Abstract
Background
Food allergy (FA) and atopic dermatitis (AD) are common conditions that often present in the first year of life. Identification of underlying mechanisms and environmental determinants of FA and AD is essential to develop and implement effective prevention and treatment strategies.
Objectives
We sought to describe the design of the Systems Biology of Early Atopy (SunBEAm) birth cohort.
Methods
Funded by the National Institute of Allergy and Infectious Diseases (NIAID) and administered through the Consortium for Food Allergy Research (CoFAR), SunBEAm is a US population-based, multicenter birth cohort that enrolls pregnant mothers, fathers, and their newborns and follows them to 3 years. Questionnaire and biosampling strategies were developed to apply a systems biology approach to identify environmental, immunologic, and multiomic determinants of AD, FA, and other allergic outcomes.
Results
Enrollment is currently underway. On the basis of an estimated FA prevalence of 6%, the enrollment goal is 2500 infants. AD is defined on the basis of questionnaire and assessment, and FA is defined by an algorithm combining history and testing. Although any FA will be recorded, we focus on the diagnosis of egg, milk, and peanut at 5 months, adding wheat, soy, cashew, hazelnut, walnut, codfish, shrimp, and sesame starting at 12 months. Sampling includes blood, hair, stool, dust, water, tape strips, skin swabs, nasal secretions, nasal swabs, saliva, urine, functional aspects of the skin, and maternal breast milk and vaginal swabs.
Conclusions
The SunBEAm birth cohort will provide a rich repository of data and specimens to interrogate mechanisms and determinants of early allergic outcomes, with an emphasis on FA, AD, and systems biology.
Key words: Food allergy, atopic dermatitis, eczema, birth cohort, systems biology, multiomics, omics
Food allergy (FA), which affects approximately 5% to 10% of young children, with the highest incidence in the first year of life, accounts for almost $25 billion per year in US costs.1, 2, 3, 4, 5 Atopic dermatitis (AD, also known as eczema) is a major risk factor for FA and other allergic diseases; in and of itself, it has major effects on the quality of life of children and families.6 AD affects approximately 13% of US children, of whom approximately one third have moderate to severe disease.6,7 Early life AD is strongly associated with subsequent FA (relative risk, ∼5-10) and asthma (relative risk, ∼3).8,9 The relationship between AD and later allergy may be causal; early life skin barrier dysfunction and inflammation may facilitate sensitization to allergens through the skin, leading to chronic systemic allergic inflammation.10
While early introduction of allergens into the diet of those with AD reduces the risk of FA, many children are already allergic at the time of introduction, and there are still no effective methods to prevent AD itself.11, 12, 13, 14, 15, 16, 17 Several potential methods of prenatal and/or early life intervention to prevent eczema and/or FA have been proposed, including introduction of allergenic foods in the neonatal period, use of emollients to enhance skin barrier (NCT01142999, NCT03376243, NCT03742414), attempts to modify the microbiome of mother, and/or infant, and various dietary interventions (NCT02286999, NCT00798226, NCT03567707). However, efforts to test these preventative methods are limited by the lack of accurate predictors to identify high-risk individuals for study. Most commonly, studies have used the presence of at least 1 family member with allergic disease to define a high-risk population, but family history has proved to be a poor predictor of FA and AD. For example, in a recent Australian study, such a definition only conferred a modest increase in risk (odds ratio, 1.4) compared to no family history.18,19 Various biomarkers that might identify high-risk newborns have been proposed, including markers of skin barrier function, in utero sensitization, and in utero immune function, but so far, there are no biomarkers with sufficient sensitivity or specificity for clinical application.20, 21, 22, 23, 24, 25, 26, 27, 28 In the absence of ways to focus studies on those at highest risk for FA, testing new interventions is inefficient, requiring large populations (eg, 2000 subjects or more) to have sufficient power to identify preventative effects. Finding biomarkers and risk factors that reliably identify a high-risk population for intervention would facilitate study of these and other approaches.
Allergic conditions are complex diseases, and parsing the biological heterogeneity hidden under their clinical umbrellas is paramount to improving prevention, diagnosis, and clinical management.29,30 While substantial research has contributed to our understanding of biological alterations underlying FA and AD, omics has the potential to advance this understanding by comprehensively capturing molecular dimensions that contribute to allergy development and disease course, thereby expanding the discovery field. Increasingly accessible high-throughput omic technologies enable systemwide characterization of biological systems at multiple levels, including the capacity to profile whole genomes, transcriptomes, proteomes, metabolomes, and microbiomes.29, 30, 31, 32 Systems approaches have been applied to tackle many challenges to date, permitting elucidation of elucidating informative details about signaling networks in breast cancer, cellular responses to cancer therapy, and drug sensitivity, toxicity, and synergy based on molecular omic backgrounds, for example.33 A systems biology approach where the biology of allergic conditions is investigated comprehensively at several levels longitudinally over time is a promising approach to identify potentially modifiable pathways of allergy development.29,30
Effective systems biology is enabled by robust multiscale data from large, well-phenotyped cohorts and integrative analytic strategies. The comprehensive modeling of molecular interactions within and across -omes that drive the behavior of complex biological systems requires rationally designed and acquired multiomic data from a well-phenotyped sample as input, plus the application of mathematical and computational tools tailored to the study of complex system architecture and behavior.29 There has been limited work done thus far on the systems biology of allergy.29
Systems Biology of Early Atopy (SunBEAm) is a general population birth cohort funded by the National Institute of Allergy and Infectious Diseases (NIAID) and spearheaded by the Consortium for Food Allergy Research, known as CoFAR, with additional support from the Atopic Dermatitis Research Network and the Immune Tolerance Network to transform approaches to studying allergy by recruiting and phenotyping a large, well-characterized multicenter cohort where systems biology can be applied to identify mechanisms and biomarkers underlying the development of FA, AD, and other allergic diseases. SunBEAm will study the role and interrelationships of genetic, clinical, biological, and environmental early life factors in the development of allergic diseases, with an emphasis on FA, AD, and their endotypes. Pregnant women, children born to them, and the children’s biological fathers are being enrolled and phenotyped with extensive longitudinal biosample collections. SunBEAm is collecting, processing, assaying, and storing environmental and biological samples for current and future use in the study of allergic disease development. Here we provide an overview of the study design and methods of SunBEAm.
