Abstract
Background
Food protein–induced enterocolitis syndrome (FPIES) is a non–IgE-mediated food hypersensitivity that affects the gastrointestinal system, especially in children, who often present with more severe clinical manifestations than adults do. Although its pathogenesis is poorly understood and biomarkers are still lacking, scientific evidence suggests that gut microbiota may play an important role in the development of FPIES.
Objective
We aimed to compare the composition of gut microbiota in children with FPIES with that in age- and sex-matched healthy controls.
Methods
We analyzed the gut microbiota profiles in fecal samples of 17 patients with FPIES (case patients) and 12 age-matched healthy children (controls) by tag sequencing of the 16S ribosomal RNA gene hypervariable V4-V5 regions. Subjects' sociodemographic, clinical, and food diary variables were described and compared between groups by using inferential statistical tests. Nonparametric linear discriminant analysis was performed for intestinal microbiota data.
Results
Patients with confirmed cases FPIES (n = 17; average patient age, 7.5 ± 3.2 years) and controls without FPIES or any atopy (n = 12, average patient age, 6.9 ± 2.7 years) were included. Fish was the main FPIES-inducing allergen in 65% of the cases. The patients with FPIES showed higher proportions of Lachnospiraceae spp (P < .0286) and a lower proportion of Ruminococcaceae spp (P < .0066), Lactobacillaceae spp (P < .0075), and Leuconostocaceae spp (P < .0173) than the controls.
Conclusions
Our data clearly show a different gut microbial signature in patients with FPIES, suggesting a new potential avenue for aiding the diagnosis and clinical management of FPIES. Larger studies are needed to confirm these results.
Key words: FPIES, gut microbiota, microbiome, Lachnospiraceae, Ruminococcaceae, Lactobacillaceae, Leuconostocaceae, non–IgE-mediated food hypersensivity
Food protein–induced enterocolitis syndrome (FPIES) is a non–IgE-mediated food hypersensitivity that affects the gastrointestinal system. Its pathogenesis is poorly understood, its diagnosis and management are challenging, and biomarkers are still lacking. Although FPIES is typically caused by cow’s milk or rice, other foods such as poultry, egg, fish, or fruit may be involved.1, 2, 3 The syndrome is characterized by lethargy, pallor, vomiting, and diarrhea leading to dehydration, hypotension, and shock if severe, which can occur in up to 15% of patients.4 Current findings suggest that acute episodes are especially dramatic in children, who often present with more severe clinical manifestations than those presented by adults.2,5, 6, 7
Altered intestinal microbiota has been associated with IgE-mediated allergic diseases8 such as asthma,9 eczema,10 and food allergy.11,12 Although a fair amount of research on IgE-mediated disease exists, much less is known about state of the microbiota in non–IgE-mediated food hypersensitivity entities and the role of the immune system in disease development.11 Previous data showed the role of gut microbiota in conditioning immune system function.13,14 In fact, microbiota may attenuate the adverse effects of food allergens and help train the immune system to maintain oral immunologic tolerance.15,16 The absence of intestinal microbiota leads to an increase in TH2 and IgE production against food antigens, and dysregulation of intestinal microbiota plays a role in IgE-mediated food allergy development.11,12 A recent review proposed that in addition to TH2 type immunity, TH1, TH17, innate immunity, and epithelial mucosal barrier defect may have a role in the pathogenesis of the condition.17 However, research on the gut microbiota alterations in non–IgE-mediated food allergy diseases such as FPIES is very limited. To the best of our knowledge, only Boyer et al18 have claimed that the microbiota from the stool of infants with FPIES was enriched with bacteria from the families Gammaproteobacteria and Porphyromonadaceae; in addition, a clearer definition of the microbiota signature of both in non–IgE-mediated FPIES is still needed.
Furthermore, other factors such as undernutrition, which is associated with immune system defects and in turn affects intestinal stability, predisposing to diarrheal illnesses, may change the status of the microbiota.19 Also, the mode of an infant's delivery has been demonstrated to have an association with the diversity and colonization pattern of his or her gut microbiota during the first 3 months of life.20 Thus, the impact of mode of delivery on the diversity and colonization levels of the infant gut microbiota and on infants' health at each stage of life should be explored.
This study aimed to compare the gut microbial profile of children with and without FPIES to identify relevant differences.
Methods
Participant selection and recruitment
A case-control study was designed and carried out in the allergy unit of the Alicante General University Hospital, where the patients were clinically referred for evaluation of gastrointestinal symptoms after ingestion of different foods. All patients diagnosed with FPIES (n = 23 case patients) were included consecutively over a period of 4 years (2011-2015) once informed consent had been provided. As the specified inclusion period was before any international consensus guidelines existed, the case patients were diagnosed according to the Sicherer criteria,21 as follows: (1) repeated exposure to the incriminated food elicited diarrhea and/or repetitive vomiting within 24 hours without any other cause for the symptoms; (2) there were no symptoms other than gastrointestinal symptoms elicited by the incriminated food; (3) removal of the offending protein from the diet resulted in resolution of the symptoms, and/or a standardized food challenge elicited diarrhea and/or vomiting within 24 hours after administration of the food. If monitored during a challenge, an increase in the absolute neutrophil count by more than 3500 per mm3 at 5 to 8 hours after the challenge was additional presumptive evidence of a positive challenge response. In any case, the authors can confirm that all the cases satisfied the current clinical diagnostic criteria.22 Oral food challenge with the implicated food was performed either when a diagnosis was not clearly established by the clinical history or to monitor tolerance a minimum of 18 months after the most recent reaction. The control group (n = 12) was selected from a nonatopic population seen in pediatric primary care. None of the controls had a history of symptoms after food ingestion. Subjects who did not provide the fecal sample or dietary records were excluded from the analysis. In all cases, the time between the last episode of FPIES and the microbiota test was no longer than 6 months. The Alicante General University Hospital ethics committee approved the study protocol.
