economic costs of food allergy mentioned in a recent publication67 vary considerably depending on the methods and parameters used and the different geographical locations. However, the studies indicate that the costs are high, exceeding several billions of dollars for individual countries. For example, a large fraction of these costs, the direct medical costs of food allergy for the population in the United States, was estimated to be $4.3 billion.
There is a significant rise in the world population suffering from allergies according to a statistic from the American Academy of Allergy, Asthma & Immunology, which estimated that more than 50 million people alone in the United States have some form of allergies. In addition, allergenic proteins are known to be cross-reactive, increasing the burden to the health care system. Data on allergenic proteins, available from multiple sources, have rapidly expanded with more efficient protein sequencing and experimental data available from clinical trials.68,69 A summary of some of the widely used allergen databases70 is shown in Table E2 (in the Online Repository available at www.jaci-global.org). The specific advantages of SDAP 2.0 are the new 3-D models of all allergens and bioinformatics tools, such as the PD tool and Cross-React, which can be used to find related allergens in the database and to generate hypotheses on potential cross-reactivities. For example, we illustrated how the PD tool can be used to identify potential cross-reactive allergens to the pollen allergen Jun a 1. We showed that several pectate lyase allergens from the common cypress (Cup s 1), Japanese cedar (Cry j 1), and Japanese cypress (Cha o 1) have a low PD value to Jun a 1, which is consistent with experimental observations from ELISA experiments. We also illustrated the usefulness of the 3-D structure to find potential conformational IgE epitopes indicating cross-reactivity by Cross-React. The Cross-React program identified 3 structurally similar proteins related to the allergen Cup s 3: Jun v 3, Jun a 3, and a thaumatin-like protein homolog from Cupressus arizonica. Cup s 3 shares sequence identity with these allergens and has a high correlation coefficient, calculated by Cross-React. We also showed that these findings from Cross-React can suggest potential conserved conformational epitopes among the PR-5 thaumatin-like allergens.
Conclusions
Our webserver SDAP 2.0 provides previously developed and novel bioinformatics tools for allergen researchers and regulatory agencies to assess the allergenic risk of novel proteins. The updated sequence and 3-D structure information allows rapid comparison of all allergens in the WHO/IUIS Allergen Nomenclature database, and many others described in the literature. SDAP 2.0 will be updated on a regular basis, especially in response to user comments and corrections. More practical examples how to use SDAP 2.0 can be found in a recent publication.71
Disclosure statement
This work is supported by the National Institutes of Health, United States (grant no. R01 AI165866).
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Key messages.
-
•
SDAP 2.0 is a novel multifunctional webserver for allergen researchers and regulatory agencies.
-
•
The SDAP 2.0 database contains more than 1600 allergenic proteins, including all those validated by the WHO/IUIS Allergen Nomenclature Sub-Committee.
-
•
Novel resources include an updated list of experimentally determined 3-D structures and a complete set of high-quality 3-D models of allergenic proteins.
-
•
Novel sequence and structure-based search tools are implemented in the web server.
Acknowledgments
We thank the Margaret Maccallum Gage and Tracy Davis Gage Professorship in Biochemistry and Allergies for financial support to W.B. and the Sealy Center for Structural Biology and Molecular Biophysics for computer resources for this project. Availability: The SDAP database is available online at https://fermi.utmb.edu/SDAP/.