Methods
Overview
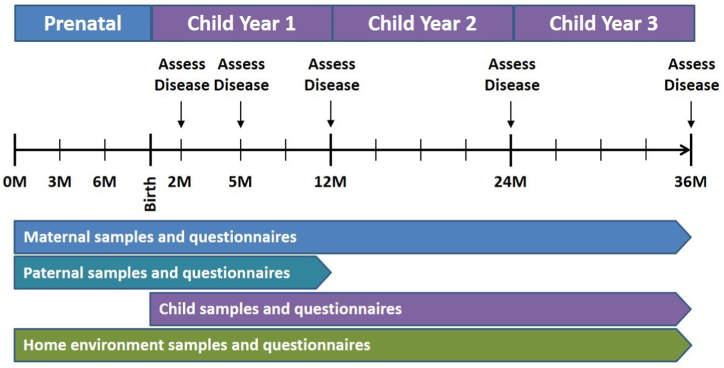
This is a prospective cohort study in which pregnant women (at any stage of pregnancy), the offspring’s biological father, and the offspring are being enrolled at 12 study sites around the United States (listed in Table E1 in this article’s Online Repository at www.jaci-global.org), and the offspring are observed from birth to age 3 years. During the study, biological and environmental samples and questionnaire information are collected from the parents and the children, and the children are assessed for allergic diseases at clinic visits at ages 2, 5, 12, 24, and 36 months. Fig 1 provides a study overview and Table I the study’s objectives.
Fig 1.
Study overview.
Table I.
Study objectives
|
|
|
Inclusion and exclusion criteria
Inclusion and exclusion criteria are defined in Table II. Briefly, women are enrolled at any stage in pregnancy without regard to personal or family history of allergic disease. The rationale is to identify risk factors in the population as a whole for the development of allergic disease, so selection of pregnant women whose children are at an elevated risk of allergic disease would prevent the study of the risk factors used to select the women and would prevent the assessment of the risk factors of children with FA and AD who do not come from backgrounds that are recognized as high risk. The biological father of their child is also enrolled, although this enrollment is optional. Inclusion and exclusion criteria are otherwise designed to include individuals likely to complete the protocol and to protect particularly vulnerable populations. Because a goal of the study is to understand the genetic transmission of FA and AD, pregnancies as a result of egg donation are excluded.
Table II.
Inclusion and exclusion criteria
| Criterion | Pregnant women | Biological fathers | Child |
|---|---|---|---|
| Inclusion |
|
|
— |
| Exclusion |
|
|
|
At screening for enrollment of either the mother or the child, if the biological mother intends to give the infant up for adoption, neither the mother nor the child should be enrolled; however, if the biological mother gives up legal guardianship of the child after the child is enrolled and the legal guardian wants the child to remain in the study, the child may remain enrolled as long as a legal guardian agrees to meet the child’s study requirements and provides written informed consent for the child’s continued participation.
Study end points
Primary clinical end points are detailed below. Secondary and exploratory end points are listed in Table III and are described below.
Table III.
Secondary and exploratory clinical end points
| Secondary clinical end points |
|
|
|
|
| Exploratory clinical end points |
|
|
|
More details are available in the Supplementary Appendix.
Food allergy
IgE-mediated, immediate-type allergy to protocol-specified foods will be assessed at each clinic visit starting at the child’s 5-month visit. The foods, which are age specific, are milk, egg, and peanut at all visits starting at the 5-month visit; and wheat, soy, tree nuts (cashew, hazel, walnut), fish (cod), shellfish (shrimp), and sesame at the 12-, 24-, and 36-month visits.
The timing of the first FA assessment at 5 months is intended to be at around the time that early introduction of peanut and other foods would be accomplished; the intention is to not delay introduction for children for whom screening would be recommended by guidelines or desired by parents. Clinical assessments and diagnostic criteria for FA are described in the FA algorithm described in the Online Repository’s Methods and Figs E1 and E2 (available at www.jaci-global.org). Briefly, determination of IgE-mediated FA status is made by a combination of exposure to foods and symptoms of FA as collected periodically by questionnaire clinical history, skin prick testing (SPT) if tolerance has not been established by clinical history, and oral challenge where necessary if the combination of clinical history and SPT are not diagnostic. If SPT cannot be completed, allergen-specific IgE is substituted in the algorithm. Ingestion and tolerance of baked forms of milk and egg are not evaluated by the algorithm.
Information on the resolution of FA is assessed periodically by questionnaire and at the 12-, 24-, and 36-month visits by the same FA algorithm used for the diagnosis of FA; however, special rules for the application of the FA algorithm to FA resolution apply (see the Methods section in the Online Repository).
Information on IgE-mediated, immediate-type allergy to foods not on the protocol-specified list is collected periodically by questionnaire starting in early infancy and assessed at each clinic visit starting at age 5 months; however, the diagnosis of allergy to these additional foods is not determined by the FA algorithm or by study-conducted oral food challenges. Allergy to a non–protocol-specified food is not included in the primary end point for FA.
Atopic dermatitis
AD is defined as the following since the last assessment (or since birth for the 2- and 5-month visits): a history of a dry or itchy rash that is (1) either continuous or intermittent lasting at least 4 weeks; or (2) requiring medicated treatment and rash was or is present in the skin creases (folds of elbows, behind the knees, fronts of ankles, or around the neck) or on the extensor aspects of the forearms or lower legs or on cheeks or trunk. Any infant fulfilling these criteria but who, on examination by a suitably trained health professional, is deemed to have a different skin disease that explains the above findings will not be classified as having AD.
In addition to the above definition, information relevant to AD classification includes collection of Scoring Atopic Dermatitis (aka SCORAD) and Eczema Area and Severity Index (aka EASI) at each clinic visit (Table IV).
Table IV.