Variables collected included age and sex, FPIES-inducing allergen, Bristol tool scale parameters to assess bowel habits (constipation, normal bowel movements, and diarrhea),23 delivery mode (cesarean vs vaginal), type of feeding during the first 6 months of life (breast-feeding exclusively, infant formula, or mixed feeding) and maternal antibiotic and/or probiotic intake during pregnancy or breast-feeding.
Sample collection and DNA isolation
Fecal samples were collected at the time of inclusion at the individuals' home and immediately stored at −20 °C before being transported to the hospital on ice in plastic cooler bags to avoid sample thaw and then stored at −80 °C until processing. DNA was isolated by using a 150-mg stool aliquot and the PowerFecal DNA Isolation Kit (MO BIO Laboratories, Carlsbad, Calif), following the manufacturer's instructions. Finally, fecal genomic DNA samples were adjusted to 20 ng/μL for 16S amplicon sequencing. The V4-V5 hypervariable from bacterial 16S ribosomal RNA gene was amplified by using the 6-mer barcoded primers S-D-Bact-0563-a-S-15 (AYTGGGYDTAAAGNG) and S-D-Bact-0907-a-A-20 (CCGTCAATTYMTTTRAGTTT).
Sequencing and OTU picking
Dual barcoded PCR products, consisting of approximately 360 bp, were purified from triplicate reactions by means of the Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare, Chicago, Ill) and quantified using Qubit 3.0 and the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, Mass). Samples were multiplexed by combining equimolar quantities of amplicon DNA (100 ng per sample) and sent to Eurofins Genomics GmbH (Ebersberg, Germany) to perform Illumina MiSeq high-throughput sequencing with a 2 × 300 phycoerythrin configuration. Raw data were assembled by using Flash software. Sample demultiplexing was carried out by using sequence information from the respective DNA barcodes and the mothur v1.36.1 suite of analysis. After assembly and barcode/primer removal, the sequences were processed to eliminate chimeras by using the UCHIME algorithm and SILVA reference set of 16S sequences.24 α-Diversity estimators such as Chao richness, Simpson evenness, Simpson reciprocal index, dominance index, and phylogenetic distance were computed by using QIIME v1.9.1 analysis suite25 and a high-quality normalized subset of 10,000 sequences per sample that were randomly selected after shuffling (×10,000) the original data set. Taxonomy assessment was performed by using the Ribosomal Database Project classifier, version12.26 The operational taxonomic unit (OTU) picking approach was performed with the normalized subset of 10,000 sequences and the UCLUST algorithm implemented in USEARCH, version 8.0.1623.
Dietary assessment
All participants were shown how to keep a 3-day food record and to complete it during the same period of the fecal sample collection. The information provided was checked for completeness, and a detailed 3-day food record was collected for 29 participants (12 controls and 17 patients with FPIES), which enabled macronutrient intake calculation. Nutrients were estimated by using the open-source DIAL software, which incorporates a large Spanish Food Composition Database (www.alceingenieria.net/nutricion/descarga.htm). The means and SDs of daily intakes were calculated for energy (kcal), protein (g), carbohydrate (g), fat (g), cholesterol (mg), and fiber (g). The t test for independent samples was used to compare group means between the case patients with FPIES and the controls.
Statistical analysis
Statistical analysis was performed by using SPSS software, version 20 for Windows (IBM, Armonk, NY). Mean values and SDs were calculated for quantitative data and compared by using the 2-tailed Student t test for independent samples. Categoric data were analyzed by using the chi-square test. P values less than .05 were considered statistically significant. For intestinal microbiota data, nonparametric linear discriminant analysis (LDA)27 was performed to compare fecal microbial communities at different taxonomy levels in the controls and case patients with FPIES. Wilcoxon rank sum or Student t tests for unpaired sample analysis were used to evaluate differences in α-diversity descriptors after Shapiro-Wilk normality test assessment. Additionally, sex, diet, delivery mode, and lactation information were explored as covariates in β-diversity approaches based on Unifrac weighted distances and permutational multivariate ANOVA (PERMANOVA) analysis, using the QIIME platform (QIIME, version 1.9.1).25 The vegan: adonis function of the R environment (version 3.6.3) was also used to explore multivariate models on microbiota structure. Graphics were created by using the ggplot2 R package.
Results
Participant characteristics
Initially, 23 patients with FPIES were included, 6 of whom were dropped from the analysis because they did not provide fecal samples (n = 5) and/or dietary records (n = 2). We included 17 patients with FPIES; their mean age was 7.5 years, and 7 of them (41%) were males. The result of testing for specific IgE to the implicated food was negative in all the patients at the time of inclusion. Patients reacted to only 1 food or to a group of similar foods. In the case of fish, most patients reacted to multiple fish species. Symptoms were induced by fish (n = 11), fruit (n = 4), milk (n = 1), and egg (n = 1). The time between food ingestion and onset of symptoms ranged from 1 hour to 4 hours. The diagnosis of FPIES was established after a median of 4 reactions (range 3-6). Twelve case patients were treated in the emergency department because of profuse vomiting, and 2 were admitted to the intensive care unit with severe symptoms (1 with hypotension and shock associated with dehydration and 1 with metabolic acidosis).
