Abstract
The nuclear receptor subfamily 5 group A member 1 (NR5A1), encoding steroidogenic factor 1 (SF-1), has been identified as a critical factor in gonadal development in animal studies. A previous study of ours suggested that upregulation of NR5A1 during early gonadal differentiation in male (46,XY) human pluripotent stem cells steers the cells into a more mature gonadal cell type. However, the detailed role of NR5A1 in female gonadal differentiation has yet to be determined. In this study, by combining the processes of gonadal differentiation and conditional gene activation, we show that NR5A1 induction predominantly upregulates the female gonadal marker inhibin subunit α (INHA) and steroidogenic markers steroidogenic acute regulatory protein (STAR), cytochrome P450 family 11 subfamily A member 1 (CYP11A1), cytochrome P450 family 17 subfamily A member 1 (CYP17A1), hydroxy-delta-5-steroid dehydrogenase (HSD3B2) and hydroxysteroid 17-beta dehydrogenase 1 (HSD17B1). In contrast, NR5A1 induction did not seem to affect the bipotential gonadal markers gata binding protein 4 (GATA4) and Wilms’ tumour suppressor 1 (WT1) nor the female gonadal markers r-spondin 1 (RSPO1) and wnt family member 4 (WNT4). Differentially expressed genes were highly associated with adrenal and ovarian steroidogenesis pathways. Moreover, time-series analysis revealed different dynamic changes between male and female induced samples, where continuously upregulated genes in female gonadal differentiation were mostly associated with adrenal steroidogenesis. Thus, in contrast to male gonadal differentiation, NR5A1 is necessary but not sufficient to steer human embryonic stem cell (hESC)-derived bipotential gonadal-like cells towards a more mature somatic, female cell fate. Instead, it seems to direct bipotential gonadal-like cells more towards a steroidogenic-like cell population. The information obtained in this study helps in elucidating the role of NR5A1 in gonadal differentiation of a female stem cell line.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13048-023-01264-5.
Keywords: CRISPR, NR5A1, SF-1, Orphan nuclear receptor, Ovary, Testis, Steroidogenesis, Gonadal development, Stem cells
Background
In humans, the gonads develop from the genital ridges, which are paired structures on the ventromedial surface of the mesonephros. Genital ridges form as a result of thickening of intermediate mesoderm and proliferation of overlaying coelomic epithelium during the latter part of the fifth week of development [1, 2]. At this stage the gonads are morphologically indistinguishable in both sexes and called bipotential gonads [3]. Sex differentiation occurs around the seventh week of development and is dependent on the chromosomal composition. A genetically male (XY) embryo has the sex-determining region on the Y chromosome (SRY), which initiates testicular development [4–6]. In the absence of SRY, thus in a genetically female (XX) embryo, ovarian development is initiated by the female-specific markers wnt family member 4 (WNT4) [7, 8], r-spondin 1 (RSPO1) and forkhead box protein L2 (FOXL2) [9–12].
The bipotential gonad expresses genes essential for gonadal development, including empty spiracles homeobox 2 (EMX2), lim homeobox 9 (LHX9), Wilms’ tumour suppressor 1 (WT1), gata binding protein 4 (GATA4) and nuclear receptor subfamily 5 group A member 1 (NR5A1) [13]. The NR5A1 gene encodes an orphan nuclear receptor, steroidogenic factor 1 (SF-1), which is a member of the nuclear receptor superfamily initially identified as a master-regulator of steroidogenic enzymes in adrenocortical cells [14–16].
NR5A1 plays an important role in the development and function of steroidogenic tissues, gonads and adrenal glands [17–19]. NR5A1 knock-out (KO) mice completely lack gonads and adrenal glands, present with female external genitalia irrespective of genetic sex and die within eight days after birth [17, 20, 21]. However, gonad-specific KO completely bypasses the lethality of NR5A1-null mice in both sexes [22, 23]. Testis-specific KO of NR5A1 causes abnormalities in the testes from very early developmental stages. The testes of adult KO mice were found to be at the level of the bladder and hypoplastic, partly due to impaired spermatogenesis [22]. In the case of ovary-specific KO of NR5A1, wild-type and KO ovaries were indistinguishable from each other during embryogenesis and at birth. However, adult females were found to be sterile, most likely due to defects in steroidogenesis [22, 23].
In humans, there are three recorded homozygous NR5A1 mutations (R92Q, D293N and R103Q). In 46,XY patients the symptoms range from primary adrenal insufficiency to complete gonadal dysgenesis and differences in sex development (DSD). Homozygous NR5A1 mutations have been found in 46,XX patients with primary adrenal insufficiency and primary ovarian failure [24]. Heterozygous NR5A1 mutations may rarely cause isolated adrenal insufficiency, but are usually accompanied by gonadal dysfunction and dysgenesis [25].
In one of our previous studies we showed that NR5A1 seems to direct male (XY) human induced pluripotent stem cells (hiPSC), differentiated into bipotential gonadal-like cells, towards more mature somatic gonadal-like cells [26]. In the present study, we conditionally activated NR5A1 in a female human embryonic stem cell (hESC) line during gonadal differentiation to study the role of NR5A1 in female gonadal differentiation. We showed that activating NR5A1 in female hESCs seems to steer bipotential gonadal-like cells towards a steroidogenic-like cell population instead of a more mature gonadal cell type, as is the case in gonadal differentiation of the male line.