Biosample collection
| Sample type | Maternal samples | Paternal samples | Child samples | Collection and processing details | Storage condition |
|---|---|---|---|---|---|
| Venous blood | At enrollment and 2 months | At enrollment | At 2 months,∗ 5 months, 12 months, 24 months, and 36 months | Collected in lavender-top tubes with EDTA. Plasma for serology, metabolomics, and proteomics, fixed whole blood cells for cytometry, and whole blood cells for RNA and DNA extraction (subject to blood volume collected and time from collection to processing) are stored frozen at −80°C after each collection and batch shipped frozen to biorepositories. PBMCs are processed and cryopreserved by the biorepository from whole blood shipped overnight at RT. | −80°C |
| Cord blood | At birth | Collected by needle from vein in cord, transferred into EDTA tubes, and transported to processing laboratory at RT. If processed within 6 hours of collection, plasma for serology, metabolomics, and proteomics, fixed whole blood cells for cytometry, and whole blood for RNA and DNA extraction (subject to blood volume collected) are stored frozen at −80°C and batch shipped to the biorepositories. PBMCs are processed and cryopreserved by the biorepository from whole blood shipped overnight at RT. For blood processed more than 6 hours after collection, plasma for serology and whole blood for DNA extraction are stored frozen at −80°C and whole blood is shipped at RT to the biorepository for PBMC and plasma collection. | −80°C | ||
| Breast milk | Up to 3 times during child’s first year | Collected by hand expression (preferred) or breast milk pump into a sterile container. May be frozen at home before return to clinic. Batch shipped frozen to the biorepository. | −80°C | ||
| Scalp hair | At enrollment | At each clinic visit | Cut hair is stored at RT and batch shipped at RT to the biorepository. | RT | |
| Home dust | In prenatal period | Between 2 and 5 months, and at 12, 24, and 36 months | Collected using a DUSTREAM collector at the end of a vacuum cleaner hose. A seat cushion in the living room and a 1 m2 floor area in the living room is each vacuumed for 1 minute. The capped dust collector is then mailed to the study site at RT, stored at −80°C, and shipped to the biorepository on dry ice. | −80°C | |
| Home water | Between 2 and 5 months | Hot tap water is collected in provided tube after allowing water to run for 30 seconds. Tube is returned to study site at RT, where the MQuant carbonate hardness test is performed. | Not stored | ||
| Nasal secretion sample | At each clinic visit | Mucosal lining fluid is sampled on strips of filter paper (fibrous hydroxylated polyester sheets) with the single-use Nasosorption FX-I device. A filter pad is inserted in the nostril, placed at the anterior part of the inferior turbinate, and left in place for 20 seconds; the device is then returned to a storage tube. Tubes are stored at −80°C and shipped frozen to the biorepository. | −80°C | ||
| Nasal swab | 1 at enrollment | 1 at enrollment | 1 at each clinic visit | An absorbent swab (HydraFlock sterile swab) is inserted into one nostril, rotated gently against the nasal mucosa, and then withdrawn; the procedure repeated in the other nostril. The swab is inserted into a DNA/RNA Shield collection tube. Tubes are stored at −80°C and batch shipped frozen to the biorepository. | −80°C |
| Saliva | At each clinic visit | Saliva is collected by swabbing inside the child’s cheeks with a collection swab (SalivaBio). Saliva is recovered via swab centrifugation and transferred into cryovials. Vials are stored at −80°C and batch shipped frozen to the biorepository. | −80°C | ||
| Skin tapes | At enrollment and at 2 months | At enrollment | At 1-2 days and at each clinic visit | A strip of sterile medical tape (D-SQAME) is applied to an area of the skin with light pressure, then slowly removed and applied to a cardboard card for storage. For adults, tape strips are collected from an adjacent site on the same anterior forearm as the skin swabs. For children, tape strips are collected from the opposite cheek and anterior forearm as the skin swabs. For children, lesional tape strips may be collected and the primary site adjusted. Cardboard cards are stored at −80°C and batch shipped frozen to a specified laboratory for processing for RNA, proteins, lipids, and metabolites. | −80°C |
| Skin swabs | At enrollment and 2 months | At enrollment | At 1-2 days and every clinic visit. | Skin swabs are collected using Puritan 20MM HydraFlock sterile swabs. Study staff swab an area of skin with a moistened swab near the area to be skin taped as follows:
|
−80°C |
| Stool | 1 sample in prenatal period; 1 sample at 2 months | 1 sample at enrollment | 1 meconium/stool sample in first 2 days of life; 1 sample at 1-2 weeks and at 1, 2, 5, 9, 12, 24, and 36 months | Mother and father collect their own stool. For the child, stool is collected from the diaper or a collection device in the toilet. One scoop of stool is placed into each of 2 Zymo DNA/RNA Shield fecal collection tubes. Tubes are stored frozen at −80°C and batch shipped frozen to the biorepository. | −80°C |
| Urine | At enrollment and at 2 months | At each clinic visit | For the child, urine is collected using a urine collection bag in the diaper or a urine collection device on the toilet. Urine is transferred into prepared cryovials, stored at −80°C, and batch shipped frozen to the biorepository. | −80°C | |
| Vaginal swabs | 1 sample in the prenatal period | Pregnant participants self-collect a vaginal swab and place it into a DNA/RNA Shield collection tube. Tubes are stored at −80°C and batch shipped frozen to the biorepository. | −80°C |
Additional study assessments are described in Table V, with additional information about timing provided in the schedules of events for the mother, father, and child in Tables E4, E5, and E6, respectively. PBMC, Peripheral blood mononuclear cell; RT, room temperature.
A heel stick may be conducted if venipuncture is not successful.
Secondary and exploratory end points
Sensitization to aeroallergens
Sensitization to aeroallergens is assessed by serum IgE and SPT at the 12-, 24-, and 36-month visits. For IgE testing at any given time point, a positive specific test is defined as an IgE concentration at or above the level of detection for the assay. For SPT at any given time point, a positive specific test is defined as a wheal ≥3 mm above the negative control wheal. For either test, different thresholds could be defined for specific analyses, and sensitization could potentially be defined by IgE alone, skin test alone, either, or both.
Recurrent wheeze
Information on wheezing episodes, other respiratory symptoms, and medication receipt is collected periodically by questionnaire starting in early infancy and assessed at each clinic visit for the interval of time since the previous visit (since birth for the 2-month visit) (see Table E6 in the Online Repository at www.jaci-global.org for the child’s schedule of events). Recurrent wheeze, assessed at age 3 years, is defined as at least 2 episodes of wheezing during the first 3 years of life, with at least 1 episode between the ages of 24 and 36 months.
