Abstract
A multiplex reverse transcription (RT)-PCR method that has been developed is capable of detecting and subtyping influenza A (H1N1 and H3N2) and B viruses as well as respiratory syncytial virus (RSV) types A and B in respiratory clinical samples taken as part of a national community-based surveillance program of influenza-like illness in England and Wales. The detection of each different pathogen depended on distinguishing five amplification products of different sizes on agarose gels following RT-PCR with multiple primer sets. The multiplex RT-PCR was tested with 65 nasopharyngeal apirates from which RSV had been isolated and 237 combined nose and throat swabs from which influenza A (H1N1 and H3N2) or B virus had been detected by virus isolation, as well as 40 respiratory samples from which other viruses including cytomegalovirus, herpes simplex virus, enteroviruses, and parainfluenza viruses had been grown. For the typing and subtyping of influenza A and B viruses and RSV types A and B, the multiplex RT-PCR gave an excellent (100%) correlation with the results of conventional typing and subtyping with specific antisera. Multiplex RT-PCR can also be used to accurately detect more than one viral template in the same reaction mixture, allowing viral coinfections to be identified with the same respiratory specimen.
Respiratory illness is the most common reason for consulting with general practitioners in all age groups (21), and seasonal respiratory infection is an important cause of hospitalization and excess mortality in the winter months. Infectious respiratory disease can be caused by several pathogens, and the clinical presentation of patients with different viral infections can be very similar, making diagnosis difficult. Prominent among the viral causes of respiratory illness are influenza A and B viruses and respiratory syncytial virus (RSV). It is estimated that influenza A virus infections are responsible for approximately 10,000 excess deaths in years of moderate epidemic activity and more than 25,000 excess deaths in years of exceptional epidemic activity (12). The excess deaths associated with influenza occur primarily in the group of individuals over the age of 65. RSV is probably the most significant respiratory pathogen of infants in the first 6 months of life and is estimated to be responsible for approximately 90,000 hospitalizations and 4,500 deaths in this age group annually in the United States (6). RSV is thus as significant a pathogen as influenza virus. Together, these viral pathogens cause a substantial burden of illness both in the community and in hospitals. Although the contribution of RSV to illness in children is understood, the role of RSV in adult respiratory illness is still largely unknown, and it is almost certainly underdiagnosed (11). As part of a national linked clinical-virological sentinel physician surveillance program in England and Wales, we are interested in assessing the contribution of RSV in the presentation of influenza-like illness to general practitioners.
Although routine diagnostic methods for influenza virus, including virus culture and antigen detection, are both sensitive and robust, PCR for the detection of influenza virus in respiratory samples taken from patients with influenza-like illness has also been shown to be useful (8) because it offers an enhanced sensitivity combined with rapid detection and subtyping ability. RSV is more labile than influenza virus, which has made conventional virus culture more difficult. As a consequence, rapid diagnostic methods for RSV have concentrated on the development of sensitive antigen detection in respiratory samples, but such tests are rarely capable of differentiating RSV A and B subtypes. These are distinguished on the basis of serological reactivity with monoclonal antibodies (22) and may be associated with illnesses of different severities (25). There is therefore a need for a sensitive test that is capable of detecting and subtyping influenza virus and RSV, preferably in a single step, and that is not dependent on the presence of infectious virus. Such a test would be useful in assessing the contribution of RSV to the overall burden of respiratory illness in the community and in diagnostic settings where subtype information might be sought. Multiplex PCR should be able to accomplish these objectives.
Multiplex PCR uses a combination of several different primer pairs in the same amplification reaction with the objective of producing different specific amplicons, depending on the targets present in the sample. Multiplex PCR has mainly been aimed at amplifying linked regions of DNA in the diagnosis of genetic disorders such as Duchenne muscular dystrophy (5) or cystic fibrosis which are associated with mutations at multiple loci (2). However, multiplex PCR has increasingly been used for the diagnosis of infectious diseases, including those caused by DNA- and RNA-containing viruses (8, 16, 18, 23), in which the starting nucleic acid template may be low in copy number and of poor quality compared with those of chromosomal DNA. In this report we describe the development and application of a multiplex reverse transcription (RT)-PCR with five nested primer sets capable of detecting and subtyping influenza virus and RSV in clinical respiratory samples.
MATERIALS AND METHODS
Virus stocks.
RSV subtype A and B strains (Long [subtype A], B908106, B3/60, and B18537) were grown in HEp-2 cells obtained from the European Cell Culture Collection (Porton Down, Salisbury, United Kingdom). Viruses were harvested when 70% of the monolayer displayed a cytopathic effect, usually 2 to 4 days postinfection. Virus stocks consisted of a mixture of mechanically disrupted cells and supernatant and were stored at −70°C immediately after harvest and until use. Influenza A (H3N2 and H1N1) and B virus strains were grown in the allantoic cavities of embryonated eggs or in Madin-Darby canine kidney (MDCK) cells in the presence of tolylsulfonyl phenylalanyl chloromethyl ketone-treated trypsin. Virus-containing tissue culture supernatant or allantoic fluid was stored at −70°C until it was required.
Titration of virus infectivity. (i) RSV.