Results
Bipotential and female gonadal markers are upregulated during gonadal differentiation
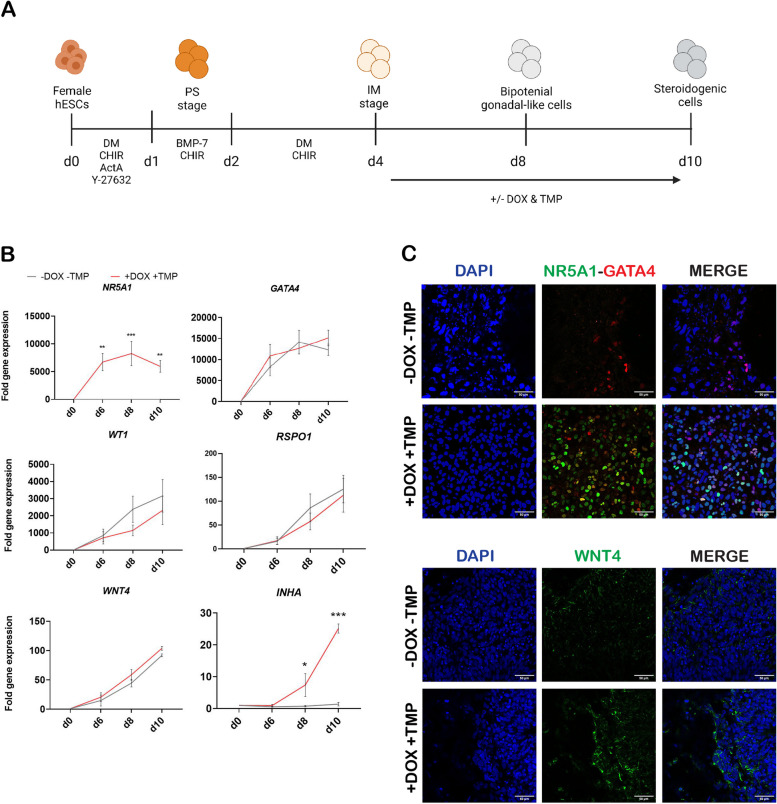
To study the role of NR5A1 in human embryonic development, a dual inducible NR5A1 activation line was established using the CRISPR/Cas9 genome editing technique, involving H9-NR5A1-DDdCas9VP192 hESC clonal line 10. This cell line was subjected to a gonadal differentiation protocol (Fig. 1A), based on a previous protocol established by Sepponen et al., 2017 [27]. NR5A1 was induced by the simultaneous administration of doxycycline hyclate (DOX) and trimethoprim (TMP) on day 4 (at the intermediate mesoderm stage) until day 10 of differentiation. RT-qPCR analyses showed that in the induced state (+ DOX + TMP), NR5A1 was significantly upregulated at days 6, 8 and 10 of differentiation, whilst being completely absent in the non-induced state (-DOX -TMP) (Fig. 1B). During the differentiation process, the bipotential markers GATA4 and WT1 were upregulated by day 8 of differentiation, but NR5A1 induction did not seem to affect their gene expression levels. A similar trend was seen for the female gonadal markers RSPO1 and WNT4 by day 10 of gonadal differentiation. However, the ovarian marker inhibin subunit α (INHA) was significantly upregulated on days 8 and 10 of gonadal differentiation by NR5A1 induction (Fig. 1B).
Fig. 1.
NR5A1 induction has a minimal effect on the upregulation of bipotential gonadal and female markers. A A schematic representation of the gonadal differentiation protocol from hESCs to steroidogenic-like cells with the matching developmental stages and administered growth factors, inhibitors, and small molecules. B RT-qPCR analysis of markers associated with gonadal development showed that NR5A1 induction was successful. Bipotential gonadal markers (NR5A1, GATA4, WT1) were upregulated by day 8, female gonadal markers (RSPO1, WNT4) were upregulated by day 10 as a result of gonadal differentiation even without NR5A1 induction and INHA was significantly upregulated after NR5A1 induction. Data are reported as mean ± SEM, n=3 biological replicates. Two-way ANOVA; 0.12 (ns), 0.033 (*), 0.002 (**), < 0.001 (***). C Confocal images of immunofluorescence staining showed expression of bipotential markers NR5A1 and GATA4, and the female gonadal marker WNT4 in non-induced (-DOX -TMP) and induced (+DOX +TMP) conditions at day 10 of gonadal differentiation. Images were taken with a ×40 objective. Scale bars 50 µm. ActA, activin A; BMP, bone morphogenetic protein; CHIR, CHIR-99021; DM, dorsomorphin; hESCs, human embryonic stem cells; IM, intermediate mesoderm; PS, primitive streak; d, day of differentiation; DOX, doxycycline hyclate; TMP, trimethoprim
Immunofluorescence co-staining of NR5A1 and GATA4 at day 10 of differentiation showed the presence of NR5A1 in the induced state, whereas it was completely undetectable in the non-induced state (Fig. 1C). GATA4, on the other hand, was present in both conditions at similar staining intensities. Likewise, WNT4 was expressed in both conditions with a similar intensity in the non-induced and induced state (Fig. 1C).
NR5A1 induces steroidogenesis during gonadal differentiation
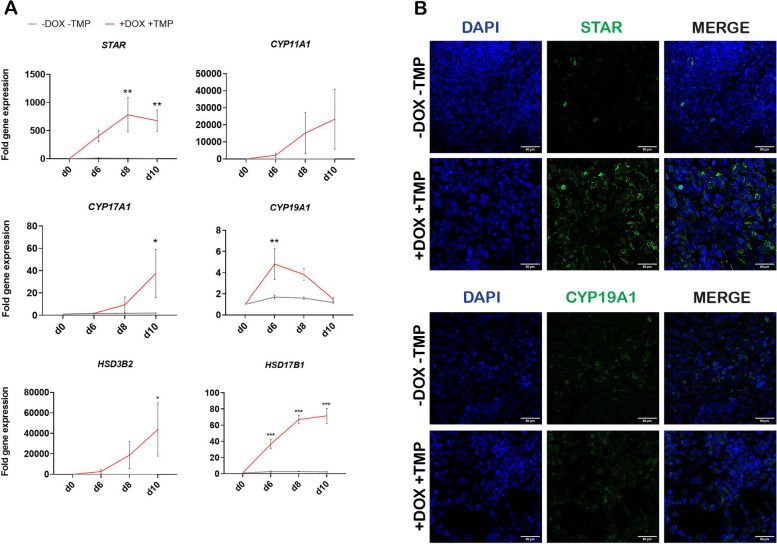
NR5A1 induction resulted in upregulation of several steroidogenesis markers. RT-qPCR analyses showed that levels of steroidogenic acute regulatory protein (STAR) mRNA were increased on day 6 and significantly increased on days 8 and 10 of differentiation in the induced state, but no STAR mRNA expression was detectable in the non-induced state (Fig. 2A). Similarly, cytochrome P450 family 11 subfamily A member 1 (CYP11A1) and cytochrome P450 family 17 subfamily A member 1 (CYP17A1) mRNA expression levels were upregulated on days 6, 8 and 10 upon NR5A1 induction, with the increase in CYP17A1 being significant on day 10. Interestingly, cytochrome P450 family 19 subfamily A member 1 (CYP19A1) levels decreased over the course of gonadal differentiation and upon NR5A1 induction, with the decrease on day 6 being statistically significant. Hydroxy-delta-5-steroid dehydrogenase (HSD3B2) showed a continuous upregulation upon NR5A1 induction, with the increase in gene expression being significant on day 10. Lastly, hydroxysteroid 17-beta dehydrogenase 1 (HSD17B1) showed a similar trend with the upregulation being significant across all timepoints (Fig. 2A).
Fig. 2.