Seasonal and perennial allergic rhinitis and conjunctivitis
Information on nasal and conjunctival signs, symptoms, and medication receipt is collected periodically by questionnaire starting in early infancy and assessed at each clinic visit for the interval of time since the previous visit (since birth for the 2-month visit). At age 36 months, a determination will be made of seasonal or perennial allergic rhinitis and allergic rhinoconjunctivitis according to diagnostic criteria that include clinical data in combination with allergic sensitization.
Non–immediate-type FA and eosinophilic esophagitis
Information on non–immediate-type FA (specifically, food protein–induced allergic proctocolitis and food protein–induced enterocolitis syndrome)34 and eosinophilic esophagitis (EoE) is collected periodically by questionnaire starting in early infancy and assessed at each clinic visit starting at age 5 month. These end points will not be included in the primary end point for FA.
For this study, the diagnosis of food protein–induced allergic proctocolitis is based on parental report of visible specks or streaks of blood mixed with mucous in the stool, clinical improvement in symptoms with dietary exclusion, and lack of systemic symptoms, vomiting, diarrhea, and poor growth,34 and we also collect parental history of physician-confirmed blood in the stool (by visual confirmation or guaiac testing). Food protein–induced enterocolitis syndrome is recorded when the diagnosis fulfills the criteria listed in Table E2 or E3 in the Online Repository at www.jaci-global.org, based on parental report. For this study, evaluations for EoE are not required or undertaken as part of the study, but the diagnosis is recorded if there is a history of a physician diagnosis of EoE and a positive biopsy result obtained during clinical care outside of the study.
Biosample collections and assessments
The schedules of events in Tables E4, E5, and E6 in the Online Repository at www.jaci-global.org detail the assessments and biosample collections for the mother, father, and child, respectively, throughout the study. Salient details of sample collections are given in Table IV and assessments in Table V.35 SunBEAm sites are collecting, processing, and barcoding biosamples according to a centralized manual of procedures. Biosamples are shipped to a central repository, where they are stored according to the conditions specified in Table IV. A laboratory information management system is used to catalog and track samples. Although there will likely be additional uses for the biosamples, we broadly expect that blood will be used to characterize immune cell identity and function, genomics and transcriptomics, sensitization status, and selected environmental exposures; breast milk to characterize microbial and other environmental exposures; hair to characterize environmental exposure; house dust to characterize allergen and other environmental exposures; house water to characterize water hardness; nasal samples to characterize nasal cytokine levels and microbiome; saliva to characterize analytes and microbiome; skin tape strips to measure proteins, lipids, metabolites, and gene expression; skin swabs to measure microbiome; stool to measure microbiome; urine to characterize metabolite levels; and vaginal swabs to characterize microbiome. Omic assays will address discovery-oriented hypotheses and yield data for system biology analyses designed to identify mechanisms and biomarkers of allergy development.
Table V.
Assessments
| Assessment type | Details |
|---|---|
| Height (or length) and weight | Study staff measure the mother’s height and weight at enrollment and at the 2- and 12-month clinic visits; the father’s at enrollment; and the child’s at each clinic visit. If the father does not come for a clinic visit, the information may be collected by questionnaire. |
| Allergen SPT | In children only, study staff conduct allergy SPT to food allergens at the 5-, 12-, 24-, and 36-month visits and to aeroallergens at the 12-, 24-, and 36-month visits.∗ |
| OFC | Starting at age 5 months, allergy to protocol-specified foods is assessed at each clinic visit. Open, graded OFCs are conducted when indicated by the FA algorithm.† Some combinations of food exposure history, symptomology, and SPT wheal size or specific IgE concentration require an OFC to determine allergy status, whereas other combinations do not. Large clinic visit windows are provided to accommodate multiple OFCs for a given child. If an OFC is indicated, before the first OFC, the mother or legal guardian will provide written informed consent. |
| SCORAD and EASI | Study staff conducts a visual assessment of the child’s skin at every clinic visit. AD is scored by SCORAD and EASI at each clinic visit. |
| TEWL | Study staff measure TEWL on the mother’s skin at enrollment and at the child’s 2-month visit; on the father’s skin at enrollment; and on the child’s skin at 1-2 days after birth (or during the first 7 days at a home visit, an unscheduled study visit, or a pediatrician visit if not collected before hospital discharge) and at every clinic visit. TEWL is measured with a GPSkin Pro device. |
| Addendum guidelines counseling | Study staff counsel parents at the 2-month visit on the Addendum Guidelines for the Prevention of Peanut Allergy35 and offer the option of the child receiving a guidelines-based assessment and recommendation at any time between ages 4 and 6 months. The ±1-month window for the 5-month visit provides for a guidelines-based assessment between ages 4 and 6 months. |
Stability studies
To optimize the management of biosamples that are being longitudinally collected and banked for SunBEAm, stability studies were initiated before the start of SunBEAm’s biosample collection. Blood and skin tape biosamples were collected from an independent sample of participants and stored, shipped, and processed under different conditions and over varying durations of storage time to assess the stability of biosamples for anticipated assay modalities within SunBEAm, including immune profiling and selected omic assays. Antibody panels were also developed, and staining conditions were tested to optimize immune profiling protocols.
Study governance
Decisions regarding the use of study data and biosamples will include input from the SunBEAm Steering Group, and final decisions are made in a cooperative manner with NIAID. The SunBEAm Steering Group, comprising the protocol chairs, the SunBEAm site investigators, the SunBEAm Analysis and Bioinformatics Center, Division of Allergy, Immunology and Transplantation/NIAID Statistical and Clinical Coordinating lead staff, a representative from the Immune Tolerance Network, and NIAID project scientists, is organized to review SunBEAm activities and to provide recommendations for study design, data and biosample collection, assays, and analyses. The SunBEAm Analysis and Bioinformatics Center is funded by an independent NIAID cooperative agreement grant and serves as SunBEAm’s center for assays, analyses, data integration, and system biology efforts. Projects seeking to generate data from SunBEAm biosamples will coordinate their activities with the SunBEAm Analysis and Bioinformatics Center.
Analysis plan
Associations between exposures (which includes risk factors and predictors) and primary, secondary, and exploratory clinical end points will be investigated in either the full cohort or the case–cohort population with bivariate and multivariate statistical techniques. Analyses will include, but will not be limited to, standard statistical techniques such as the following:
-
•
Chi-square and t tests for categorical and continuous data, respectively.