Supplementary data
References
- 1.Pomes A., Davies J.M., Gadermaier G., Hilger C., Holzhauser T., Lidholm J., et al. WHO/IUIS Allergen Nomenclature: providing a common language. Mol Immunol. 2018;100:3–13. doi: 10.1016/j.molimm.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolkhir P., Elieh-Ali-Komi D., Metz M., Siebenhaar F., Maurer M. Understanding human mast cells: lesson from therapies for allergic and non-allergic diseases. Nat Rev Immunol. 2022;22:294–308. doi: 10.1038/s41577-021-00622-y. [DOI] [PubMed] [Google Scholar]
- 3.Ring J. History of allergy: clinical descriptions, pathophysiology, and treatment. Handb Exp Pharmacol. 2022;268:3–19. doi: 10.1007/164_2021_509. [DOI] [PubMed] [Google Scholar]
- 4.Dreskin S.C., Koppelman S.J., Andorf S., Nadeau K.C., Kalra A., Braun W., et al. The importance of the 2S albumins for allergenicity and cross-reactivity of peanuts, tree nuts, and sesame seeds. J Allergy Clin Immunol. 2021;147:1154–1163. doi: 10.1016/j.jaci.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nesbit J.B., Schein C.H., Braun B.A., Gipson S.A.Y., Cheng H., Hurlburt B.K., et al. Epitopes with similar physicochemical properties contribute to cross reactivity between peanut and tree nuts. Mol Immunol. 2020;122:223–231. doi: 10.1016/j.molimm.2020.03.017. [DOI] [PubMed] [Google Scholar]
- 6.Schein C.H., Negi S.S., Braun W. Still SDAPing along: 20 years of the Structural Database of Allergenic Proteins. Front Allergy. 2022;3 doi: 10.3389/falgy.2022.863172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanciuc O., Schein C.H., Braun W. Data mining of sequences and 3D structures of allergenic proteins. Bioinformatics. 2002;18:1358–1364. doi: 10.1093/bioinformatics/18.10.1358. [DOI] [PubMed] [Google Scholar]
- 8.Ivanciuc O., Schein C.H., Garcia T., Oezguen N., Negi S.S., Braun W. Structural analysis of linear and conformational epitopes of allergens. Regul Toxicol Pharmacol. 2009;54:S11–S19. doi: 10.1016/j.yrtph.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Ree R., Sapiter Ballerda D., Berin M.C., Beuf L., Chang A., Gadermaier G., et al. The COMPARE Database: a public resource for allergen identification, adapted for continuous improvement. Front Allergy. 2021;2 doi: 10.3389/falgy.2021.700533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudharson S., Kalic T., Hafner C., Breiteneder H. Newly defined allergens in the WHO/IUIS Allergen Nomenclature Database during 01/2019-03/2021. Allergy. 2021;76:3359–3373. doi: 10.1111/all.15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schein C.H., Ivanciuc O., Midoro-Horiuti T., Goldblum R.M., Braun W. An allergen portrait gallery: representative structures and an overview of IgE binding surfaces. Bioinform Biol Insights. 2010;4:113–125. doi: 10.4137/BBI.S5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu W., Negi S.S., Schein C.H., Maleki S.J., Hurlburt B.K., Braun W. Distinguishing allergens from non-allergenic homologues using Physical-Chemical Property (PCP) motifs. Mol Immunol. 2018;99:1–8. doi: 10.1016/j.molimm.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters B., Sidney J., Bourne P., Bui H.H., Buus S., Doh G., et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Negi S.S., Braun W. Cross-React: a new structural bioinformatics method for predicting allergen cross-reactivity. Bioinformatics. 2017;33:1014–1020. doi: 10.1093/bioinformatics/btw767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiers M.W., Kleter G.A., Nijland H., Peijnenburg A.A., Nap J.P., van Ham R.C. Allermatch, a webtool for the prediction of potential allergenicity according to current FAO/WHO Codex alimentarius guidelines. BMC Bioinformatics. 2004;5:133. doi: 10.1186/1471-2105-5-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittag D., Batori V., Neudecker P., Wiche R., Friis E.P., Ballmer-Weber B.K., et al. A novel approach for investigation of specific and cross-reactive IgE epitopes on Bet v 1 and homologous food allergens in individual patients. Mol Immunol. 2006;43:268–278. doi: 10.1016/j.molimm.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Sharma N., Patiyal S., Dhall A., Pande A., Arora C., Raghava G.P.S. AlgPred 2.0: an improved method for predicting allergenic proteins and mapping of IgE epitopes. Brief Bioinform. 2021:22. doi: 10.1093/bib/bbaa294. [DOI] [PubMed] [Google Scholar]