NR5A1 induction upregulates steroidogenesis markers but downregulates the aromatase enzyme CYP19A1. A RT-qPCR analysis of markers associated with steroidogenesis showed that NR5A1 induction upregulated the markers STAR, CYP11A1, CYP17A1, HSD3B2 and HSD17B1, but downregulated the aromatase enzyme CYP19A1. Data are reported as mean ± SEM, n=3 biological replicates. Two-way ANOVA; 0.12 (ns), 0.033 (*), 0.002 (**), < 0.001 (***). B Confocal images of immunofluorescence staining showed expression of steroidogenic markers STAR and CYP19A1 in non-induced (-DOX -TMP) and induced (+DOX +TMP) conditions at day 10 of gonadal differentiation. Images were taken with a ×40 objective. Scale bars 50 µm. d, day of differentiation; DOX, doxycycline hyclate; TMP, trimethoprim
Immunofluorescence staining at day 10 of gonadal differentiation showed minimal presence of STAR in the non-induced state, whereas its staining intensity was increased in the induced condition. In contrast, there was little difference in CYP19A1 expression between the two conditions (Fig. 2B).
NR5A1 induces transcriptional changes during differentiation of bipotential gonadal-like cells
To further study the effect of NR5A1 expression during differentiation, we performed bulk RNA sequencing (RNA-seq) analyses on NR5A1-induced (IND, +DOX + TMP) and non-induced (CTRL, -DOX -TMP) samples taken on days 4, 6, 8 and 10 of differentiation. Principal Component Analysis (PCA) of the top 500 most variable genes showed clear separate clustering of the induced and non-induced samples, as well as clear divergence between the different days of differentiation (Suppl. Figure 1 A). According to the PCA results, in gonadal differentiation of the female line, about 52% of the variance observed between the samples could be attributed to the differentiation protocol itself (Principal Component 1, PC1) and 34% of the variance was due to NR5A1 induction (Principal Component 2, PC2) (Suppl. Figure 1 A).
Furthermore, we performed pairwise differential expression (DE) comparisons between samples at different timepoints and between induced (IND, +DOX + TMP) and non-induced (CTRL, -DOX-TMP) conditions at a specific timepoint during differentiation. Additionally, we compared the number of differentially expressed genes (DEGs) (coding and non-coding, upregulated and downregulated) in the female line and the male line, the latter having been previously reported [26] (Suppl. Figure 1B). Two main differences were found between the female and male lines undergoing gonadal differentiation as regards DEGs. Firstly, in the female line the highest number of DEGs was found at day 6 of differentiation (3,496), whilst in the male line this was at day 10 of differentiation (5,721). Secondly, there were more upregulated DEGs at each timepoint in the female line subjected to gonadal differentiation, whilst in the male line at each timepoint there were more downregulated DEGs. However, one similarity was found: at each timepoint in both the female and male lines the coding DEGs dominated over the non-coding DEGs in conditions of both upregulated and downregulated DEGs (Suppl. Figure 1B).
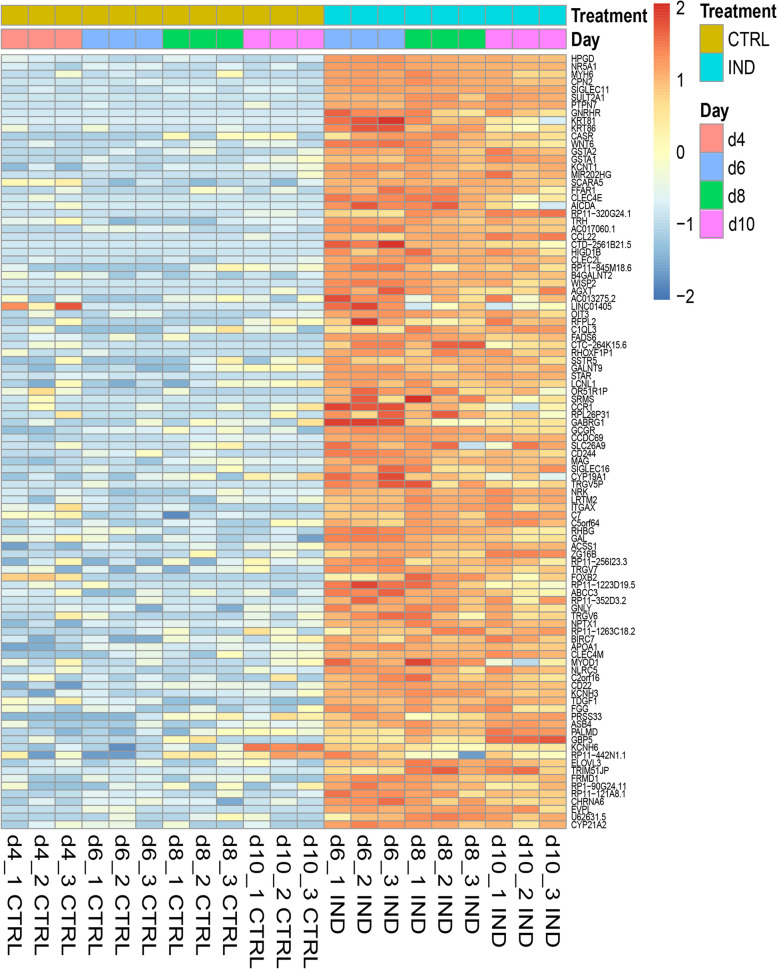
Next, we plotted a heatmap to represent the expression profile of the top 100 upregulated DEGs with the highest fold change at day 6 of gonadal differentiation in the female line. NR5A1 induction was followed by clear upregulation of this set of genes at all timepoints studied (Fig. 3). We selected several genes from this list that were known to be associated with female somatic gonadal cells or steroidogenesis and performed RT-qPCR analyses to validate the RNA-seq results (Suppl. Figure 2 A). The results showed that indeed 15-hydroxyprostaglandin dehydrogenase (HPGD), glutathione S-transferase A1 (GSTA1), glutathione S-transferase A2 (GSTA2), wnt family member 6 (WNT6), beta-1,4-N-acetyl-galactosaminyltransferase 2 (B4GALNT2) and gonadotropin releasing hormone receptor (GNRHR) mRNA expression levels were induced upon NR5A1 induction with DOX and TMP (Suppl. Figure 2 A).
Fig. 3.
Expression pattern of the top 100 upregulated DE genes at day 6 of differentiation upon NR5A1 induction
The heat map shows comparison of non-induced (CTRL, -DOX-TMP) and induced (IND, +DOX + TMP) RNA-sequenced samples across all timepoints. The top 100 DEGs at day 6 of differentiation are displayed and each rectangle represents the expression of a specific gene within a technical replicate. The intensity of gene expression is indicated by a colour scale based on row z-scores (red, highest expression levels; blue, lowest expression levels). d, day.