-
•
Logistic regression for dichotomous outcomes, such as peanut allergy by age 12 month (yes or no).
-
•
Ordinal or multinomial logistic regression for categorical outcomes, such as AD by age 12 months categorized as severe, moderate, mild, or none.
-
•
Linear regression for continuous outcomes, such as SPT wheal sizes or IgE concentrations.
-
•
Poisson regression for end points that are counts, such as number of food allergies or number of positive SPTs.
-
•
Cox regression for time-to-event analyses, such as diagnosis of shellfish allergy by age 36 months.
-
•
Mixed linear models for multilevel, longitudinal, or correlated data.
-
•
Nested case–cohort design.
Because of the high cost of analyzing biological and environmental samples for the entire cohort, some samples will be collected, processed, and stored for later analysis. Many questions of interest that involve associations between a disease outcome and information derived from samples can be investigated with a nested case–control design in which stored samples are analyzed only for disease cases and selected controls. This design can result in substantial cost savings to the study; however, a significant limitation of the nested case–control design is that it requires the selection of a separate control group for each disease under study, and this study has multiple disease end points. To overcome this limitation, SunBEAm will utilize a case–cohort design that will provide a common control group across multiple case–control analyses and reduce the number of samples that have to be assayed. The design was first described in 1986 by Prentice,36 and since then, it has become a standard design in prospective cohort studies. Twenty percent of study participants will be randomly enrolled onto the subcohort without respect to their disease outcome. For the analysis of a given disease, such as AD, all cases will be identified and selected from the entire birth cohort (inside and outside the subcohort); however, noncases will only be selected from inside the subcohort. Thus, the sample size for the analysis (ie, the case–cohort sample) will be the number of participants in the subcohort (which will consist of cases and noncases) plus all cases that arise outside of the subcohort. For selected analyses, only samples collected from the subcohort and the cases that arise outside of the subcohort have to be assayed. This subcohort can be utilized for the investigation of any number of disease outcomes.
Although it is anticipated that the case–cohort design will be used for most investigations involving stored samples, a nested case–control design may be utilized for a given investigation if the situation warrants. Decisions about which study questions will be investigated with the full cohort, a case–cohort sample, or a nested case–control sample will be made throughout the life of the study and will be dependent on resources available, assay costs, suitability of samples for storage, time to complete assays, and statistical considerations. Various weighting methods have been proposed to make results for the case–cohort sample representative of the full cohort.36, 37, 38, 39 At the time of the analysis, the lead statistician and scientist will make recommendations to the SunBEAm Steering Group on a weighting method. Sensitivity analyses may be conducted to evaluate the effects of the different weighting methods. A variety of statistical, mathematical, and modeling approaches will be used for systems biology analyses, including network models, machine learning, and causal inference.30,33,40,41
Sample size considerations
Sample size of full cohort
The sample size for a prospective cohort design is a function of the desired statistical power and the proportion of participants in the study sample with the exposure (or risk factor or predictor) of interest. Sample size is optimized at a risk factor proportion of 0.50 (ie, 1:1). Sample sizes increase sharply at risk factor proportions of <0.30 and >0.70. The overall risk of disease in the study sample was also considered; less prevalent diseases require a larger sample size. For example, investigations of FA questions will require a larger sample size than investigations of AD. The size of the relative risk was also taken into account; smaller relative risks require larger sample sizes.
The accrual objective is to enroll at least 2500 pregnant women and their offspring. Fig E3 in the Online Repository at www.jaci-global.org shows power curves for 8 disease prevalences at 3 different cohort sizes. The tables embedded within the panels show the minimum exposure prevalences that can be investigated at 80% power. For a sample size of 2500, a prevalence of FA of 6% (the clinical end point with the lowest risk), a relative risk of 2.0, and a 20% dropout rate, the study could investigate exposures with a prevalence as low as 9.4% at 80% power. It is important to note that for exposures that are rarer than 9.4%, the study will not have sufficient power to find statistically significant associations unless the relative risks are greater than 2.0 or the sample size is increased.
Sample size of subcohorts
As above, the subcohort is used for case–cohort analyses when the cost of analyzing all subjects is prohibitive. Fig E4 in the Online Repository at www.jaci-global.org shows power curves for 8 disease prevalences at 3 subcohort sizes. The tables embedded within the panels show the minimum risk factor proportion that can be investigated at 80% power. Sample size selection for the subcohort is a compromise between a smaller sample size that minimizes the number of biological samples that have to be assayed in a case–cohort analysis and a larger sample size that allows for the investigation of rare exposures among rare diseases. For this study, a subcohort sample size of 500 was selected because it allows for investigations of exposures with the smallest proportions among the 3 sample size examples while staying within budgetary constraints for conducting sample assays. A subcohort size larger than 500 (not shown) does not provide sufficient gains in power to offset the additional assays that would be required. We note that analyses using the full cohort of 2500 will have greater statistical power than analyses using the case–cohort design. For example, for the full cohort of 2500 and a disease prevalence of 6%, the minimum risk factor proportion that can be investigated at a relative risk of 2.0 and 80% power is 0.094, compared to 0.158 for a case–cohort analysis based on a subcohort of 500.
Summary
The SunBEAm birth cohort is a large-scale effort to understand the mechanisms of early life AD and FA and to identify risk factors that can be used with the ultimate goal of preventing these diseases. The comprehensive clinical phenotyping combined with the collection of multiple simultaneous biosamples allows for a systems biology approach to these diseases. The collection and storage of these biosamples for future use will provide an invaluable resource to the scientific community to explore the contribution of environmental factors and biological processes to the development of these diseases.
Disclosure statement
Funded by the following grants from NIAID/National Institutes of Health (NIH): UM2AI130836, UM1AI130838, UM1AI130839, UM1AI130781, UM1AI130936, UM1AI130936, UM1AI130780, UM1AI130934, UM1AI130570, UM1AI173380, UM1AI151958, and UM1AI109565.
The authorships of P.C.F., A.T., W.D., S.H., and S.P. do not constitute endorsement by the National Institute of Allergy and Infection Diseases, the National Institutes of Health, or any other agency of the United States government.