- 18.Maurer-Stroh S., Krutz N.L., Kern P.S., Gunalan V., Nguyen M.N., Limviphuvadh V., et al. AllerCatPro—prediction of protein allergenicity potential from the protein sequence. Bioinformatics. 2019;35:3020–3027. doi: 10.1093/bioinformatics/btz029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimitrov I., Bangov I., Flower D.R., Doytchinova I., AllerTOP v. 2--a server for in silico prediction of allergens. J Mol Model. 2014;20:2278. doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 20.Negi S.S., Braun W. Automated detection of conformational epitopes using phage display peptide sequences. Bioinform Biol Insights. 2009;3:71–81. doi: 10.4137/bbi.s2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith S.A., Chruszcz M., Chapman M.D., Pomes A. Human monoclonal IgE antibodies—a major milestone in allergy. Curr Allergy Asthma Rep. 2023;23:53–65. doi: 10.1007/s11882-022-01055-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanciuc O., Schein C.H., Braun W. SDAP: database and computational tools for allergenic proteins. Nucleic Acids Res. 2003;31:359–362. doi: 10.1093/nar/gkg010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oezguen N., Zhou B., Negi S.S., Ivanciuc O., Schein C.H., Labesse G., et al. Comprehensive 3D-modeling of allergenic proteins and amino acid composition of potential conformational IgE epitopes. Mol Immunol. 2008;45:3740–3747. doi: 10.1016/j.molimm.2008.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Power T.D., Ivanciuc O., Schein C.H., Braun W. Assessment of 3D models for allergen research. Proteins. 2013;81:545–554. doi: 10.1002/prot.24239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nesbit J.B., Hurlburt B.K., Schein C.H., Cheng H., Wei H., Maleki S.J. Ara h 1 structure is retained after roasting and is important for enhanced binding to IgE. Mol Nutr Food Res. 2012;56:1739–1747. doi: 10.1002/mnfr.201100815. [DOI] [PubMed] [Google Scholar]
- 27.Maleki S.J., Teuber S.S., Cheng H., Chen D., Comstock S.S., Ruan S., et al. Computationally predicted IgE epitopes of walnut allergens contribute to cross-reactivity with peanuts. Allergy. 2011;66:1522–1529. doi: 10.1111/j.1398-9995.2011.02692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barre A., Sénéchal H., Nguyen C., Granier C., Rougé P., Poncet P. Identification of potential IgE-binding epitopes contributing to the cross-reactivity of the major cupressaceae pectate-lyase pollen allergens (group 1) Allergies. 2022;2:106–118. [Google Scholar]
- 29.Pichler U., Hauser M., Wolf M., Bernardi M.L., Gadermaier G., Weiss R., et al. Pectate lyase pollen allergens: sensitization profiles and cross-reactivity pattern. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barre A., Sénéchal H., Nguyen C., Granier C., Poncet P., Rougé P. Structural basis for the IgE-binding cross-reacting epitopic peptides of Cup s 3, a PR-5 thaumatin-like protein allergen from common cypress (Cupressus sempervirens) pollen. Allergies. 2023;3:11–24. [Google Scholar]
- 31.Soman K.V., Midoro-Horiuti T., Ferreon J.C., Goldblum R.M., Brooks E.G., Kurosky A., et al. Homology modeling and characterization of IgE binding epitopes of mountain cedar allergen Jun a 3. Biophys J. 2000;79:1601–1609. doi: 10.1016/S0006-3495(00)76410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson W.R. Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics. 1991;11:635–650. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- 33.Altschul S., Madden T., Schaffer A., Zhang J.H., Zhang Z., Miller W., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Faseb J. 1998;12 doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aalberse R.C. Assessment of sequence homology and cross-reactivity. Toxicol Appl Pharmacol. 2005;207:149–151. doi: 10.1016/j.taap.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 35.Breiteneder H., Mills C. Structural bioinformatic approaches to understand cross-reactivity. Mol Nutr Food Res. 2006;50:628–632. doi: 10.1002/mnfr.200500274. [DOI] [PubMed] [Google Scholar]
- 36.Mathura V.S., Schein C.H., Braun W. Identifying property based sequence motifs in protein families and superfamilies: application to DNase-1 related endonucleases. Bioinformatics. 2003;19:1381–1390. doi: 10.1093/bioinformatics/btg164. [DOI] [PubMed] [Google Scholar]