To further understand the effect of NR5A1 induction on the differentiation of bipotential gonadal-like cells in the female line, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses on the DEGs at days 6, 8 and 10 of differentiation using the online ENRICHR tool (Suppl. Figure 2B). ‘Cortisol synthesis and secretion’ was the dominant associated pathway on days 6, 8 and 10 of differentiation. ‘Aldosterone synthesis and secretion’ was the second most associated pathway with DEGs on day 6, whilst on day 8 this was ‘cell adhesion molecules’, and ‘neuroactive ligand-receptor interaction’ on day 10. Across all three timepoints, the KEGG pathway ‘ovarian steroidogenesis’ was also associated with upregulated DEGs (Suppl. Figure 2B).
Lastly, we looked at the expression profiles of genes known to be associated with ovarian, testicular or adrenal steroidogenesis from previously published papers by Mamsen et al. [28], del Valle et al. [29] and Lecluze et al. [30]. Most of the genes associated with steroidogenesis were clearly upregulated in our datasets upon NR5A1 activation (Suppl. Figure 3).
Different dynamic changes in expression pattern were induced by NR5A1 in the female and male human pluripotent stem cell (hPSC) lines
In normal gonadal development the future fate of bipotential gonadal cells is largely dependent on the presence or absence of SRY. Moreover, ovaries and testes are inherently different organs that develop differently. Therefore, the sequencing data from gonadal differentiation of the female line was further compared with the previously published sequencing data from gonadal differentiation of the male line [26] in both control and induced conditions. We performed time-series analysis on the NR5A1-induced DEGs to identify the genes that presented dynamic changes in their expression paths during differentiation. These paths were grouped first according to their expression in either female or male samples and induced and non-induced conditions. Secondly, they were grouped according to their direction of expression at each timepoint. For example, genes showing continuous upregulation over the timepoints (d4 > d6 > d8 > d10) were labelled as up-up-up.
Table 1 shows an overview of the number of dynamic DEGs in induced and non-induced conditions in the female and male lines that were subjected to gonadal differentiation. Genes with a MaxPP value greater than 0.5 were considered to be high-confidence dynamic DEGs. In the induced condition of the female line about 8,193 genes were dynamic DEGs, of which 2,441 were high-confidence, whereas in the non-induced condition the figures were 7,127 and 2,595, respectively. In comparison, the induced condition of the male line showed 12,581 dynamic DEGs, of which 3,620 were high-confidence and in the non-induced condition there were 12,963 dynamic DEGs, of which 3,983 were high-confidence. The table also shows the overlap in dynamic DEGs between female and male lines in induced and non-induced time-series. We focused specifically on the DEGs induced in only female or male samples.
Table 1.
Differential expression – time-series analysis comparison of the female and male lines
| Induced | Non-induced | |||
|---|---|---|---|---|
| Female | All dynamic DEGs (high confidence MaxPP > 0.5) | 8193 (2441) | 7127 (2595) | |
| Male | All dynamic DEGs (high confidence MaxPP > 0.5) | 12,581(3620) | 12,963 (3983) | |
| Induced time-series | Non-induced time-series | |||
| Female-Male overlap | Only in female induced (high confidence) | 2004 (607) | Only in female non-induced (high confidence) | 3808 (1513) |
| In females, induced or non-induced but not male induced (high confidence) | 4423 (1484) | |||
| In females, induced AND non-induced (high confidence) | 2419 (877) | |||
| Only in males (high confidence) | 2296 (834) | Only in males non-induced (high confidence) | 9613 (2998) | |
| In males, induced or non-induced but not female induced (high confidence) | 8779 (2682) | |||
| In males, induced AND non-induced (high confidence) | 6483 (1848) | |||
| Shared in females and males, induced, same pattern | 770 (294) | Shared in females and males, non-induced, same pattern | 647 (313) | |
| Shared in females and males, induced, different pattern | 2968 (738) | Shared in females and males, non-induced, different pattern | 2641 (801) |
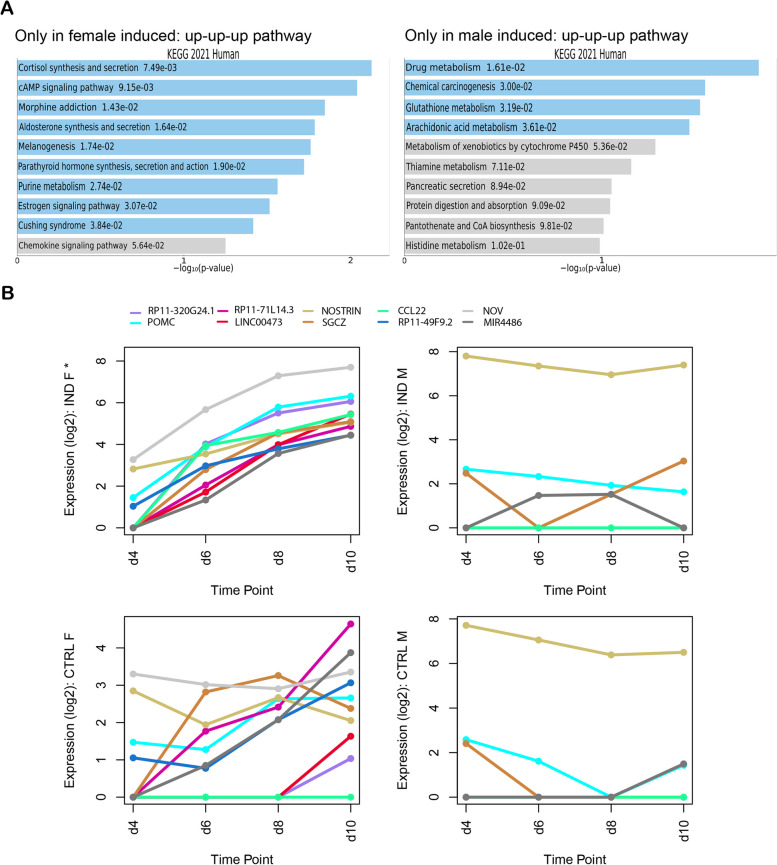
About 2,004 genes were expressed in ‘Only in female induced’ samples, meaning that these genes showed dynamic changes in their expression pattern during gonadal differentiation of female cells upon NR5A1 activation. Of these genes, 96 could be grouped in the up-up-up pathway and 40 of them showed significant upregulation (MaxPP > 0.5). These genes were mostly associated with ‘cortisol synthesis and secretion’ and ‘cAMP signalling’ KEGG pathways. Several of these genes, including POMC, CCL22, NOV (CCN3) and ADCY4 have previously been associated with expression or action in the adrenal gland/adrenocortical cells [31–34]. The gene with the highest MaxPP in the female induced up-up-up pathway was RP11-320G24.1, followed closely by POMC. RP11-320G24.1 is a long non-coding RNA (lncRNA) that is part of a tissue-specific co-expression gene regulatory network in the adrenal gland. It has been reported to have a positive correlation with NR5A1, amongst other genes [35]. Pro-opiomelanocortin (POMC), on the other hand, is precursor peptide of adrenocorticotrophic hormone (ACTH) [31].