Disclosure of potential conflict of interest: C. Keet reports royalties from UpToDate. S. H. Sicherer reports royalty payments from UpToDate and Johns Hopkins University Press; grants to his institution from NIAID, Food Allergy Research and Education, and Pfizer; personal fees from the American Academy of Allergy, Asthma & Immunology; and deputy editorship of the Journal of Allergy and Clinical Immunology: In Practice, outside of the submitted work. C. M. Davis reports grant support from NIAID/NIH, Immune Tolerance Network, DBV Technologies, Regeneronm Pfizer, and Allergenis; and consultant for Aimmune. S. Anvari reports research support from NIAID/NIH and DBV Technologies; and serves on the advisory board for Sanofi. M. Groetch receives royalties from UpToDate, FARE, and the Academy of Nutrition and Dietetics; serves on the medical advisory board of IFPIES, as senior advisor to FARE, and as health sciences advisor for APFED; and sits on the editorial board of Journal of Food Allergy. W. Shreffler reports royalty payments from UpToDate; grants to his institution from NIAID and Food Allergy Research and Education; sponsored research funds from Aimune, Angany Therapeutics, Moderna, Regeneron, Vedanta Biosciences and consultant fees from Aimmune, ALK, Allergy Therapeutics, Novartis, Regeneron, and Sanofi. Dr. Leung reports support from Sanofi, Genentech, Leo Pharma, and Incyte. H. Kim reports advisory board for ALK, Kenota Health, and Ukko Inc; consultant for Allergy Therapeutics Ltd, Belhaven Biopharma, Duke Clinical Research Institute, Genentech, and Nutricia. J. Wang receives research support from NIAID, Aimmune, DBV Technologies, and Regeneron; and consultancy fees from ALK Abello and Jubilant HollisterStier. R. A. Wood reports research support from NIAID, Aimmune, Astellas, DBV, FARE, Genentech, HAL-Allergy, Novartis, and Regeneron; and royalties from UpToDate. The rest of the authors declare that they have no relevant conflicts of interest.
Acknowledgments
We acknowledge the following individuals who made this work possible: Acara Carter; Adam Glawe, MS; Ajay Eapen, MD; Alana Snowden, BS; Alexander Grishin, PhD; Alexis Gemelas, CRC; Alysar Bittar, RN; Amanda Cyrus, BA; Amanda Vega, BA; Amber Ali, BS; Amit Singh; Amy Caulum, RD; Amy Dresen, BS; Amy Eapen, MD; Ana P. Aguirre, BA; Andrea Gasparoto De Medeiros Amarante, RN, MS; Andrew Long, PharmD; Andrew Winslow, MD; Anna Anderson, MD; Anna-Liisa Vockell, MSN, BSN/RN; Asma Bazzy, RN; Berenice Alfaro, RN; Beth A. Mattucci, RN; Bin Su, PhD; Briahnna Austin, MS; Bruce J. Lanser, MD, MPH; Bryanna Oakley, BS; Bryle Barrameda, BA; Carissa Keung, BS; Caroline Bronchick, RN; Cassandra Thomas, RN; Catherine Feight; Cecilia Berin, PhD; Channon Campbell, RN; Chivon McMullen Jackson, RN, MS; Christen M. Hillenbrand, BS; Christine Johnson, PhD; Claire Hillier, BS; Daisy Tran Vita, RN, BSN; Daniel Jackson, MD; Danielle Whiteside, BA; Deanna Hamilton, RN; Deiny DeLaCerda, RN; Edward Zoratti, MD; Elika Eshghi, BS; Elizabeth Wash, RN, MSN, FNP-BC; Emily English, RN, MSN, CPNP; Emily Gallagher, BSN, MPH; Emily Seminara, BS; Eric Schauberger, DO, PhD; Erin Kane, BA; Estrella Guerrero, BS; Fabian Rivera, MBA; Fernanda Ochoa Toro; Galina Grishina, MS; Gerald Nepom, MD, PhD; Gina Crisafi, BS; Gledson Hanelt, RN; Harold Ames, MS; Holly Semble, BSN, RN; Inga Peter, PhD; Irina Burd, MD; Jane Rice, RN ; Janelle Aby; Janine Westra; Jennifer Bagley, BSN; Jennifer Bump, MD, MBA; Jennifer Fishman, RN; Jennifer Martin, CCRP; Jennifer Smith, BS; Jenny Nguyen, BS; Jeri Wolven, RN; Jessica Macdougall, MD; Joanna Grabowska, MS; Jocelyn Chang, RD, MS; Jourdon Robinson, MS; Julia Santarosa, BA; Julie Seung; Jyothi Tirumalasetty, MD; Kaci Elrod, BS; Karen Dorman, RN, MS; Kari Nadeau, MD, PhD; Karla Ramirez, MD; Kathy Pitts, PNP, PhD; Katie Cruz; Katrina Bakhl, BA; Kimberly Kersey, CRC; Kjersti Aagaard, MD; Latha Satish, PhD; Latina Robinson, RD; Laura Susick, PhD; Lauren Herlihy, RN, MSN, CPNP; Lindsay Stewart; Lucia Costanza, BA; Mackenzie Keil, MS; Mae Yefimov; Makeda Pinnock, RN; Marco Ramirez-Gama, BS; Margaret E. Shannon, MA; Margaret Warren; Meg Hunter, BS; Melissa Hearrell, FNP, MSN; Melissa Yaeger, BS; Michael G. Sherenian, MD; Michael Marget, BA; Michele Cootauco, CRNP; Michelle Backer; Michelle Huffaker, MD; Michelle Mishoe, MS; Mikaela Hairston, RN; Mike Kulis, PhD; Mollie Schrodi, BSN; Molly Boone, RD; Morgan McGee; Nelly Hernandez, BS; Olisabueze E. Dimbo, BS; Olivia Raeber, PhD; Pamela Groh, RN/CRC; Patricia Taylor, NP; Peter K. Amezcua, CRC; Raul Giron, BS; Rebekah L. Epstein, BS; Rediet Bayou; Rida Saeed; Robert Rossi, MD; Rose Madison, BA; Safia Nawaz, MD; Samantha Henry, MS; Samuel J. Arbes, DDS, MPH, PhD; Sarah Bennick, RN, MSN, CPNP; Scott Boyd, MD, PhD; Shannon Garcia, BS; Shelley Randall; Sima Ramratnam, MD; Sky Strange, CRC; Stacey Bellemore, MS; Susan Leung, RN; Susan Raine, MD, JD; Suzanne Barshow, MD; Suzanne House, BS; Tiffany Narine, MSC; Timothy Sun; Valentina Vannoni, LLC; Veronica Chacko, BS; Warren Blackmon, BS; William Taylor, BA; Zeping Wang, CRC.