As shown in Table I, the sociodemographic and clinical characteristics of the patients with FPIES were compared with those of 12 control donors. A higher percentage of case patients than controls had been delivered via cesarean section (62.5% vs 25.0%). Because delivery mode influences the microbiota composition, further analyses were performed to include this parameter as a potential factor in microbiota changes such as microbial community level (see the β-diversity analyses). Also, the percentage of controls who were breast-fed exclusively for the first 6 months of life was higher than the percentage of children with FPIES (45.5% vs 12.5%) although the difference did not reach statistical significance.
Table I.
Demographic and clinical characteristics of patients with FPIES and control donors
| Patients with FPIES (n = 17) | Controls (n = 12) | |||||
|---|---|---|---|---|---|---|
| Variable | No. | % | No. | % | P value∗ | |
| Sex | ||||||
| Female | 10 | 58.8 | 8 | 66.7 | .67 | |
| Male | 7 | 41.2 | 4 | 33.3 | ||
| Age, y (SD) | 7.5 (3.2) | 6.9 (2.7) | .60 | |||
| FPIES-inducing allergen | ||||||
| Fruit | 4 | 23.5 | — | |||
| Fish | 11 | 64.7 | — | |||
| Egg | 1 | 5.9 | — | |||
| Milk | 1 | 5.9 | — | |||
| Delivery mode | ||||||
| Cesarean | 10 | 62.5 | 3 | 25.0 | .05 | |
| Vaginal | 6 | 37.5 | 9 | 75.0 | ||
| Type of feeding | ||||||
| Breast-feeding exclusively | 2 | 12.5 | 5 | 45.5 | .06 | |
| Infant formula | 1 | 6.3 | 0 | 0.0 | .40 | |
| Mixed feeding | 13 | 81.3 | 7 | 54.6 | .14 | |
| Bristol stool scale parameters to assess bowel habits | ||||||
| Normal | 11 | 73.3 | 8 | 66.7 | .71 | |
| Constipation and diarrhea | 4 | 26.7 | 4 | 33.3 | ||
| Maternal antibiotic intake during pregnancy or breast-feeding | 5 | 31.3 | 2 | 16.7 | .38 | |
| Maternal probiotic intake during pregnancy or breast-feeding | 10 | 71.4 | 8 | 66.7 | .79 | |
P values calculated using the chi-square test.
Table II shows the mean (SD) daily intake of energy (kcal), protein, fat, and carbohydrates in the patients and the controls. We observed a slightly lower intake of energy, protein, and carbohydrates in the FPIES group; however, none of the differences were statistically significant.
Table II.
Means and SDs of energy, protein, carbohydrates, and fat intake in the study participants
| Nutrient | Patients with FPIES (n=17) |
Controls (n=12) |
P value | ||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| Energy (kcal) | 2017.5 | 487.2 | 2211.0 | 373.6 | .25 |
| Protein (g) | 80.1 | 21.4 | 92.3 | 26.4 | .21 |
| Protein (%) | 16.0 | 14.9 | |||
| Carbohydrates (g) | 227.0 | 59.2 | 252.7 | 62.2 | .29 |
| Carbohydrates (% of daily intake) | 47.2 | 47.5 | |||
| Fat (g) | 83.7 | 26.9 | 88.0 | 16.1 | .60 |
| Fat (% of daily intake) | 36.9 | 35.9 | |||
Gut microbiota analysis in children with and without FPIES
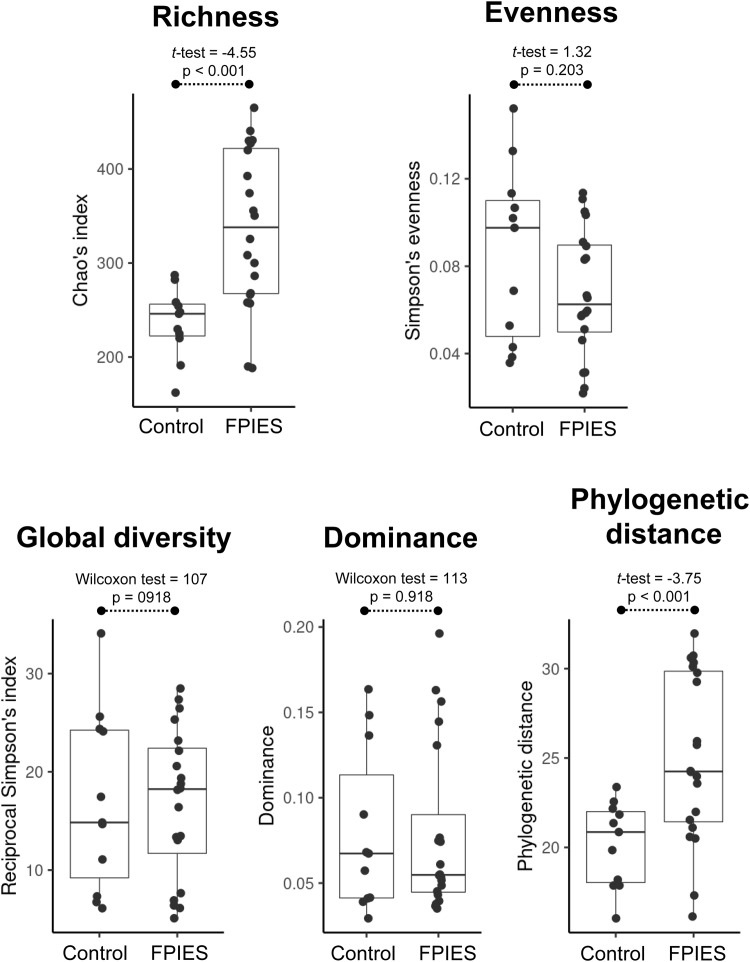
Microbial diversity and microbiota composition were analyzed in the fecal samples of 17 infants and children with FPIES and 12 control donors. With regard to the 5 different α-diversity descriptors evaluated, we found that richness and phylogenetic distance differed between the groups (Fig 1), indicating that subjects with FPIES have more bacterial species inhabiting their intestinal tract and those species are more diverse and genetically distinct from one another.