HEp-2 cells were seeded at 5 × 104 cells/ml in minimal essential medium (MEM) (Gibco-BRL) containing 10% fetal calf serum in 96-well microtiter plates (Greiner Labortechnik) and were incubated overnight at 37°C in a CO2 atmosphere. One hundred microliters of each virus dilution was added in duplicate to wells containing cell monolayers. The plates were sealed and spun at 1,500 × g for 45 min at 37°C. Samples were aspirated from the wells and replaced with MEM containing 2% fetal calf serum and were further incubated for 24 h at 37°C in 5% CO2. Medium was expelled from the plates, and the cells were washed with phosphate-buffered saline (PBS) prior to fixation for 20 min with absolute methanol with 2% (vol/vol) hydrogen peroxide. Virus-infected cells were detected by the addition of polyclonal goat anti-RSV antibody (Biogenesis, Dorset, United Kingdom) diluted 1:800 in PBS–0.05% Tween 20 (PBST) for 1 h. The subtype was determined with an RSV subtype B-specific monoclonal antibody (clone 7858; National Bacteriological Laboratory, Stockholm, Sweden) diluted 1:1,000 in PBST in parallel infected wells. Following three washes with PBST, rabbit anti-goat or anti-mouse horseradish peroxidase conjugate (Chemicon) was added to each well and the plates were incubated for a further hour at 37°C. After washing three times in PBST, a freshly prepared insoluble horseradish peroxidase substrate (3-amino-ethyl-carbazole) was added to each well, and the plates were incubated at room temperature for 45 min in the dark. The wells were examined under a microscope for the presence of pink-stained cells. Each pink cell was considered to represent infection by a plaque-forming unit of RSV.
(ii) Influenza virus.
Titration of influenza virus infectivity was performed as described previously (8, 24). Briefly, confluent MDCK cells were washed with PBS and incubated for 1 h at room temperature with virus inoculum diluted in MEM. The inoculum was removed, and the cells were overlaid with medium containing 0.5% indubiose, nonessential amino acids, and 3 μg of tolylsulfonyl phenylalanyl chloromethyl ketone-treated trypsin/ml and incubated at 37°C in 5% CO2 for 48 h. The cells were fixed with 5% (vol/vol) glutaraldehyde and stained with 2% (vol/vol) carbol fuschin.
Nucleic acid extraction and cDNA synthesis.
RNA was extracted from a 100- to 150-μl volume of sample (egg fluids, tissue culture material, clinical specimens, and water controls) by a guanidinium thiocyanate-silica binding method (3, 13). Briefly, specimen was added to a tube containing 840 to 890 μl of lysis buffer (120 g of guanidinium thiocyanate, 100 ml of 0.1 M Tris-HCl [pH 6.4], 22 ml of 0.2 M EDTA [pH 8.0], 2.6 g of Triton X-100) and 10 μl of silica suspension, mixed, and incubated for 10 min at room temperature; and the RNA that bound to the silica was washed twice with 1 ml of buffer L2 (120 g of guanidinium thiocyanate, 100 ml of 0.1 M Tris-HCl [pH 6.4]), twice with 1 ml of 70% (vol/vol) ethanol, and once with 1 ml of acetone and then dried at 56°C for 10 min. It was then resuspended in 30 μl of RNase-free water and converted into cDNA by RT-PCR. For RT 22.2 μl of RNA was added to a reaction mixture (17.8 μl) containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 7.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 1.5 mM, 25 ng of each random primer [(pdN)6; Pharmacia], 1.6 U of RNasin (Promega), and 200 U of Moloney murine leukemia virus reverse transcriptase (Gibco BRL). The reaction mixture was incubated at room temperature for 10 min, 37°C for 45 min, and 95°C for 5 min and quenched on ice.
PCR.
The primers used in the study are described in Table 1. The properties of the primers were analyzed with OLIGO 5 primer analysis software (National Biosciences Inc.). Each primer pair was used at 5 pmol in the primary amplification and 25 pmol in the secondary amplification. For the primary PCR 20 μl of cDNA was added to 80 μl of a reaction mixture containing 10 mM Tris-HCl (pH 8.8), 3.5 mM MgCl2, 2.5 mM KCl, and 1.5 U of Taq polymerase. Amplification with a DNA Engine thermocycler (MJ Research) consisted of 1 cycle at 94°C for 2 min, followed by 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min. Two microliters of primary product was then transferred to 48 μl of the secondary amplification mixture as above (plus each deoxynucleoside triphosphate at a concentration of 0.2 mM). The samples were then incubated for 1 cycle at 94°C for 2 min and then 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min. Amplicons were visualized by ethidium bromide staining following electrophoresis on 2.25% NuSieve (FMC BioProducts) agarose gels.
TABLE 1.