Supplementary data
References
- 1.Osborne N.J., Koplin J.J., Martin P.E., Gurrin L.C., Lowe A.J., Matheson M.C., et al. Prevalence of challenge-proven IgE-mediated food allergy using population-based sampling and predetermined challenge criteria in infants. J Allergy Clin Immunol. 2011;127:668–676.e1-2. doi: 10.1016/j.jaci.2011.01.039. [DOI] [PubMed] [Google Scholar]
- 2.Nwaru B.I., Hickstein L., Panesar S.S., Roberts G., Muraro A., Sheikh A., et al. Prevalence of common food allergies in Europe: a systematic review and meta-analysis. Allergy. 2014;69:992–1007. doi: 10.1111/all.12423. [DOI] [PubMed] [Google Scholar]
- 3.Acker W.W., Plasek J.M., Blumenthal K.G., Lai K.H., Topaz M., Seger D.L., et al. Prevalence of food allergies and intolerances documented in electronic health records. J Allergy Clin Immunol. 2017;140:1587–1591.e1. doi: 10.1016/j.jaci.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta R.S., Warren C.M., Smith B.M., Blumenstock J.A., Jiang J., Davis M.M., et al. The public health impact of parent-reported childhood food allergies in the United States. Pediatrics. 2018;142 doi: 10.1542/peds.2018-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta R., Holdford D., Bilaver L., Dyer A., Holl J.L., Meltzer D. The economic impact of childhood food allergy in the United States. JAMA Pediatr. 2013;167:1026–1031. doi: 10.1001/jamapediatrics.2013.2376. [DOI] [PubMed] [Google Scholar]
- 6.Silverberg J.I. Public health burden and epidemiology of atopic dermatitis. Dermatol Clin. 2017;35:283–289. doi: 10.1016/j.det.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Silverberg J.I., Simpson E.L. Associations of childhood eczema severity: a US population-based study. Dermatitis. 2014;25:107–114. doi: 10.1097/DER.0000000000000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran M.M., Lefebvre D.L., Dharma C., Dai D., Lou W.Y.W., Subbarao P., et al. Predicting the atopic march: results from the Canadian Healthy Infant Longitudinal Development Study. J Allergy Clin Immunol. 2018;141:601–607.e8. doi: 10.1016/j.jaci.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 9.Martin P.E., Eckert J.K., Koplin J.J., Lowe A.J., Gurrin L.C., Dharmage S.C., et al. Which infants with eczema are at risk of food allergy? Results from a population-based cohort. Clin Exp Allergy. 2015;45:255–264. doi: 10.1111/cea.12406. [DOI] [PubMed] [Google Scholar]
- 10.Sampson H.A., O’Mahony L., Burks A.W., Plaut M., Lack G., Akdis C.A. Mechanisms of food allergy. J Allergy Clin Immunol. 2018;141:11–19. doi: 10.1016/j.jaci.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Du Toit G., Roberts G., Sayre P.H., Bahnson H.T., Radulovic S., Santos A.F., et al. Randomized trial of peanut consumption in infants at risk for peanut allergy. N Engl J Med. 2015;372:803–813. doi: 10.1056/NEJMoa1414850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perkin M.R., Logan K., Marrs T., Radulovic S., Craven J., Flohr C., et al. Enquiring About Tolerance (EAT) study: feasibility of an early allergenic food introduction regimen. J Allergy Clin Immunol. 2016;137:1477–1486.e8. doi: 10.1016/j.jaci.2015.12.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellach J., Schwarz V., Ahrens B., Trendelenburg V., Aksunger O., Kalb B., et al. Randomized placebo-controlled trial of hen’s egg consumption for primary prevention in infants. J Allergy Clin Immunol. 2017;139:1591–1599.e2. doi: 10.1016/j.jaci.2016.06.045. [DOI] [PubMed] [Google Scholar]
- 14.Natsume O., Kabashima S., Nakazato J., Yamamoto-Hanada K., Narita M., Kondo M., et al. Two-step egg introduction for prevention of egg allergy in high-risk infants with eczema (PETIT): a randomised, double-blind, placebo-controlled trial. Lancet. 2017;389(10066):276–286. doi: 10.1016/S0140-6736(16)31418-0. [DOI] [PubMed] [Google Scholar]
- 15.Palmer D.J., Metcalfe J., Makrides M., Gold M.S., Quinn P., West C.E., et al. Early regular egg exposure in infants with eczema: a randomized controlled trial. J Allergy Clin Immunol. 2013;132:387–392.e1. doi: 10.1016/j.jaci.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Palmer D.J., Sullivan T.R., Gold M.S., Prescott S.L., Makrides M. Randomized controlled trial of early regular egg intake to prevent egg allergy. J Allergy Clin Immunol. 2017;139:1600–1607.e2. doi: 10.1016/j.jaci.2016.06.052. [DOI] [PubMed] [Google Scholar]
- 17.Wei-Liang Tan J., Valerio C., Barnes E.H., Turner P.J., Van Asperen P.A., Kakakios A.M., et al. A randomized trial of egg introduction from 4 months of age in infants at risk for egg allergy. J Allergy Clin Immunol. 2017;139:1621–1628.e8. doi: 10.1016/j.jaci.2016.08.035. [DOI] [PubMed] [Google Scholar]
- 18.Koplin J.J., Allen K.J., Gurrin L.C., Peters R.L., Lowe A.J., Tang M.L., et al. The impact of family history of allergy on risk of food allergy: a population-based study of infants. Int J Environ Res Public Health. 2013;10:5364–5377. doi: 10.3390/ijerph10115364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keet C., Pistiner M., Plesa M., Szelag D., Shreffler W., Wood R., et al. Age and eczema severity, but not family history, are major risk factors for peanut allergy in infancy. J Allergy Clin Immunol. 2021;147:984–991.e5. doi: 10.1016/j.jaci.2020.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nissen S.P., Kjaer H.F., Host A., Nielsen J., Halken S. Can family history and cord blood IgE predict sensitization and allergic diseases up to adulthood? Pediatr Allergy Immunol. 2015;26:42–48. doi: 10.1111/pai.12264. [DOI] [PubMed] [Google Scholar]