Fig 1.
α-Diversity on fecal microbiota. The α-diversity, including study of the Chao index, Simpson evenness, Simpson reciprocal index, dominance, and phylogenetic distance descriptors were assessed and compared between sample groups (controls and case patients with FPIES). Distributions of the respective metrics are presented as box plots. Results of statistical assessment are shown on top of box plots, respectively.
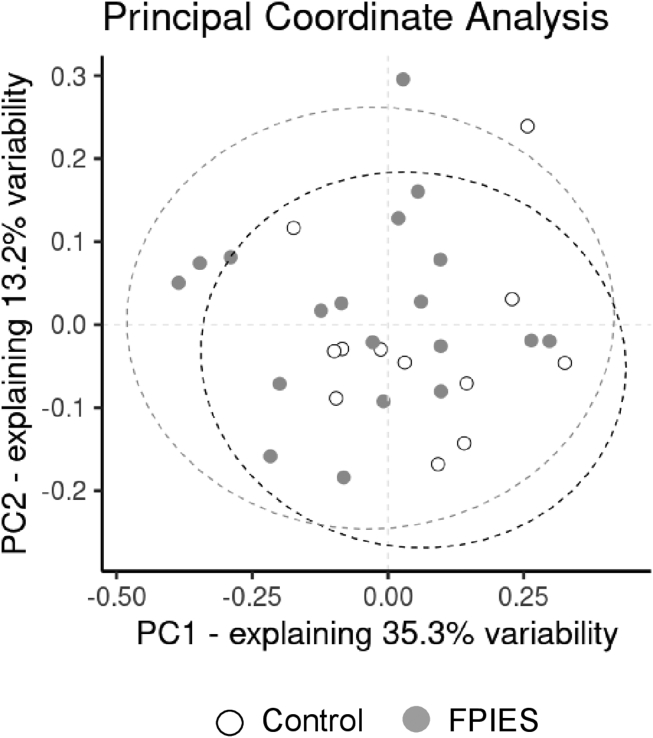
We also performed a β-diversity analysis based on abundance and phylogenetic relationships among the 623 different OTUs compiled after the processing of more than 430,000 DNA reads. After calculating the distances between samples by using Unifrac weighted metrics, we performed nonparametric and permutation tests (PERMANOVA). This approach was useful to assess the impact of different variables on the microbial community structure in addition to the disease status. Consequently, we analyzed gut microbiota profile according to the different variables compiled for all participants, including diet, sex, delivery mode, type of milk feeding, and FPIES or health status. According to this analysis, only FPIES status (PERMANOVA result = 2.08 [P < .046]) seems to be related to the observed changes in the fecal microbial community (Fig 2), whereas other important and common covariates regularly found to shape human intestinal microbiota seem to influence microbiota structure in this study's subjects to a lesser extent (PERMANOVA testDelivery_mode = 0.65 [P = .817]; PERMANOVA testFeeding type = 1.21, [P = .243], PERMANOVA testSex = 1.80 [P = .087]). We further tried to set a multivariable model explaining microbiota structure by using an Adonis (permutation-based) approach. Combining FPIES condition likely shaping microbiota structure with sex condition exhibited no significant influence on the intestinal microbiota structure. The resulting model did not support an interaction between both variables to determine the microbiota composition in both groups (F test = 1.07; R2 = 0.11; P = .330).
Fig 2.
Microbial community structure. β-Diversity evaluation of the fecal microbial community structure is provided using principal coordinate (PC) analysis. Filled points indicate distribution of samples from the controls (white) and patients with FPIES (gray) across bidimensional space (the 2 main PCs exhibiting the highest variation explained). Ellipses, delimiting the 95% CI of data distribution, are shown accordingly.