Properties of primers used for RSV and influenza virus multiplex PCR
| Amplification steps and primera | Sequence (5′→3′) | Gene position | Melting temp (°C) | G+C content (%) | Optimal annealing temp (°C) | Maximum annealing temp (°C) | Amplicon size (bp) | Sensitivity (PFU) |
|---|---|---|---|---|---|---|---|---|
| Primary | ||||||||
| AHI A | CAGATGCAGACACAATATGT | HA | 55 | 40 | 52 | 63 | 1,015 | NAb |
| AHI FII | AAACCGGCAATGGCTCCAAA | HA | 72 | 50 | ||||
| AH3 A | CAGATTGAAGTGACTAATGC | HA | 55 | 40 | 52 | 62 | 883 | NA |
| AH3 DII | GTTTCTCTGGTACATTCCGC | HA | 62 | 50 | ||||
| BHA A | GTGACTGGTGTGATACCACT | HA | 56 | 50 | 53 | 64 | 900 | NA |
| BHA DII | TGTTTTCACCCATATTGGGC | HA | 65 | 45 | ||||
| RSVAB F | GTCTTACAGCCGTGATTAGG | N | 63 | 50 | 52 | 66 | 838 | NA |
| RSVAB R | GGGCTTTCTTTGGTTACTTC | P | 64 | 45 | ||||
| Secondary | ||||||||
| AHI B | ATAGGCTACCATGCGAACAA | HA | 63 | 45 | 52 | 62 | 944 | <1 |
| AHI EII | CTTAGTCCTGTAACCATCCT | HA | 55 | 45 | ||||
| AH3 B | AGCAAAGCTTTCAGCAACTG | HA | 63 | 45 | 54 | 69 | 591 | <1 |
| AH3 CII | GCTTCCATTTGGAGTGATGC | HA | 65 | 50 | ||||
| BHA B | CATTTTGCAAATCTCAAAGC | HA | 61 | 35 | 54 | 67 | 767 | <1 |
| BHA CII | TGGAGGCAATCTGCTTCACC | HA | 68 | 55 | ||||
| RSVA F | GATGTTACGGTGGGGAGTCT | N | 64 | 45 | 48 | 51 | 334 | <1 |
| RSVA R | GTACACTGTAGTTAATCACA | N | 51 | 35 | ||||
| RSVB F | AATGCTAAGATGGGGAGTTC | N | 64 | 45 | 50 | 61 | 183 | <1 |
| RSVB R | GAAATTGAGTTAATGACAGC | N | 58 | 35 |
F, forward; R, reverse.
NA, not applicable.
Clinical specimens.
Nasopharyngeal aspirates, bronchoalveolar lavages, endotracheal aspirates, and untyped isolates of RSV were obtained from clinical diagnostic material collected between October 1995 and May 1996 from hospitals in the London area. Combined nose and throat swabs (made of rayon) were obtained from patients with community-acquired respiratory illness investigated during clinical virological surveillance of influenza in England and Wales in the winter season of 1995 and 1996 (15). Aliquots of material for PCR testing were made immediately on receipt of the specimen and were stored at −70°C until use. All other clinical material was also stored at −70°C.
RESULTS
The oligonucleotide primers designed to amplify RSV were placed in the N and P regions of the genome, because they are highly conserved and are regions of the RSV genome which allow subtyping of RSV strains into RSV A and B types (1, 17, 22). The G+C contents, melting temperatures, and lengths of the primers were chosen and analyzed by using OLIGO 5 primer design software to ensure that they not only met the essential criteria for optimal PCR primers (7) but also could be used together in a multiplex PCR under similar conditions already determined to be effective for the detection and subtyping of influenza A and B viruses (Table 1) (8). Moreover, the primers were designed to ensure that the final reaction products could easily be differentiated on the basis of size from each other and from the reaction products from amplification of influenza virus templates (Table 1). All of the primers chosen were 20mers and had G+C contents of less than or equal to 55% (Table 1). Annealing temperatures of 50 and 60°C in the first and second rounds of amplification, respectively, were selected to give maximum product yield and specificity. All of the primers were also analyzed by using OLIGO 5 software for the formation of dimers either within or between pairs; no significant theoretical mispriming was identified on any template.
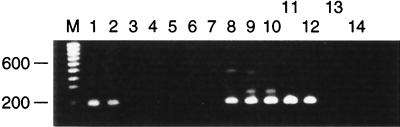
Biochemical optimization of the amplification conditions for each primer set was performed, and final concentrations of 10 mM Tris-HCl (pH 8.8), 3.5 mM MgCl2, 2.5 mM KCl (Optiprime buffer 7), and 1.5 U of Taq polymerase (Gibco-BRL) were found to be optimal for the maximum yield of the specific product of each nested primer set in a multiplex reaction mixture. The previously published biochemical conditions (1.5 mM MgCl2, 25 mM KCl [pH 8.8]) selected for multiplex RT-PCR detection of influenza A and B viruses were found to be suboptimal for the detection of RSV type B (Fig. 1). Modifications of the PCR assay protocol included an increase in the concentration of Taq polymerase in the secondary reaction mixture and an increase in the concentration of MgCl2 in both the primary and the secondary reaction mixtures. An increase in the concentration of Taq in the secondary reaction mixture significantly increased the level of product formation (data not shown). Lowering of the pH of the reaction mixture to below 8.5 was found to substantially decrease the sensitivity of detection (data not shown). Wide ranges of cycling conditions and primer concentrations were tested. The final amplification protocol included initial denaturation of 94°C for 2 min and then 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min for the primary reaction and 94°C for 2 min and then 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min for the secondary reaction.
FIG. 1.