In the ‘Only in male induced’ samples, 2,296 genes were expressed that showed dynamic changes in their expression patterns during male gonadal differentiation after NR5A1 activation. A total of 133 genes could be grouped into the up-up-up pathway, 98 of which showed significant upregulation (MaxPP > 0.5). These genes were largely associated with ‘drug metabolism’ and ‘chemical carcinogenesis’ KEGG pathways. The gene with the highest MaxPP value in the up-up-up pathway was tescalcin (TESC), a transcript expressed during the early stages of testicular differentiation in mice [36, 37] (Fig. 4A).
Fig. 4.
Time-series analysis demonstrated different dynamic changes in gene expression between female and male samples. A Bar charts showing the correlated KEGG 2021 human pathways of the continuously upregulated (up-up-up) genes in female induced samples across the different timepoints in gonadal differentiation. The bar charts show the top 10 enriched terms in the chosen sample set, along with their corresponding p-values. Coloured bars correspond to terms with significant p-values (< 0.05). B Log2 plots showing the top 10 continuously upregulated genes (RP11-320G24.1, POMC, RP11-71L14.3, LINC00473, NOSTRIN, SGCZ, CCL22, RP11-491F9.2, NOV and MIR4486) in the female induced samples (IND F) compared with their expression in control female (CTRL F), induced male (IND M) and control male (CTRL M) samples. The asterisk (*) symbolizes significant expression of these genes
Figure 4B shows the expression patterns across the different timepoints of differentiation of the top 10 (10 highest MaxPP values) up-up-up genes from the ‘Only in female induced’ samples. The genes (RP11-320G24.1, POMC, RP11-71L14.3, LINC00473, NOSTRIN, SGCZ, CCL22, RP11-491F9.2, NOV and MIR4486) are plotted in log2 values and compared against their expression in female control, male induced and male control samples. As these genes belong to the up-up-up pathway of the ‘Only in female induced’ samples, they showed an upward trend in the female induced samples and a slight upward trend in the female control samples, but their expression remained constant and low in the male samples. Moreover, some of the genes were not even expressed in the male samples (Fig. 4B).
Taken together, different genes showed continuous upregulation in the female induced and male induced samples.
Discussion
In this study, we showed that inducing NR5A1 in a female (46,XX) hESC line, which has been differentiated to bipotential gonadal-like cells, steered these cells towards a steroidogenic-like cell population, but did not seem to promote ovarian differentiation. Bipotential gonadal markers were upregulated by day 8 of differentiation, which is a result of the differentiation process rather than the effect of NR5A1 induction. By day 10 of gonadal differentiation, we saw upregulation of early ovarian markers such as RSPO1, WNT4 and INHA, but for RPSO1 and WNT4 this upregulation was not due to NR5A1 induction but again a result of the differentiation protocol itself. NR5A1 induction, however, significantly upregulated the expression of the steroidogenic genes STAR, CYP11A1, CYP17A1, HSD3B2 and HSD17B1. Interestingly, expression of the aromatase gene (CYP19A1), which is expressed by the granulosa cells [38], was relatively low and was decreased significantly on day 6 of differentiation due to NR5A1 induction and continued to decrease over the course of differentiation. These are the first indicators that NR5A1 directs the bipotential gonadal-like cells towards a steroidogenic-like cell population in a female cell line.
These interpretations of the results are also supported by our bulk RNA-seq data. According to the KEGG pathway analyses, NR5A1-induced pathways seem to be predominantly associated with ‘cortisol and aldosterone synthesis and secretion’ and not with ovarian-type steroidogenic pathways. Moreover, the top 20 DEGs in the non-induced versus the induced female samples at day 6 of differentiation were HPGD, NR5A1, MYH6, CPN2, SIGLEC11, SULT2A1, PTPN7, GNRHR, KRT81, KRT86, CASR, WNT6, GSTA1, GSTA2, KCNT1, MIR202HG, SCARA5, FFAR1, CLEC4E and AICDA. Among these, HPGD, SULT2A1, GNRHR and GSTA1 have previously been associated with adrenal steroidogenesis [29, 39]. Time-series analysis further confirmed the presence of a steroidogenic cell-like population in the female induced samples, as continuously upregulated genes were again mostly associated with ‘cortisol synthesis and secretion’. Together, these results suggest that NR5A1 induction in a female stem-cell line promotes the generation of a predominantly steroidogenic-like cell population possibly resembling an adrenal cell type instead of a more mature female gonadal-like cell type, as is the case in gonadal differentiation of the male cell line [26].
RNA-seq further highlighted some key differences observed between gonadal differentiation of the female and male lines. The PCAs showed that PC2 in gonadal differentiation of the female line explains the greater variation in the data than PC2 in gonadal differentiation of the male line [26] (PC2 values 34% and 18%, respectively). As both PCs depicted similar levels of total variation in the respective data sets, these data suggest that NR5A1 induction has a wider impact on the female line. Pairwise DE analysis showed that in the female cell line, the highest number of DEGs could be found at day 6 of gonadal differentiation, while in the male cell line this was on day 10. Thus, the largest effect due to NR5A1 induction seems to occur earlier during gonadal differentiation of the female line than of the male line. The pairwise DE analysis also showed that there were in general more upregulated DEGs during gonadal differentiation of the female line, whilst downregulated DEGs dominated in the male. Next, DE time-series analysis was performed and compared with the corresponding male DE time-series analysis. Interestingly, different genes showed dynamic changes in their expression patterns in female versus male induced samples. This further demonstrates the divergence in differentiation patterns in female and male hPSCs. Whereas in female induced samples RP11-320G24 and POMC were the two most highly continuously upregulated genes, TESC was the top continuously upregulated gene in the male induced samples. As mentioned previously, RP11-320G24.1 is part of an adrenal gland tissue-specific co-expression gene regulatory network and has been shown to have a positive correlation with NR5A1 [35]. POMC is generally found in the hypothalamus, yet mRNA transcripts have also been found in the normal adrenal gland and the ovary [40]. POMC can be cleaved and produces ACTH, which regulates adrenal steroidogenesis. While ACTH production mainly takes place within the corticotrophic cells of the anterior pituitary, its expression has also been found in the adrenal medulla, the brain, the skin and the placenta [31]. TESC, on the other hand, is a known testicular transcript expressed during the early stages of testicular differentiation [36, 37].