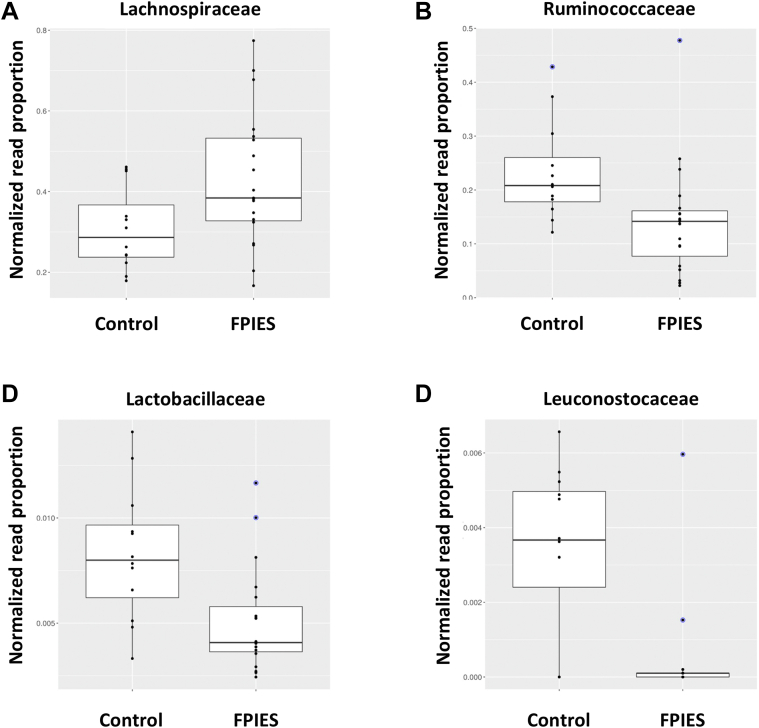
As major changes observed at the microbial community level seem to be primarily associated with the disease condition, thus discarding the presence of confounding variables, we then further assessed the distribution of microbial species present in feces, performing taxonomy analysis over high-quality DNA reads. Taxonomy distribution at phylum level showed no differences in microbiota composition between groups; however, comparisons at the family level indicated that the microbiota communities in patients with FPIES contained a higher proportion of Lachnospiraceae spp (P < .0286; LDA score = 4.79) and a lower proportion of Ruminococcaceae spp (P < .0066; LDA score = 4.67), Lactobacillaceae spp (P < .0075; LDA score = 3.81), and Leuconostocaceae spp (P < .0173; LDA score = 3.72) than the microbiota communities in the controls (Fig 3, A-D).
Fig 3.
Taxonomy changes. Normalized proportions of DNA reads for the Lachnospiraceae (A), Ruminococcaceae (B), Lactobacillaceae (C), and Leuconostocaceae (D) bacterial families. Differential abundance between sample groups (samples from controls and patients with FPIES) was found. Relative proportions of DNA reads for each taxonomy category are shown as box plots. Blue points indicate outliers.
An OTU analysis to determine differences in microbiota species between groups generated a list of putative species differentially associated with each, as indicated by an LDA score higher than 3 (Table III).
Table III.
List of top OTUs associated with health status
| OTU | Health status∗ | Blast (%)† | LDA‡ | P value§ |
|---|---|---|---|---|
| OTU2 | Controls | Faecalibacterium prautsnitzii (98) | 4.57 | .011 |
| OTU241 | Controls | Bacteroides ovatus (99) | 4.08 | .015 |
| OTU34 | Patients with FPIES | Eubacterium hallii (98) | 4.03 | .032 |
| OTU50 | Controls | Parabacteroides distasonis (100) | 3.52 | .032 |
| OTU46 | Patients with FPIES | Spiroplasmas abaudiense (84) | 3.49 | .006 |
| OTU42 | Patients with FPIES | Ruminococcus albus (93) | 3.47 | <.001 |
| OTU37 | Controls | Intestinimonas spp (95) | 3.38 | .043 |
| OTU540 | Controls | Bacteroides caccae (99) | 3.35 | .029 |
| OTU23 | Controls | Enterococcus hirae (100) | 3.33 | .023 |
| OTU26 | Controls | Parabacteroides merdae (100) | 3.25 | .026 |
| OTU47 | Controls | Lactobacillus hominis (100) | 3.22 | .009 |
| OTU59 | Controls | Weisella confusa (100) | 3.19 | .007 |
| OTU68 | Controls | Ruminococcus spp (96) | 3.18 | .002 |
| OTU83 | Patients with FPIES | Parabacteroides goldsteinii (100) | 3.13 | .034 |
| OTU73 | Patients with FPIES | Lactococcus taiwanensis (100) | 3.00 | .006 |
| OTU105 | Patients with FPIES | Blautia spp (96) | 3.00 | .032 |
Association based on the higher abundance of the respective OTU (in either the controls or patients with FPIES).
Taxonomy assignment on the basis of best hit against the National Center for Biotechnology Information 16S reference database, taking into account the complete alignment of the OTU sequence.
Taxonomy assignment based on a sequence identity. Only scores higher than 97% are shown, indicating reliable taxonomy identifications.
LDA and P value according to LDA effect size analysis.
The included list of probable microbial species associated with health or disease status matches with the data obtained from comparisons at family level (P < .05). Accordingly, species belonging to the families Ruminococcaceae (OTU2, OTU68, and OTU37), Lactobacillaceae (OTU47), and Leuconostocaceae (OTU59) were found in higher proportions in the controls, whereas Lachnospiraceae spp such as Eubacterium hallii (OTU34) and Blautia spp (OTU105) were more abundant in the FPIES group. Additionally, some Bacteroidaceae spp (OTU241 and OTU540) and Porphyromonadaceae spp (OTU50 and OTU26) also seemed to be associated with a status of health.
Discussion
FPIES is a non–IgE-dependent allergy characterized by delayed gastrointestinal reactions to foods. Current knowledge on gastrointestinal immune events during FPIES comes from research on milk enteropathies, whereas new data on other FPIES immune mechanisms are also emerging.2,3,28 There is some evidence for a systemic innate immune activation affecting monocytes, neutrophils, and eosinophils in FPIES.29 Moreover, the microbiota may be involved in the syndrome's pathogenesis, and its modulation can have an effect on the FPIES.30 To our knowledge, there are no previous original research articles on gut microbiome composition in FPIES. Here, our results showed that subjects with FPIES subjects have a significantly higher diversity of bacteria in their intestine and that those species seem to be genetically distant from one another. This led to us to hypothesize that certain bacteria species could be found in a specific manner in FPIES. The taxonomy analysis partially supported this notion because the subjects with FPIES exhibited a higher proportion of species of Lachnospiraceae, which is a very complex bacterial clade with a vast array of traits linked to health and disease states.31,32 On the other hand, lower proportions of Ruminococcaceae spp, Lactobacillaceae spp, and Leuconostocaceae spp were detected in the subjects with FPIES than in the control subjects.