Serial 10-fold dilutions of tissue culture-grown RSV type B VS1039 were prepared in virus transport medium (starting concentration of RSV, 105 PFU/ml [lanes 1 and 8]). Each dilution was subjected to multiplex RT-PCR with all of the primer sets. Lanes 1 to 6; 10 mM Tris-HCl (pH 8.4), 1.5 mM MgCl2, 25 mM KCl, and 1.5 U of Taq in the primary reaction mixture and 0.75 U of Taq in the secondary reaction mixture; lanes 8 to 13, 10 mM Tris (pH 8.8), 3.5 mM MgCl2, 2.5 mM KCl, and 1.5 U of Taq in the primary reaction mixture and 1.5 U of Taq in the secondary reaction mixture.; lanes 7 and 14, water controls; lane M, molecular size marker. Numbers on the left are in base pairs.
Mechanical means of hot start for this multiplex RT-PCR were not attempted, because this assay was designed for use with large numbers of samples, which would make such an approach impractical. Nonmechanical means of hot start were tested; these included the use of Taq-start Antibody (CLONTECH) and Ampli-Taq Gold (Perkin-Elmer). No improvement in sensitivity or specificity was seen with either of these hot-start methods, and no form of chemical hot start was incorporated into the multiplex RT-PCR.
Specificity.
The multiplex RT-PCR was tested for its specificity for all of the viral targets (influenza A [H1N1 and H3N2] and B viruses and RSV types A and B) by first using the RSV primers and then adding each of the influenza virus primer pairs sequentially to simulated clinical specimens. No mispriming was observed when all of the primer sets were present with either an influenza A or B virus or an RSV type A or B template. A product of the expected size was obtained for each viral template by the multiplex RT-PCR with all of the primer sets present. The specific products could all be clearly separated and identified on a 2.25% NuSieve agarose gel. This was true for both laboratory-adapted virus control material (tissue culture-grown virus, influenza virus and RSV, or egg-grown influenza virus) and for clinical samples containing wild-type strains (Fig. 2). The product specificities of the amplicons obtained from the multiplex RT-PCRs were also confirmed by sequence analysis. There was no detectable PCR product following nucleic acid extraction and multiplex RT-PCR amplification from 40 clinical samples (nasopharyngeal aspirates, nose and throat swabs, or broncholalveolar lavage material) containing human parainfluenza virus types 1 to 3 (n = 15 samples), human cytomegalovirus (n = 7), herpes simplex virus type 1 (n = 4), untyped enteroviruses (n = 3), and rhinoviruses (n = 11) (data not shown). The multiplex RT-PCR was tested with a blind panel of 65 nasopharyngeal aspirate, broncheoaveolar lavage, or endotracheal aspirate specimens; 40 contained RSV type A and 20 contained RSV type B, as determined by enzyme-linked immunosorbent assay (ELISA) with RSV type-specific monoclonal antibodies; 5 of these specimens consisted of negative nasopharyngeal aspirates from which no virus was recovered. There was a 100% correlation between the RSV subtype determined by PCR and that determined by the subtype-specific ELISA. There was also a 100% correlation between the type determined by PCR and antigenic type for 237 nose and throat swab specimens containing influenza A virus (H1N1 and H3N2) and influenza B virus, for which antigenic typing of influenza virus grown in tissue culture from the original specimen was performed by hemagglutination-inhibition with postinfection ferret antisera as described previously (4). Twenty nose and throat swab specimens which were negative both by culture and by immunofluorescence for any virus were also included as a negative control panel to assess specificity. No detectable PCR products were seen.
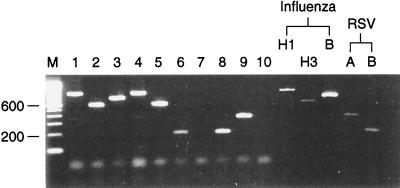
FIG. 2.
Typing and subtyping of influenza virus and RSV in a panel of clinical specimens by multiplex RT-PCR. Lanes 1 to 6, 8, and 9, combined nose and throat swabs; lane 1, influenza A (H1N1) virus; lane 2, influenza A (H3N2) virus; lane 3, influenza B virus; lane 4, influenza A (H1N1) virus; lane 5, influenza A (H3N2) virus; lane 6, RSV type B; lane 7, virus transport medium; lane 8, RSV type B; lane 9, RSV type A; lane 10, water control. Influenza virus controls were A/Taiwan/1/86 H1N1, A/Thessaloniki/1/95 H3N2, and B/Harbin 7/94, and RSV controls were RSV type A Long and RSV type B VS1039. Amplicons were analyzed by electrophoresis on a 2.25% NuSieve agarose gel stained with ethidium bromide. Lane M, molecular size marker. Numbers on the left are in base pairs.
Sensitivity.