As for potential targets of NR5A1, gonadotrophin releasing hormone receptor (GNRHR) might be a putative target for the processes of gonadal differentiation of the female line and steroidogenesis. GNRHR has been known to be expressed in normal human reproductive tissues such as the ovary [41] and additionally its mRNA was highly expressed in our NR5A1 induced cells. Moreover, gonadotrophin releasing hormone (GNRH) has been known to directly regulate the activity of steroidogenic enzymes such as CYP19A1 [42]. Thus, it is possible that GNRH can mediate the steroidogenic effect of NR5A1. In the male hPSC model, scavenger receptor class a member 5 (SCARA5) had already been recognised as a putative target of NR5A1 by Sepponen et al. [26]. Our data supports the finding of SCARA5 being a putative target of NR5A1, as SCARA5 is among the top 20 DEGs in the non-induced and induced genes at day 6 of differentiation of the female line.
As the focus of our study was to investigate the impact of NR5A1 expression on female bipotential gonadal-like cells, the identity of these steroidogenic-like cells was not thoroughly elucidated. The low CYP19A1 mRNA and protein expression, together with the general increase in steroidogenic gene expression, might be an indication of the progression of bipotential gonadal-like cells towards an adrenocortical population, as expression of CYP19A1 has been reported to be relatively low in the normal adult adrenal gland [43]. However, our cells represent early fetal cells and not adult cells, and therefore certain genes (such as CYP19A1) could be expressed at levels different to those reported in adults. Moreover, a significant upregulation of INHA following NR5A1 induction was observed. INHA is known to be produced by human granulosa cells, but also by the steroidogenic theca cells [44, 45]. Furthermore, ‘ovarian steroidogenesis’ is one of the KEGG pathways that was significantly expressed during DE analysis on days 6, 8 and 10 of differentiation. Therefore, the presence of some female steroidogenic cells, theca cells, in the heterogenous pool of cells, cannot be excluded. Additionally, in male gonadal differentiation the male pre-gonadal cells also strongly express steroidogenic enzymes and secrete steroid hormones [26]. Thus, it might be possible that the steroidogenic cells observed here could represent (at least partly) pre-granulosa cells in the early stages of development. Most likely, the steroidogenic-like cell population might be a mixture of adrenocortical and thecal cells amongst other cell types.
Our results are in line with those reported by Smith et al. [46], where they created a CYP17A1 conditional knock-out mouse (cKO) model, where NR5A1 was depleted in the steroidogenic Leydig cells and theca cells of mature mice. With this CYP17A1 cKO model they showed that NR5A1 depletion in Leydig cells of mature male mice can result in male infertility, whilst depletion of NR5A1 in theca cells of mature female mice mainly caused impaired steroidogenesis, with fertility being preserved. Moreover, in 2019 Stévant et al. [47] showed that NR5A1-positive progenitors in fetal female (XX) mice give rise to both pre-granulosa cells and steroidogenic precursor cells. This suggests that indeed NR5A1 might not be as essential in female gonadal development as it is for male gonadal development but has a more prominent role in steroidogenesis.
Limitations of this study include the unclear identity of the cell type obtained by the end of our gonadal differentiation protocol. Although it is fairly certain that the generated cells are steroidogenic, it remains uncertain what type of steroidogenic cells they actually are (ovarian, testicular or adrenal). Moreover, it must be noted that there was a lack in upregulation of specific ovarian markers during gonadal differentiation. Thus, despite generating bipotential gonadal-like and steroidogenic cells, our protocol (including NR5A1 activation) did not seem to completely reflect the developmental process of female gonadal cells. Additionally, it is very clear that the cell population was relatively heterogeneous even though the cells were directed towards a specific cell lineage. As a technical factor, the expression levels of NR5A1 differed in the female and male lines, with levels in the female line being 5–10 times higher, which may have contributed to the gene expression observed. Another limitation of this study, and related to expression levels, is the use of artificial induction of NR5A1. As yet, we have not been able to establish a differentiation protocol where NR5A1 could be induced endogenously without the use of genome-editing techniques. Thus, we do not know how high the natural gene expression level of NR5A1 during gonadal differentiation might be. This makes it difficult to compare the in vitro with the in vivo situation and thus our current protocol remains unsuitable for clinical applications where patient lines could be reprogrammed and differentiated.
Conclusions
We conclude that during gonadal differentiation of a female hPSC line, NR5A1 seems to steer hESC-derived bipotential gonadal-like cells towards a heterogeneous, steroidogenic-like cell population. This differs from gonadal differentiation of a male hPSC line, where NR5A1 directs the hiPSCs towards a more mature, gonadal-like cell type. Thus, NR5A1 is a necessary factor in gonadal differentiation of a female cell line yet does not seem to be sufficient to direct the gonadal-like cells towards a granulosa cell-like state. The findings in this study could aid in elucidation of the mechanisms underlying human fetal gonadal development.
Materials and methods
Cell culture
All cells were cultured in a humidified incubator supplied with 5% CO2 at 37 °C. H9-NR5A1-DDdCas9VP192 (clone 10) hESCs were cultured on Geltrex® LDEV-free human embryonic stem cell-qualified-reduced growth factor basement membrane matrix (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) or growth factor-reduced Matrigel (Corning, NY, USA) in tissue culture-treated dishes (Corning, NY, USA) in Essential 8 (E8) medium (Thermo Fisher Scientific, Waltham, MA, USA). The cells were routinely passaged every 4–5 days. For routine passaging, the cells were incubated in 0.5 mM EDTA (Invitrogen™, Thermo Fisher Scientific, CA, USA) in phosphate-buffered saline (PBS) for 4 min.