Our results showed that the proportion cesarean section deliveries was higher among the patients with FPIES than among the controls. In this regard, it was reported previously that patients with FPIES related to cow’s milk are more likely to be delivered by cesarean section33 and that this fact could disrupt microbiota composition and predispose to eczema.34 Within the first months of life, the infant microbiota undergoes substantial reorganization, which is driven primarily by body site35 and also associated with mode of delivery.20 It was intriguing to see that the percentage of controls who were breast-fed exclusively was 3-fold higher than the percentage of patients with FPIES (45.5% vs 12.5% [P = .06])—a percentage that would likely to be statistically significant with a larger sample size. In the same way, a nonsignificant 2-fold increase in maternal antibiotic use in the group of those with FPIES was found. Recently, maternal antibiotic use was found to be significantly higher in infants with FPIES than in allergy-free infants.36 Although the aforementioned factors are thought to influence the microbiota, for patients with FPIES, controls, and both groups together, no significant differences were observed, suggesting that the study was underpowered for this specific analysis. Nevertheless, we should note that it was sufficient to find differences between patients with FPIES and the controls. In view of this fact, larger cohorts of patients with FPIES might merit further attention.
The incompletely understood pathophysiology of FPIES, lack of diagnostic biomarkers, and poor knowledge of its natural history have recently marked this illness as an important target for study. A review of the literature shows that frequent features of the pathologic microbiota among different types of non–IgE-mediated food allergy can be observed.30 In particular, an increase of Proteobacteria and Porphyromonadaceae in patients with FPIES has been observed at phylum level.18 Alterations in gut microbiota have also been described in infants with atopic eczema and food sensitization.12 A microbial signature has been proposed for atopic dermatitis in children with and without food allergy.10 Evidence from IgE-mediated allergic disorders suggests a possible role of the microbiota in the pathogenesis of the non–IgE-mediated food allergies, such as FPIES, and the need to understand the effects of its modulation on these disorders.30 In our data, composition of the gut microbiota in children with FPIES did not differ drastically from that in the controls in terms of the number or composition of bacterial species. We did detect more Lachnospiraceae spp and fewer Ruminococcaceae spp in patients with FPIES than in the healthy controls. Moreover, different authors have detected a reduction of cells such as TH17 lymphocytes under Lachnospiraceae supplementation in inflammatory bowel disease.37,38 As TH17 cells are responsible for neutrophil activation and improvement of epithelial cell barrier function, their reduction may provoke susceptibility to infection in intestinal mucosa and an increase in allergen permeability.
Furthermore, Lachnospiraceae spp and Ruminococcaceae spp aid in dietary carbohydrate fermentation and production of short-chain fatty acids (propionate, butyrate, and acetate), which are natural fuel for gastrointestinal colonocytes and anti-inflammatory effectors.39 A reduction of Ruminococcaceae in patients with FPIES can contribute to a proinflammatory profile in their response, as previously also observed for patients with atopic dermatitis.40 In any case, their impact on host physiology is often inconsistent across studies, so it cannot be assured that the increase in Lachnospiraceae would be enough to support the decrease in Ruminococcaceae-derived effects.
We also observed a higher abundance of Bacteroidaceae spp and Porphyromonadaceae spp in the healthy controls than in the patients with FPIES. These data agree with those in previous reports regarding the reduced presence of these families in pediatric allergies,10,11 and they open the door to possible strategies for preventing or treating FPIES by modulating the microbiota. Also, a lower Lactobacillaceae presence in the FPIES group was found, which is particularly interesting because studies from Berni Canani et al have shown that Lactobacillus GG supplementation in infants with cow’s milk allergy, including in a group of infants with non–IgE-mediated allergy, accelerated the development of tolerance.41 The findings here are therefore in line with the hypothesis that a dysregulated microbiota may be involved in the pathogenesis of and contribute to the development of allergic symptoms, which can be therapeutically modulated. Other factors affecting the composition of the microbiota also need to be taken into account, as a recent publication has revealed the importance not only of the geographic location of the patient, but also of underappreciated sex- or ethnicity/race-based differences in microbiota composition, disease risk, and therapeutic responses.42 Further studies should investigate the potential role of the specific microbiota signature in patients with FPIES in development and/or progression of the non-IgE food protein allergy.
Sample size should be noted as 1 important limitation in this study. Although our conclusions are strongly supported by the experimental data, higher numbers of patients and control donors should be included in larger, prospective studies to confirm the findings. Although the association with maternal antibiotic and probiotic intake during pregnancy or breast-feeding were analyzed, patients' previous antibiotic or probiotic use, and type of infant formula used were not recorded. These data, together with the concomitant illness at time of specimen collection, may influence the intestinal microbiome. Thus, the interest in analyzing these factors should be considered in future and larger studies. A caveat of the study is that no correction for multiple comparisons was made. It should be noted that our study cohort was also characterized by a high proportion of fish-induced FPIES, probably because seafood is predominant in the study population's dietary habits. How the food specificity of FPIES might make a difference in microbiota composition should be further explored. Finally, although associations of FPIES with bacterial species were found, immune-related data were not included in the original design of the study. Therefore, in future research efforts should be made to obtain larger sample sizes so as to understand the mechanism of the disease.