The sensitivity of detection of influenza virus and RSV with the nested primer sets used individually and in a multiplex reaction was determined. Serial 10-fold dilution series of freshly harvested tissue culture fluid (RSV or influenza virus) or egg-grown material (influenza virus only) were prepared in viral transport medium. From 100 μl of each dilution, nucleic acid was immediately extracted for cDNA synthesis. An equivalent volume of each dilution was taken for infectivity assays for either RSV or influenza virus, which were set up on the same day. cDNA synthesis was followed by PCR with primer sets used individually and in a multiplex reaction. Thus, the endpoint of detection of infectious virus could be directly compared with the endpoint of detection of viral RNA by multiplex RT-PCR. Because only 50% of the cDNA obtained from each extraction was amplified in each PCR, the PCR endpoint was described as a PCR 50% dose (PCR D50) endpoint. In practice, for both influenza virus and RSV, RT-PCR detected viral nucleic acid at 1 to 2 10-fold dilutions below the last dilution at which infectious virus particles could be identified in the presence or absence of all five primer sets. Therefore, multiplex RT-PCR was capable of reliably detecting 1 or fewer PFU of influenza A virus (H1N1 or H3N2) or influenza B virus and RSV types A and B. In all cases, the endpoint of multiplex RT-PCR detection was unaltered by the presence of all the primer sets in a multiplex reaction (Fig. 3).
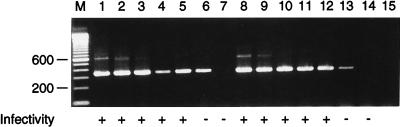
FIG. 3.
Serial 10-fold dilutions of tissue culture-grown RSV type A (Long) were prepared in virus transport medium (starting dilution, 105 PFU/ml [lane 1]). Detection of RSV PFU in 100 μl of each dilution by the infectivity assay is indicated by the plus signs. Material equivalent to 75 μl of each dilution was subjected to either multiplex RT-PCR (with all the primer sets [lanes 1 to 7]) or uniplex RT-PCR (containing only the RSV type A primer sets [lanes 8 to 14]). Lane 15, water control; lane M, molecular size marker. Numbers on the left are in base pairs.
Reagent preparation.
Preparation of individual reaction mixtures containing five primer sets for the primary and secondary PCRs for a large number of tests was found to be very time-consuming and prone to error in terms of the omission of reagents. To overcome this problem and to facilitate the performance of multiplex RT-PCR testing in diagnostic settings, we investigated the preparation and storage of reagent master mixtures. Solutions containing all components of the primary or secondary reaction mixtures, excluding Taq polymerase, were aliquoted in amounts sufficient to complete 15 PCRs and were stored in sterile screw-top Sarstedt tubes at −20°C. The sensitivity of detection of the largest PCR product, derived from the influenza A virus (H1N1) virus template (944 bp), was compared by using reagent mixtures which had been stored at −20°C for between 1 and 6 months and a freshly prepared reaction mixture. The frozen reaction mixtures were defrosted either at room temperature or in a 37°C heating block prior to the addition of Taq polymerase, and the reaction was completed as usual. No significant difference in the sensitivity of detection of the H1N1 reaction product could be seen, either qualitatively in the last dilution at which H1N1 could be detected or quantitatively in the amount of product produced in the PCR (assessed by densitometry) (Fig. 4). Low-copy-number controls for influenza A (H3N2) virus and influenza B virus as well as RSV types A and B were also detected as reliably by the frozen reagent mixture as by the freshly made reagents.
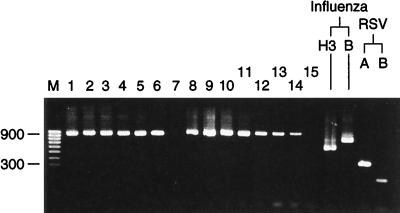
FIG. 4.
Comparison of freshly prepared and stored reagents. Multiplex RT-PCR (with all the primer sets) was performed with a 10-fold dilution series of A/Taiwan/1/86 (H1N1) starting at 10−1 with both freshly prepared amplification mixtures (lanes 1 to 7) and amplification mixtures frozen for 6 months at −20°C (lanes 8 to 14). Results for influenza virus controls (A/Thessaloniki/1/95 H3N2; B/Harbin/7/94 B) and RSV controls (RSV type A Long, RSV type B VS1039) are shown. Lane 15, water control; Lane M, molecular size marker. Numbers on the left are in base pairs.
Dual infections.
The ability of the multiplex reaction mixture to detect the presence of more than one viral template in the same starting material was assessed by the preparation of nose and throat swab material spiked with various combinations of viral templates. The multiplex reaction was capable of detecting all five templates simultaneously, as well as various combinations of templates both in spiked material and in clinical specimens (Fig. 5). This indicated that coinfections could be detected by multiplex RT-PCR.
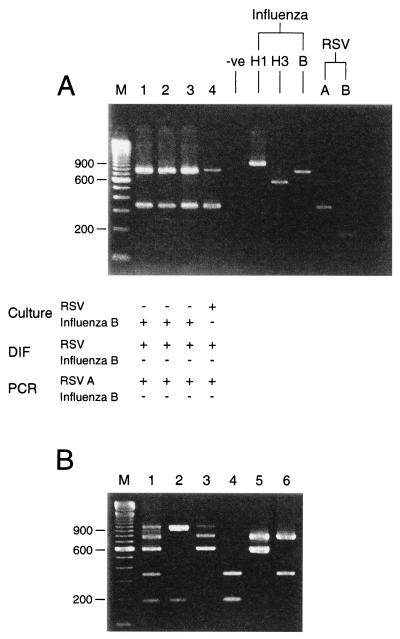
FIG. 5.