Differentiation of the hESCs was performed as previously described (condition M, Sepponen et al., 2017). Undifferentiated hESCs were dissociated with 0.5 mM EDTA, counted, and seeded at a density of 2.24 × 105 cells/cm2 on tissue culture-treated 12-well plates (Corning, NY, USA) for the collection of RNA, or on µ-slide 4 wells (IBIDI, Gräfelfing, Germany) for immunofluorescence staining. Both 12-well culture plates and µ-slides were coated with human placental-derived type I collagen (Corning, NY, USA). Prior to seeding the cells were centrifuged at 60 × g for 5 min and resuspended in intermediate mesoderm (IM) medium [DMEM/F12 (Gibco™, Thermo Fisher Scientific, Waltham, MA, USA) + Glutamax (Thermo Fisher Scientific, Waltham, MA, USA) supplemented (2%) with B-27™ Supplement (Thermo Fisher Scientific, Waltham, MA, USA)]. The medium was additionally supplemented with 10 µM rho kinase inhibitor (ROCKi, Y-27,632 2HCL, Selleckchem, Houston, TX, USA), activin A (Q-kine, Cambridge, UK) at 100 ng/mL, 5 µM GSK-3α/β inhibitor (CHIR-99,021, Selleckchem, Houston, TX, USA) and 2 µM BMP inhibitor (dorsomorphin, Selleckchem, Houston, TX, USA). On the following day, the media were changed to IM medium supplemented with BMP7 (Peprotech, Cranbury, NJ, USA) at 10 ng/mL and 3 µM CHIR-99,021. After 24 h, the medium was replaced with IM medium containing 2 µM dorsomorphin and 3 µM CHIR-99,021, which was kept on the cells for 48 h. Between media changes, the cells were washed once with PBS containing Mg2+ and Ca2+ (Gibco™, Thermo Fisher Scientific, Waltham, MA, USA). On day 4 until day 10 of differentiation, NR5A1 was induced by supplementing IM medium with DOX (Sigma-Aldrich, Saint Louis, MO, USA) at 1 µg/mL and 1 µM TMP (Sigma-Aldrich, Saint Louis, MO, USA). Most of the induction medium was changed daily.
Generating H9-NR5A1-DDdCas9Vp192 hESCs
The H9 cell line (WiCell Research Institute, Madison, WI, USA) was engineered to express a DDdCas9-VP192 construct under the control of a DOX-inducible promoter.
PiggyBac plasmids PB-GG-NR5A1-PGK-Puro (containing four guides targeting NR5A1 promoter, described by Sepponen et al., 2022), PB-tight-DDdCas9VP192-T2A-GFP-IRES-Neo (RRID:Addgene_ 102,889, Weltner et al., 2018) and PB-CAG-rtTA (kindly provided by the Biomedicum Stem Cell Centre, University of Helsinki, Helsinki, Finland), together with a PB transposase (kindly provided by the Biomedicum Stem Cell Centre, University of Helsinki, Helsinki, Finland), were simultaneously introduced to H9 hESCs. Cells were electroporated as described previously (Sepponen et al., 2022). Briefly, H9 cells were incubated with 10 µM Y-27,632 2HCl in E8 Basal Medium for 4 h prior to dissociation with StemPro® Accutase® (GibcoTM, Thermo Fisher Scientific, Waltham, MA, USA). They were rinsed with PBS and trypsinised with Accutase for 5 min at 37 ˚C. Subsequently, dissociated cells were resuspended in 5% fetal bovine serum (FBS) in PBS and counted. The cells were pelleted and resuspended in R buffer (Invitrogen™, Thermo Fisher Scientific, CA, USA). Two million cells, 1 µg PB-GG-NR5A1-PGK-Puro, 1 µg PB-tight-DDdCas9VP192-T2A-GFP-IRES-Neo, 0.5 µg PB-CAG-rtTA and 0.5 µg PB transposase were used for each transfection, using the NeonTM Transfection System (Invitrogen™, Thermo Fisher Scientific, CA, USA). The electroporation program settings have been described previously (Balboa et al., 2015 [48]; Sepponen et al., 2022 [26]). The cells were plated on Geltrex® (Gibco, Thermo Fisher Scientific, Waltham, MA, USA)-coated dishes for 24 h, followed by a change of medium (E8).
Selection of colonies with guides targeting NR5A1 was started 72 h after transfection with puromycin at 0.5 µg/mL for 2 days and at 0.25 µg/mL for 4 days. Cells were cultured in fresh E8 medium without antibiotics for 4 days and passaged. Following this, colonies containing DDdCas9 were selected with G418 sulphate (Merck Millipore, Darmstadt, Germany) at 0.25 mg/mL and DOX at 0.5 µg/mL for 3 days, followed by selection with G418 at 0.1 mg/mL and DOX at 0.5 µg/mL for an additional 3 days. To obtain colonies from single clones, cells were sorted with Influx equipment (BD, Biosciences) into microwell 96-well plates (Nunc, Thermo Fisher Scientific, Rochester, NY, USA), picked and expanded as described previously (Sepponen et al., 2022 [26]).
Immunofluorescence and confocal microscopy
At day 10 of differentiation, cells on µ-slide 4 wells were washed once with PBS + Mg2+, +Ca2+ and fixed with 4% paraformaldehyde at room temperature for 15 min, after which they were washed three times with PBS (Medicago, Uppsala, Sweden). Next, the cells were permeabilised with 0.5% Triton® X-100 (Fisher Scientific, Waltham, MA, USA), washed three times with 0.1%Tween® (Fisher Scientific, Waltham, MA, USA) in PBS and blocked with UltraVision Protein Block (Fisher Scientific, Waltham, MA, USA) at room temperature for 10 min. The cells were then incubated with primary antibodies diluted in 0.1% Tween in PBS overnight at 4 °C: mouse monoclonal anti-SF1 (R and D Systems, Cat# PP-N1665-00, RRID:AB_2251509, 1:250), goat polyclonal anti-FOXL2 (Novus Biologicals, Cat# NB100-1277, RRID:AB_2106187, 1:250), goat polyclonal anti-GATA4 (Santa Cruz Biotechnology, Cat# sc-1237, RRID:AB_2108747, 1:250), mouse monoclonal anti-WNT4 (Santa Cruz Biotechnology, Cat# sc-376,279, RRID:AB_10986273, 1:250), mouse monoclonal anti-CYP19A1 (Santa Cruz Biotechnology, Cat# sc-374,176, RRID:AB_10986411, 1:500), and mouse monoclonal anti-STAR (Santa Cruz Biotechnology, Cat# sc-166,821, RRID:AB_2197661, 1:250). Subsequently, the cells were washed three times with 0.1% Tween in PBS. Next, cells were incubated with the following secondary antibodies diluted 1:1000 in 0.1% Tween in PBS: Alexa Fluor 488 donkey anti-mouse immunoglobulin G (IgG) (Molecular Probes, Cat# A-21,202, RRID:AB_141607) or Alexa Fluor® 594 donkey anti-goat IgG (Thermo Fisher Scientific, Cat# A-11,058, RRID:AB_2534105). Incubation was carried out for 45 min at room temperature in the dark. Next, the cells were washed twice with 0.1% Tween in PBS. Nuclei were labelled with 4’,6-diamidino-2-phenylindole (DAPI) dilactate (Invitrogen, Thermo Fisher Scientific, Cat#D3571, RRID:AB_2307445) at a 1:1000 ratio in 0.1% Tween in PBS for 10 min in the dark and washed twice with 0.1% Tween in PBS.