- 37.Ivanciuc O., Midoro-Horiuti T., Schein C.H., Xie L., Hillman G.R., Goldblum R.M., et al. The property distance index PD predicts peptides that cross-react with IgE antibodies. Mol Immunol. 2009;46:873–883. doi: 10.1016/j.molimm.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivanciuc O., Mathura V., Midoro-Horiuti T., Braun W., Goldblum R.M., Schein C.H. Detecting potential IgE-reactive sites on food proteins using a sequence and structure database, SDAP-Food. J Agr Food Chem. 2003;51:4830–4837. doi: 10.1021/jf034218r. [DOI] [PubMed] [Google Scholar]
- 39.Schein C.H., Ivanciuc O., Braun W. Common physical-chemical properties correlate with similar structure of the IgE epitopes of peanut allergens. J Agric Food Chem. 2005;53:8752–8759. doi: 10.1021/jf051148a. [DOI] [PubMed] [Google Scholar]
- 40.Schein C.H., Ivanciuc O., Braun W. Bioinformatics approaches to classifying allergens and predicting cross-reactivity. Immunol Allergy Clin North Am. 2007;27:1–27. doi: 10.1016/j.iac.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanciuc O., Oezguen N., Mathura V.S., Schein C.H., Xu Y., Braun W. Using property based sequence motifs and 3D modeling to determine structure and functional regions of proteins. Curr Med Chem. 2004;11:583–593. doi: 10.2174/0929867043455819. [DOI] [PubMed] [Google Scholar]
- 42.Schein C.H., Zhou B., Oezguen N., Mathura V.S., Braun W. Molego-based definition of the architecture and specificity of metal-binding sites. Proteins Struct Funct Bioinform. 2005;58:200–210. doi: 10.1002/prot.20253. [DOI] [PubMed] [Google Scholar]
- 43.Braun B.A., Schein C.H., Braun W. DGraph clusters flaviviruses and beta-coronaviruses according to their hosts, disease type, and human cell receptors. Bioinform Biol Insights. 2021;15 doi: 10.1177/11779322211020316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mariani V., Biasini M., Barbato A., Schwede T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics. 2013;29:2722–2728. doi: 10.1093/bioinformatics/btt473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ozden B., Kryshtafovych A., Karaca E. Assessment of the CASP14 assembly predictions. Proteins. 2021;89:1787–1799. doi: 10.1002/prot.26199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rego N., Koes D. 3Dmol.js: molecular visualization with WebGL. Bioinformatics. 2015;31:1322–1324. doi: 10.1093/bioinformatics/btu829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radauer C., Bublin M., Wagner S., Mari A., Breiteneder H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J Allergy Clin Immunol. 2008;121:847–852.e7. doi: 10.1016/j.jaci.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 49.Ivanciuc O., Garcia T., Torres M., Schein C.H., Braun W. Characteristic motifs for families of allergenic proteins. Mol Immunol. 2009;46:559–568. doi: 10.1016/j.molimm.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mistry J., Bateman A., Finn R.D. Predicting active site residue annotations in the Pfam database. BMC Bioinform. 2007;8:298. doi: 10.1186/1471-2105-8-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vieths S., Scheurer S., Ballmer-Weber B. Current understanding of cross-reactivity of food allergens and pollen. Ann N Y Acad Sci. 2002;964:47–68. doi: 10.1111/j.1749-6632.2002.tb04132.x. [DOI] [PubMed] [Google Scholar]
- 53.Geroldinger-Simic M., Zelniker T., Aberer W., Ebner C., Egger C., Greiderer A., et al. Birch pollen-related food allergy: clinical aspects and the role of allergen-specific IgE and IgG4 antibodies. J Allergy Clin Immunol. 2011;127:616–622.e1. doi: 10.1016/j.jaci.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 54.Masthoff L.J., van Hoffen E., Mattsson L., Lidholm J., Andersson K., Zuidmeer-Jongejan L., et al. Peanut allergy is common among hazelnut-sensitized subjects but is not primarily the result of IgE cross-reactivity. Allergy. 2015;70:265–274. doi: 10.1111/all.12554. [DOI] [PubMed] [Google Scholar]
- 55.Midoro-Horiuti T., Mathura V., Schein C.H., Braun W., Yu S., Watanabe M., et al. Major linear IgE epitopes of mountain cedar pollen allergen Jun a 1 map to the pectate lyase catalytic site. Mol Immunol. 2003;40:555–562. doi: 10.1016/s0161-5890(03)00168-8. [DOI] [PubMed] [Google Scholar]