Simultaneous detection of multiple templates. (A) Lanes 1 to 4, sequential clinical specimens from an immunosuppressed child. Corresponding culture and direct immunofluorescence (DIF) results are shown. Sample types were bronchoalveolar lavages (lane 1), endotracheal aspirates (lane 2), and nasopharyngeal aspirates (lanes 3 and 4). Specimens in all four lanes were positive for RSV by direct immunofluorescence, the samples in lanes 1 to 3 were negative for RSV by culture, the sample in lane 4 was positive for RSV by culture, the samples in all four lanes were positive for RSV type A by PCR, the samples in all four lanes were negative for influenza virus by direct immunofluorescence, the samples in lanes 1 to 3 were positive for influenza virus by culture, the sample in lane 4 was negative for influenza virus by culture, and the samples in all four lanes were positive for influenza B virus by PCR. (B) Multiplex RT-PCR (with all primer sets) was performed with combined nose and throat swabs spiked with various combinations of influenza and RSV templates. Lanes 1 to 6 were positive (+) or negative (−) for influenza A virus H1N1, influenza A virus H3N2, influenza B virus, RSV type A, and RSV type B, as follows: lane 1, +, +, +, +, and +, respectively; lane 2, +, −, −, −, and +, respectively; lane 3, +, +, +, −, and −, respectively; lane 4, −, −, −, +, and +, respectively; lane 5, −, +, +, −, and −, respectively; lane 6, −, +, −, +, and −, respectively. (A and B) Lanes M, molecular size markers. Numbers on the left are in base pairs.
DISCUSSION
Multiplex PCRs capable of detecting 13 or more separate regions of chromosomal DNA and of detecting and typing several bacterial pathogens have been described previously (5, 20). However, the use of multiplex RT-PCR for the detection of multiple pathogens with RNA genomes has been much more limited, possibly due to the difficulties of overcoming the inherent inefficiency of the RT step in the RT-PCR or of nucleic acid extraction when the starting material is of poor quality.
We have used a multiplex RT-PCR strategy based on the detection of the HA1 portion of the hemagglutinin gene of influenza A and B viruses and on the NP-P region of the RSV genome. Both of these regions of the respective genomes contain genetic information important in allowing the antigenic subtyping of the viruses, and thus, the PCR products of the reactions can potentially be used to provide additional information about strain variation, either by sequencing or by PCR restriction analysis, as has been demonstrated for influenza virus (9).
Primer design.
The extraction method chosen uses guanidinium thiocyanate and silica binding. This has been shown to be both sensitive for clinical samples containing RNA viruses and very effective at removing the inhibitors to PCR which may be contained in the starting material (14). The primer design strategy for the detection of RSV type A or B was slightly different from that for the detection of influenza virus in that the primary reaction product from amplification of the RSV genomic template does not discriminate between RSV types A and B but the amplified products of the secondary PCR do. This is in contrast to the amplification of the influenza virus template, in which discrimination between templates is achieved with the primary reaction products.
The RSV primers were designed to match as closely as possible the primers which had already been designed and optimized for a multiplex RT-PCR detection of influenza virus (8) with respect to G+C content, length, and melting temperature. Constraints on the detection of PCR products from amplification of RSV included the need to produce amplicons which could be distinguished on the basis of size from those already designed for influenza virus (Table 1; Fig. 1) and the relatively small portions of the RSV genome in which subtype information resides (NP-P or F genes). We wished to design a PCR which was capable of distinguishing RSV type A from RSV type B so that the PCR could be used in epidemiological studies of respiratory infection to test the hypothesis that the severity of community-acquired disease due to RSV might correlate with strain type (25).
Several different sets of RSV primers were evaluated both theoretically and practically. This included primers located in the F gene, which proved to be unsuitable because of either theoretical excess primer base pairing, the production of misprimed products in a multiplex reaction, or the failure to amplify template in a multiplex reaction mixture (data not shown). The RSV primer sets used in this work were the only evaluated set which were found to amplify RSV templates in a multiplex reaction mixture containing all primer sets. Initial evaluation of the sensitivity of detection of equivalent concentrations of RSV type A and B templates indicated that the level of amplification of the RSV type B template was substantially reduced in the presence of all primer sets compared to that of the RSV type A template and compared to that in the presence of the RSV type B primers alone, although the reasons for this could not be deduced. Biochemical optimization of both the primary and the secondary PCR mixtures included alterations of the MgCl2 concentration and the salt and buffer formulations but not the buffer pH. Increasing the concentration of Taq polymerase in the secondary reaction mixture produced a substantial increase in sensitivity for RSV type B, when the multiplex reaction mixture was used, to a sensitivity equivalent to that for RSV type A detection (data not shown), and these conditions were subsequently used.
Comparison of the types and subtypes obtained by PCR of RSV isolates collected during the winter of 1995 and 1996 gave 100% correlation with the types of the same isolates obtained serologically with monoclonal antibodies specific for the F region of the RSV genome, indicating that typing of RSV by PCR is feasible that the types and correlate with the types obtained in more classical assays.