In summary, a different microbiota profile was found in children with FPIES, who presented a higher proportion of Lachnospiraceae spp and a lower proportion of Ruminococcaceae spp, Lactobacillaceae spp, and Leuconostocaceae spp. We believe that these findings should encourage further investigation into the microbiota as a contributor to FPIES disease pathology.
Clinical implications.
The identification of differences in gut microbiota between subjects with FPIES and control subjects should encourage further investigation into the microbiota as a contributor to FPIES disease pathology.
Acknowledgments
We are grateful to Ms Megan Harris for her assistance in language editing.
Footnotes
Supported by the Health Institute Carlos III, Ministry of Science, Innovation and Universities, Spain (grant AGL2017-88801-P) and the network ARADYAL (grant RD16/06/32). The contract to A.B.-P. is supported by the European Union’s Seventh Framework Programme under the grant agreement no. 613979 (MyNewGut), and the contract to M.C. is supported by the Sara Borrell programme (CD14/00237).
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
References
- 1.Online information center of medicines of the Spanish Agency of Medicines and Health Products (AEMPS-CIMA): Spanish Agency of Medicines and Health Products (AEMPS-CIMA); [cited 2016]. Available at: https://cima.aemps.es/cima/publico/home.html
- 2.González-Delgado P., Caparrós E., Moreno M.V., Clemente F., Flores E., Velásquez L., et al. Clinical and immunological characteristics of a pediatric population with food protein-induced enterocolitis syndrome (FPIES) to fish. Pediatr Allergy Immunol. 2016;27:269–275. doi: 10.1111/pai.12529. [DOI] [PubMed] [Google Scholar]
- 3.Moreno M.V., Caparrós E., Fernández J., González-Delgado P. Immunologic study of two fruit-induced FPIES cases. Pediatr Allergy Immunol. 2017;28:713–715. doi: 10.1111/pai.12769. [DOI] [PubMed] [Google Scholar]
- 4.Nowak-Węgrzyn A., Katz Y., Mehr S.S., Koletzko S. Non-IgE-mediated gastrointestinal food allergy. J Allergy Clin Immunol. 2015;135:1114–1124. doi: 10.1016/j.jaci.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 5.Lee E., Barnes E.H., Mehr S., Campbell D.E. Differentiating acute food protein-induced enterocolitis syndrome from its mimics: a comparison of clinical features and routine laboratory biomarkers. J Allergy Clin Immunol Pract. 2019;7:471–478.e3. doi: 10.1016/j.jaip.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Guibas G.V., Tsabouri S., Makris M., Priftis K.N. Food protein-induced enterocolitis syndrome: pitfalls in the diagnosis. Pediatr Allergy Immunol. 2014;25:622–629. doi: 10.1111/pai.12237. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez-Delgado P., Caparrós E., Moreno M.V., Cueva B., Fernández J. Food protein-induced enterocolitis-like syndrome in a population of adolescents and adults caused by seafood. J Allergy Clin Immunol Pract. 2019;7:670–672. doi: 10.1016/j.jaip.2018.07.028. [DOI] [PubMed] [Google Scholar]
- 8.Simonyté Sjödin K., Hammarström M.L., Rydén P., Sjödin A., Hernell O., Engstrand L., et al. Temporal and long-term gut microbiota variation in allergic disease: a prospective study from infancy to school age. Allergy. 2019;74:176–185. doi: 10.1111/all.13485. [DOI] [PubMed] [Google Scholar]
- 9.Hevia A., Milani C., López P., Donado C.D., Cuervo A., González S., et al. Allergic patients with long-term asthma display low levels of bifidobacterium adolescentis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fieten K.B., Totté J.E.E., Levin E., Reyman M., Meijer Y., Knulst A., et al. Fecal microbiome and food allergy in pediatric atopic dermatitis: a cross-sectional pilot study. Int Arch Allergy Immunol. 2018;175(1-2):77–84. doi: 10.1159/000484897. [DOI] [PubMed] [Google Scholar]
- 11.Ling Z., Li Z., Liu X., Cheng Y., Luo Y., Tong X., et al. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol. 2014;80:2546–2554. doi: 10.1128/AEM.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C.C., Chen K.J., Kong M.S., Chang H.J., Huang J.L. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr Allergy Immunol. 2016;27:254–262. doi: 10.1111/pai.12522. [DOI] [PubMed] [Google Scholar]
- 13.Mazmanian S.K., Liu C.H., Tzianabos A.O., Kasper D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Zheng D., Liwinski T., Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dethlefsen L., McFall-Ngai M., Relman D.A. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephen-Victor E., Chatila T.A. Regulation of oral immune tolerance by the microbiome in food allergy. Curr Opin Immunol. 2019;60:141–147. doi: 10.1016/j.coi.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su K.W., Shreffler W.G., Yuan Q. Gastrointestinal immunopathology of food protein-induced enterocolitis syndrome and other non-immunoglobulin E-mediated food allergic diseases. Ann Allergy Asthma Immunol. 2021;126:516–523. doi: 10.1016/j.anai.2021.02.024. [DOI] [PubMed] [Google Scholar]
- 18.Boyer J., Scuderi V. Comparison of the gut microbiome between food protein-induced enterocolitis sydrome (FPIES) infants and allergy-free infants. Ann of Allergy Asthma Immunol. 2017;119:e3. [Google Scholar]