- 56.Midoro-Horiuti T., Goldblum R.M., Kurosky A., Wood T.G., Schein C.H., Brooks E.G. Molecular cloning of the mountain cedar (Juniperus ashei) pollen major allergen, Jun a 1. J Allergy Clin Immunol. 1999;104:613–617. doi: 10.1016/s0091-6749(99)70332-5. [DOI] [PubMed] [Google Scholar]
- 57.Fraczkiewicz R., Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comp Chem. 1998;19:319. [Google Scholar]
- 58.Poncet P., Senechal H., Charpin D. Update on pollen-food allergy syndrome. Expert Rev Clin Immunol. 2020;16:561–578. doi: 10.1080/1744666X.2020.1774366. [DOI] [PubMed] [Google Scholar]
- 59.Cortegano I., Civantos E., Aceituno E., del Moral A., Lopez E., Lombardero M., et al. Cloning and expression of a major allergen from Cupressus arizonica pollen, Cup a 3, a PR-5 protein expressed under polluted environment. Allergy. 2004;59:485–490. doi: 10.1046/j.1398-9995.2003.00363.x. [DOI] [PubMed] [Google Scholar]
- 60.Suarez-Cervera M., Castells T., Vega-Maray A., Civantos E., del Pozo V., Fernandez-Gonzalez D., et al. Effects of air pollution on Cup a 3 allergen in Cupressus arizonica pollen grains. Ann Allergy Asthma Immunol. 2008;101:57–66. doi: 10.1016/S1081-1206(10)60836-8. [DOI] [PubMed] [Google Scholar]
- 61.Leone P., Menu-Bouaouiche L., Peumans W.J., Payan F., Barre A., Roussel A., et al. Resolution of the structure of the allergenic and antifungal banana fruit thaumatin-like protein at 1.7-A. Biochimie. 2006;88:45–52. doi: 10.1016/j.biochi.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 62.Dreskin S.C., Germinaro M., Reinhold D., Chen X., Vickery B.P., Kulis M., et al. IgE binding to linear epitopes of Ara h 2 in peanut allergic preschool children undergoing oral immunotherapy. Pediatr Allergy Immunol. 2019;30:817–823. doi: 10.1111/pai.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X., Negi S.S., Liao S., Gao V., Braun W., Dreskin S.C. Conformational IgE epitopes of peanut allergens Ara h 2 and Ara h 6. Clin Exp Allergy. 2016;46:1120–1128. doi: 10.1111/cea.12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao S., Patil S.U., Shreffler W.G., Dreskin S.C., Chen X. Human monoclonal antibodies to Ara h 2 inhibit allergen-induced, IgE-mediated cell activation. Clin Exp Allergy. 2019;49:1154–1157. doi: 10.1111/cea.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Otsu K., Guo R., Dreskin S.C. Epitope analysis of Ara h 2 and Ara h 6: characteristic patterns of IgE-binding fingerprints among individuals with similar clinical histories. Clin Exp Allergy. 2015;45:471–484. doi: 10.1111/cea.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mueller G.A., Gosavi R.A., Pomes A., Wunschmann S., Moon A.F., London R.E., et al. Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity. Allergy. 2011;66:878–885. doi: 10.1111/j.1398-9995.2010.02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bilaver L.A., Chadha A.S., Doshi P., O’Dwyer L., Gupta R.S. Economic burden of food allergy: a systematic review. Ann Allergy Asthma Immunol. 2019;122:373–380.e1. doi: 10.1016/j.anai.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 68.Langlois A., Lavergne M.H., Leroux H., Killer K., Azzano P., Paradis L., et al. Protocol for a double-blind, randomized controlled trial on the dose-related efficacy of omalizumab in multi-food oral immunotherapy. Allergy Asthma Clin Immunol. 2020;16:25. doi: 10.1186/s13223-020-00419-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fleischer D.M., Greenhawt M., Sussman G., Begin P., Nowak-Wegrzyn A., Petroni D., et al. Effect of epicutaneous immunotherapy vs placebo on reaction to peanut protein ingestion among children with peanut allergy: the PEPITES randomized clinical trial. JAMA. 2019;321:946–955. doi: 10.1001/jama.2019.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goodman R.E., Breiteneder H. The WHO/IUIS Allergen Nomenclature. Allergy. 2019;74:429–431. doi: 10.1111/all.13693. [DOI] [PubMed] [Google Scholar]
- 71.Schein CH. Identifying Similar Allergens and Potentially Cross-Reacting Areas Using Structural Database of Allergenic Proteins (SDAP) Tools and D-Graph Beatriz Cabanillas (ed.), Food Allergens: Methods and Protocols, Methods in Molecular Biology, vol. 2717, chapter 18. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.