We have compared the PCR endpoints in PCR D50 with those of infectivity testing (in PFU per milliliter) for both influenza virus and RSV). We are satisfied that the PCR procedures used were very sensitive and were much more sensitive than the infectivity tests (Fig. 3). In the case of influenza virus, formal titration for determination of the sensitivity of detection of purified influenza virus RNA indicated that PCR had the ability to detect reliably 40 or fewer genome equivalents of influenza virus (8). We have demonstrated that PCR for the detection of freshly grown, infectious RSV was at least as sensitive as PCR for the detection of infectious influenza virus in a multiplex reaction mixture. We would expect that the sensitivity of detection of a purified RSV RNA template would be approximately equivalent to this, although the virus particle-to-infectivity ratios may differ slightly between orthomyxoviridae and paramyxoviridae, and formal particle counts were not determined in this study.
Coinfection.
The frequency of coinfection with RSV and influenza virus or coinfection with different subtypes of influenza virus or different subtypes of RSV is not well documented in the literature and is essentially unknown, although it is likely to be low, on the order of 3 to 4% of total infections with either pathogen (10, 19). However, for patients with viral respiratory infections in whom several pathogens have been tested for and in whom coinfection has been detected, one of the pathogens present is often RSV (26). It is of interest that the multiplex RT-PCR developed in the present investigation, which was designed to amplify a single pathogen from a clinical sample, is capable of amplifying more than one pathogen in the event of coinfection. The multiplex RT-PCR was clearly capable of detecting the presence of at least two pathogens simultaneously when they were present at both high and low copy numbers, by using all possible combinations of RSV and influenza virus templates, in both spiked, simulated clinical specimens and genuine clinical specimens in which the presence of two viruses had been demonstrated by culture of a specimen from an immunocompromised child with a persistent infection (Fig. 5). Successful amplification of all five templates in the same reaction mixture with a spiked simulated clinical sample required additional Taq polymerase in the secondary reaction. There did not appear to be a selective loss of a particular amplicon when the Taq concentration was limiting because in different reaction mixtures containing different copy numbers of various templates, different separate amplicons were not synthesized. Failure to amplify a product also did not appear to relate to product size. When the concentration of Taq was doubled for the secondary PCR, all five amplicons could be clearly visualized. This suggests that the catalytic action of Taq is a rate-limiting factor in achieving simultaneous amplification of multiple templates in the same reaction mixture. Work is in progress to investigate this observation and compare the catalytic properties of a variety of different polymerases for their contribution to improvement in product yield in simultaneous template amplifications.
Preparation of the reagent mixture for multiplex RT-PCR containing five nested primer sets is laborious, and in order to investigate whether this part of the procedure could be streamlined, we attempted to evaluate the preparation of master mixtures in which reagent combinations containing all the necessary primers, buffers, and deoxynucleoside triphosphates could be stored prior to use. Performance of the multiplex PCR would therefore require only the preparation of nucleic acid template, the addition of the mixture to a preprepared tube, and the addition of Taq polymerase. It was clear that storage of preprepared reaction mixtures did not alter the sensitivity of detection of virus, even when the mixtures were stored for up to 6 months (Fig. 4), nor did it increase the level of production of nonspecific products. Thus, prior preparation of reagents is feasible and can increase the throughput of specimens for multiplex RT-PCR.
We have demonstrated that multiplex RT-PCR can be used for the detection and subtyping of RSV and influenza A and B viruses in clinical respiratory samples. The assay described here is both highly sensitive and specific for each individual pathogen and is capable of detecting coinfections in both clinical samples and simulated specimens. It should prove to be useful in studies of viral respiratory illness in both surveillance and diagnostic settings.
REFERENCES
- 1.Anderson L J, Hierholzer J C, Tsou C, Hendry R M, Fernie B F, Stone Y, McIntosh K. Antigenic characterisation of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis. 1985;163:626–633. doi: 10.1093/infdis/151.4.626. [DOI] [PubMed] [Google Scholar]
- 2.Axton R A, Brock D J. A single tube multiplex system for the simultaneous detection of 10 common cystic fibrosis mutations. Hum Mutat. 1995;5:260–262. doi: 10.1002/humu.1380050311. [DOI] [PubMed] [Google Scholar]
- 3.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillion P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraverty P. Antigenic relationships between influenza B viruses. Bull W H O. 1971;45:755–766. [PMC free article] [PubMed] [Google Scholar]