- 21.Pesonen M., Kallio M.J., Siimes M.A., Elg P., Bjorksten F., Ranki A. Cord serum immunoglobulin E as a risk factor for allergic symptoms and sensitization in children and young adults. Pediatr Allergy Immunol. 2009;20:12–18. doi: 10.1111/j.1399-3038.2008.00736.x. [DOI] [PubMed] [Google Scholar]
- 22.Tariq S.M., Arshad S.H., Matthews S.M., Hakim E.A. Elevated cord serum IgE increases the risk of aeroallergen sensitization without increasing respiratory allergic symptoms in early childhood. Clin Exp Allergy. 1999;29:1042–1048. doi: 10.1046/j.1365-2222.1999.00594.x. [DOI] [PubMed] [Google Scholar]
- 23.Ferguson A., Dimich-Ward H., Becker A., Watson W., DyBuncio A., Carlsten C., et al. Elevated cord blood IgE is associated with recurrent wheeze and atopy at 7 yrs in a high risk cohort. Pediatr Allergy Immunol. 2009;20:710–713. doi: 10.1111/j.1399-3038.2009.00869.x. [DOI] [PubMed] [Google Scholar]
- 24.Vogt H., Braback L., Zetterstrom O., Zara K., Falth-Magnusson K., Nilsson L. Asthma heredity, cord blood IgE and asthma-related symptoms and medication in adulthood: a long-term follow-up in a Swedish birth cohort. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah P.S., Wegienka G., Havstad S., Johnson C.C., Ownby D.R., Zoratti E.M. The relationship between cord blood immunoglobulin E levels and allergy-related outcomes in young adults. Ann Allergy Asthma Immunol. 2011;106:245–251. doi: 10.1016/j.anai.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eiriksson T.H., Sigurgeirsson B., Ardal B., Sigfusson A., Valdimarsson H. Cord blood IgE levels are influenced by gestational age but do not predict allergic manifestations in infants. Pediatr Allergy Immunol. 1994;5:5–10. doi: 10.1111/j.1399-3038.1994.tb00211.x. [DOI] [PubMed] [Google Scholar]
- 27.Lilja G., Johansson S.G., Kusoffsky E., Oman H. IgE levels in cord blood and at 4-5 days of age: relation to clinical symptoms of atopic disease up to 18 months of age. Allergy. 1990;45:436–444. doi: 10.1111/j.1398-9995.1990.tb01094.x. [DOI] [PubMed] [Google Scholar]
- 28.Chang W.T., Sun H.L., Lue K.H., Chou M.C. Predictability of early onset atopic dermatitis by cord blood IgE and parental history. Acta Paediatr Taiwan. 2005;46:272–277. [PubMed] [Google Scholar]
- 29.Irizar H., Kanchan K., Mathias R.A., Bunyavanich S. Advancing food allergy through omics sciences. J Allergy Clin Immunol Pract. 2021;9:119–129. doi: 10.1016/j.jaip.2020.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunyavanich S., Schadt E.E. Systems biology of asthma and allergic diseases: a multiscale approach. J Allergy Clin Immunol. 2015;135:31–42. doi: 10.1016/j.jaci.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang H.H.F., Sly P.D., Holt P.G., Holt K.E., Inouye M. Systems biology and big data in asthma and allergy: recent discoveries and emerging challenges. Eur Respir J. 2020;55:1900844. doi: 10.1183/13993003.00844-2019. [DOI] [PubMed] [Google Scholar]
- 32.Dhondalay G.K., Rael E., Acharya S., Zhang W., Sampath V., Galli S.J., et al. Food allergy and omics. J Allergy Clin Immunol. 2018;141:20–29. doi: 10.1016/j.jaci.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Meyer P., Saez-Rodriguez J. Advances in systems biology modeling: 10 years of crowdsourcing DREAM challenges. Cell Syst. 2021;12:636–653. doi: 10.1016/j.cels.2021.05.015. [DOI] [PubMed] [Google Scholar]
- 34.Boyce J.A., Assa’ad A., Burks A.W., Jones S.M., Sampson H.A., Wood R.A., et al. Guidelines for the diagnosis and management of food allergy in the United States: summary of the NIAID-sponsored expert panel report. J Allergy Clin Immunol. 2010;126:1105–1118. doi: 10.1016/j.jaci.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Togias A., Cooper S.F., Acebal M.L., Assa’ad A., Baker J.R., Jr., Beck L.A., et al. Addendum guidelines for the prevention of peanut allergy in the United States: report of the National Institute of Allergy and Infectious Diseases–sponsored expert panel. J Allergy Clin Immunol. 2017;139:29–44. doi: 10.1016/j.jaci.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prentice R.L. A case–cohort design for epidemiologic cohort studies and disease prevention trials. Biometrika. 1986;73:1–11. [Google Scholar]
- 37.Self S.G., Prentice R.L. Asymptotic distribution theory and efficiency results for case–cohort studies. Ann Stat. 1988;16:64–81. [Google Scholar]
- 38.Barlow W.E. Robust variance estimation for the case–cohort design. Biometrics. 1994;50:1064–1072. [PubMed] [Google Scholar]
- 39.Onland-Moret N.C., van der A.D., van der Schouw Y.T., Buschers W., Elias S.G., van Gils C.H., et al. Analysis of case–cohort data: a comparison of different methods. J Clin Epidemiol. 2007;60:350–355. doi: 10.1016/j.jclinepi.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 40.Cohain A., Schadt E., Bunyavanich S. In: Middleton’s allergy. 9th ed. Burks A.W., Holgate S.T., O’Hehir R.E., Bacharier L., Broide D.H., Hershey G.K.K., et al., editors. Vol. 1. Elsevier; Amsterdam: 2019. Systems biology; pp. 352–361. [Google Scholar]
- 41.Villaverde A.F., Banga J.R. Reverse engineering and identification in systems biology: strategies, perspectives and challenges. J R Soc Interface. 2014;11 doi: 10.1098/rsif.2013.0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.