- 19.Kau A.L., Ahern P.P., Griffin N.W., Goodman A.L., Gordon J.I. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rutayisire E., Huang K., Liu Y., Tao F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants' life: a systematic review. BMC Gastroenterol. 2016;16:86. doi: 10.1186/s12876-016-0498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sicherer S.H. Clinical aspects of gastrointestinal food allergy in childhood. Pediatrics. 2003;111:1609–1616. [PubMed] [Google Scholar]
- 22.Nowak-Węgrzyn A., Chehade M., Groetch M.E., Spergel J.M., Wood R.A., Allen K., et al. International consensus guidelines for the diagnosis and management of food protein-induced enterocolitis syndrome: executive summary-workgroup report of the Adverse Reactions to Foods Committee, American Academy of Allergy, Asthma & Immunology. J Allergy Clin Immunol. 2017;139:1111–1126.e4. doi: 10.1016/j.jaci.2016.12.966. [DOI] [PubMed] [Google Scholar]
- 23.Lane M.M., Czyzewski D.I., Chumpitazi B.P., Shulman R.J. Reliability and validity of a modified Bristol Stool Form Scale for children. J Pediatr. 2011;159:437–441.e1. doi: 10.1016/j.jpeds.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goswami R., Blazquez A.B., Kosoy R., Rahman A., Nowak-Węgrzyn A., Berin M.C. Systemic innate immune activation in food protein-induced enterocolitis syndrome. J Allergy Clin Immunol. 2017;139:1885–1896.e9. doi: 10.1016/j.jaci.2016.12.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berin M.C. Advances in understanding immune mechanisms of food protein-induced enterocolitis syndrome. Ann Allergy Asthma Immunol. 2021;126:478–481. doi: 10.1016/j.anai.2021.01.033. [DOI] [PubMed] [Google Scholar]
- 30.Mennini M., Fierro V., Di Nardo G., Pecora V., Fiocchi A. Microbiota in non-IgE-mediated food allergy. Curr Opin Allergy Clin Immunol. 2020;20:323–328. doi: 10.1097/ACI.0000000000000644. [DOI] [PubMed] [Google Scholar]
- 31.Sorbara M.T., Littmann E.R., Fontana E., Moody T.U., Kohout C.E., Gjonbalaj M., et al. Functional and genomic variation between human-derived isolates of lachnospiraceae reveals inter- and intra-species diversity. Cell Host Microbe. 2020;28:134–146.e4. doi: 10.1016/j.chom.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vacca M., Celano G., Calabrese F.M., Portincasa P., Gobbetti M., De Angelis M. The controversial role of human gut lachnospiraceae. Microorganisms. 2020;8:573. doi: 10.3390/microorganisms8040573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz Y., Goldberg M.R., Rajuan N., Cohen A., Leshno M. The prevalence and natural course of food protein-induced enterocolitis syndrome to cow's milk: a large-scale, prospective population-based study. J Allergy Clin Immunol. 2011;127:647–653.e1-3. doi: 10.1016/j.jaci.2010.12.1105. [DOI] [PubMed] [Google Scholar]
- 34.Penders J., Gerhold K., Stobberingh E.E., Thijs C., Zimmermann K., Lau S., et al. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J Allergy Clin Immunol. 2013;132:601–607.e8. doi: 10.1016/j.jaci.2013.05.043. [DOI] [PubMed] [Google Scholar]
- 35.Chu D.M., Ma J., Prince A.L., Antony K.M., Seferovic M.D., Aagaard K.M. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23:314–326. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyer J., Sgambelluri L., Yuan Q. Association of antibiotic usage with food protein-induced enterocolitis syndrome development from a caregiver's survey. JPGN Rep. 2021;2:e132. doi: 10.1097/PG9.0000000000000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang W., Su J., Zhang X., Cheng X., Zhou J., Shi R., et al. Elevated levels of Th17 cells and Th17-related cytokines are associated with disease activity in patients with inflammatory bowel disease. Inflamm Res. 2014;63:943–950. doi: 10.1007/s00011-014-0768-7. [DOI] [PubMed] [Google Scholar]
- 38.Natividad J.M., Pinto-Sanchez M.I., Galipeau H.J., Jury J., Jordana M., Reinisch W., et al. Ecobiotherapy rich in firmicutes decreases susceptibility to colitis in a humanized gnotobiotic mouse model. Inflamm Bowel Dis. 2015;21:1883–1893. doi: 10.1097/MIB.0000000000000422. [DOI] [PubMed] [Google Scholar]
- 39.Arpaia N., Campbell C., Fan X., Dikiy S., van der Veeken J., deRoos P., et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee M.J., Kang M.J., Lee S.Y., Lee E., Kim K., Won S., et al. Perturbations of gut microbiome genes in infants with atopic dermatitis according to feeding type. J Allergy Clin Immunol. 2018;141:1310–1319. doi: 10.1016/j.jaci.2017.11.045. [DOI] [PubMed] [Google Scholar]
- 41.Berni Canani R., Nocerino R., Terrin G., Coruzzo A., Cosenza L., Leone L., et al. Effect of Lactobacillus GG on tolerance acquisition in infants with cow's milk allergy: a randomized trial. J Allergy Clin Immunol. 2012;129 doi: 10.1016/j.jaci.2011.10.004. :580-2, 2.e1-5. [DOI] [PubMed] [Google Scholar]
- 42.Porras A.M., Shi Q., Zhou H., Callahan R., Montenegro-Bethancourt G., Solomons N., et al. Geographic differences in gut microbiota composition impact susceptibility to enteric infection. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109457. [DOI] [PMC free article] [PubMed] [Google Scholar]