- 5.Chamberlain J S, Gibbs R A, Rainer J E, Nguyen P N, Caskey C T. Deletion screening of the muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988;16:11141–11156. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connor E, Top F, Kramer A the PREVENT Study Group. Reduction of respiratory syncytial virus hospitalisation among premature infants and infants with bronchopulmonary dysplasia using respiratory syncytial virus immune globulin prophylaxis. Pediatrics. 1997;99:93–99. doi: 10.1542/peds.99.1.93. [DOI] [PubMed] [Google Scholar]
- 7.Dieffenbach C W, Lowe T M J, Dveksler G S. General concepts for primer design. PCR Methods Appl. 1993;3:S30–S37. doi: 10.1101/gr.3.3.s30. [DOI] [PubMed] [Google Scholar]
- 8.Ellis J S, Fleming D M, Zambon M C. Multiplex reverse transcription-PCR for surveillance of Influenza A and B viruses in England and Wales in 1995 and 1996. J Clin Microbiol. 1997;35:2076–2082. doi: 10.1128/jcm.35.8.2076-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis J S, Sadler C J, Laidler P, Rebelo de Andrade H, Zambon M C. Analysis of influenza A H3N2 strains isolated in England during 1995-1996 using polymerase chain reaction restriction. J Med Virol. 1997;51:234–241. [PubMed] [Google Scholar]
- 10.Falsey A R, Cunningham C K, Barker W H, Kouides R W, Yuen J B, Menegus M, Weiner L B, Bonville C A, Betts R F. Respiratory syncytial virus and Influenza A infections in the hospitalised elderly. J Infect Dis. 1995;172:389–394. doi: 10.1093/infdis/172.2.389. [DOI] [PubMed] [Google Scholar]
- 11.Fleming D M, Cross K W. Respiratory syncytial virus or influenza? Lancet. 1993;342:1507–1510. doi: 10.1016/s0140-6736(05)80082-0. [DOI] [PubMed] [Google Scholar]
- 12.Fleming D M, Chakraverty P, Sadler C J, Litton P. Combined clinical and virological surveillance of influenza in winters of 1992 and 1993. Br Med J. 1995;311:290–291. doi: 10.1136/bmj.311.7000.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson K M, Mori J, Clewley J P. Detection of HIV-1 in serum, using reverse transcription and the polymerase chain reaction (RT-PCR) J Virol Methods. 1993;43:101–109. doi: 10.1016/0166-0934(93)90093-7. [DOI] [PubMed] [Google Scholar]
- 14.Hale A D, Green J, Brown D W G. Comparison of four RNA extraction methods for the detection of small round structured viruses in faecal specimens. J Virol Methods. 1996;57:195–201. doi: 10.1016/0166-0934(95)01966-9. [DOI] [PubMed] [Google Scholar]
- 15.Hutchinson E J, Joseph C A, Zambon M, Fleming D M, Watson J M. Influenza surveillance in England and Wales: October 1995 to June 1996. Commun Dis Rep Rev. 1996;6:R163–R169. [PubMed] [Google Scholar]
- 16.Jin L, Richards A, Brown D W. Development of a dual target-PCR for detection and characterisation of measles virus in clinical specimens. Mol Cell Probes. 1996;10:191–200. doi: 10.1006/mcpr.1996.0027. [DOI] [PubMed] [Google Scholar]
- 17.Johnson P R, Collins P L. The 1B (NS2), 1C (NS1) and N proteins of human respiratory syncytial virus (RSV) of antigenic subgroups A and B: sequence conservation and divergence within RSV genomic RNA. J Gen Virol. 1989;70:1539–1547. doi: 10.1099/0022-1317-70-6-1539. [DOI] [PubMed] [Google Scholar]
- 18.Lazarus P, Caruana S. Typing of common human papilloma virus strains by multiplex PCR. Anal Biochem. 1996;243:198–201. doi: 10.1006/abio.1996.0506. [DOI] [PubMed] [Google Scholar]
- 19.Lina B, Valette M, Foray S, Luciani J, Stagnara J, See D M, Aymard M. Surveillance of community-acquired viral infections due to respiratory viruses in Rhone-Alpes (France) during winter 1995 to 1995. J Clin Microbiol. 1996;34:3007–3011. doi: 10.1128/jcm.34.12.3007-3011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahony J B, Luinstra K E, Tyndall M, Sellors J W, Krepel J, Chernesky M. Multiplex PCR for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in genitourinary specimens. J Clin Microbiol. 1995;33:3049–3053. doi: 10.1128/jcm.33.11.3049-3053.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCormick A, Fleming D, Charlton J. Morbidity statistics from General Practice: fourth national study 1991–1992. Series MB5, no. 3: London, United Kingdom: Her Majesty’s Stationery Office; 1995. [Google Scholar]
- 22.Mufson M A, Örvell C, Rafnar B, Norby E. Two distinct subtypes of human respiratory syncytial virus. J Gen Virol. 1985;66:2111–2124. doi: 10.1099/0022-1317-66-10-2111. [DOI] [PubMed] [Google Scholar]
- 23.Read S J, Jeffery K J M, Bangham C R M. Aseptic meningitis and encephalitis: the role of PCR in the diagnostic laboratory. J Clin Microbiol. 1997;35:691–696. doi: 10.1128/jcm.35.3.691-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tobita K, Siguira A, Enomoto C, Furuyama M. Plaque assay and primary isolation of influenza A viruses in an established line of canine kidney cells (MDCK) in the presence of trypsin. Med Microbiol Immunol. 1975;162:9–14. doi: 10.1007/BF02123572. [DOI] [PubMed] [Google Scholar]
- 25.Walsh E E, McConnochie K M, Long C E, Hall C B. Severity of respiratory syncytial virus infection is related to virus strain. J Infect Dis. 1997;175:814–820. doi: 10.1086/513976. [DOI] [PubMed] [Google Scholar]
- 26.Yun B-Y, Kim M-R, Park J Y, Choi E H, Lee H J, Yun C K. Viral etiology and epidemiology of acute lower respiratory tract infections in Korean children. Pediatr Infect Dis. 1995;14:1054–1059. doi: 10.1097/00006454-199512000-00005. [DOI] [PubMed] [Google Scholar]