Images were captured with a TCS SP8 confocal microscope with a white laser (Leica Microsystems, Mannheim, Germany) using an 812 × 812 format and an HC PL APO CS2 40x/1.30 oil objective. Images were processed using Fiji version 2.3.0 equipment (http://fiji.sc).
Real time quantitative polymerase chain reactions (RT-qPCRs)
Total RNA was isolated using Nucleospin® RNA kits (Macherey-Nagel, Düren, Germany). Genomic DNA was removed separately by using ribonuclease-free deoxyribonuclease (Promega, Madison, WI, USA), after which RNA was again purified (Nucleospin® RNA clean-up kits). Reverse transcription into complementary DNA was carried out with the use of Moloney murine leukaemia virus reverse transcriptase (Promega. Madison, WI, USA), oligo(dt)18 primers, random hexamer primers, RiboLock RNAse Inhibitor, and a mixture of four deoxynucleotide triphosphates (all from Thermo Fisher Scientific, Waltham, MA, USA). cDNA was combined with a mixture of 5× HOT FIREPol® EvaGreen® qPCR Mix Plus (no ROX) (Solis Biodyne, Tartu, Estonia) and a primer mix containing forward and reverse primers (Metabion, Planegg, Germany) (primer sequences are listed in Table 2). Relative messenger RNA (mRNA) expression levels were analysed with Lightcycler® 96 equipment (Roche Diagnostics, Basel, Switzerland). For relative quantification of gene expression, the ΔΔCt method (Livak and Schmittgen, 2001) was employed. Expression levels were normalized to those of the housekeeping gene peptidylprolyl isomerase G (PPIG) and presented relative to expression in undifferentiated H9 cells.
Table 2.
RT-qPCR primer pair sequences
| Primer Name | Sequence of 5′ Primer (Forward) | Sequence of 3′ Primer (Reverse) |
|---|---|---|
| NR5A1 | CAGGAGTTTGTCTGCCTCAA | GCACAGGGTGTAGTCAAGCA |
| GATA4 | CAGGCGTTGCACAGATAGTG | CCCGACACCCCAATCTC |
| WT1 | GGCAGCACAGTGTGTGAACT | CCAGGCACACCTGGTAGTTT |
| FOXL2 | TTTGTCCCCTCAGTTTATGTCC | TGAATTTGGGCAGGAGACG |
| WNT4 | GATGTGCGGGAGAGAAGCAA | ATTCCACCCGCATGTGTGT |
| RSPO1 | GCAACCCCGACATGAACAAG | CAAGCCCTCCTTACACTTGG |
| STAR | GAGTCAGCAGGACAATGGGG | CGCTCCACGAGCTCTTCATA |
| CYP11A1 | TGGTGACAATGGCTGGCTAAA | ATAAACCGACTCCACGTTGC |
| CYP17A1 | TCAGCCGCACACCAACTATC | GCAAACTCACCGATGCTGGA |
| CYP19A1 | GTGGCCCATGGCATTTTATA | GAGCTCTACTGGGGAACCAG |
| HPGD | CAGCCGGTTTATTGTGCTTC | CAGTCTCACACCACTGTTCATA |
| HSD3B2 | GAGGCAGTAAGGACTTGGACT | TGACCCAGAAGAGGGCGTAA |
| HSD17B1 | TGATGGGGCTGCCTTTCAAT | ATCAGGCTCAAGCTGTGGAC |
| GSTA1 | GATGTTCCAGCAAGTGCCAAT | CAGGTGGACATACGGGCA |
| GSTA2 | ATCCTTCTTCTGCCCTTTAGTC | GTCTTGTCCGTGGCTCTTTA |
| WNT6 | AACAGGACATTCGGGAGACG | CAGCTCGCCCATAGAACAGG |
| B4GALNT2 | TACCAAGACTTTCCTCCGCC | AGCAAACCAACCCTTCCCAA |
| GNRHR | AGTTTTTCACAATGGTGGCATCAA | TGGCAAATGCAACCGTCATT |
| PPIG | TCTTGTCAATGGCCAACAGAG | GCCCATCTAAATGAGGAGTTG |
RNA sequencing (RNA-seq)
Cells cultured in triplicate in 12-well culture plates were lysed with QlAzol Lysis Reagent (Qiagen, Hilden, Germany) at days 4, 6, 8 and 10 of differentiation. Total RNA was isolated using miRNeasy Mini Kits (Qiagen, Hilden, Germany). Additional on-column DNase Digestion with the RNase-Free DNase Set (Qiagen, Hilden, Germany) was performed. Determination of RNA quality, library preparation and sequencing were performed by the Sequencing unit of the Institute for Molecular Medicine Finland FIMM Technology Centre, University of Helsinki, Finland. Total RNA quality checks were carried out by using LabChip GX Touch RNA assays (PerkinElmer, USA) for RNA integrity checks and Qubit BR RNA assays (Thermo Fisher Scientific, USA) for total RNA quantification. Library preparation from 800 ng of total RNA was performed according to Illumina methodology (stranded total RNA prep with Ribo-Zero Plus Reference Guide, Illumina, San Diego, CA, USA). Library quality checks were performed using LabChip GX Touch HT High Sensitivity assays (PerkinElmer, USA). The libraries were quantified for sequencing using KAPA Library Quantification Kits (KAPA Biosystems, Wilmington, MA, USA). Sequencing was performed by using Illumina equipment (NovaSeq system using an S1 flow cell with lane divider, Illumina, San Diego, CA, USA). Read length for the paired-end run was 2 × 151 bp.
Data pre-processing was performed at FIMM. Quality control of raw sequence data was performed with FastQC v0.11.9. Trim-Galore v0.6.5 was used for adapter trimming and quality filtering. RNA-seq reads were then aligned against the human reference genome (GRCh38 release 82) using STAR-2.7.6a. Then, the quality of RNA-Seq quality matrices was inspected with RNA-SeQC v2.4.2 and the quality of RNA-Seq data was assessed with RSeQC v4.0.0. FeatureCounts software was then used to summarise the read distribution over several genomic features.
Data analysis
The gene-by-sample expression data matrix, prepared by merging the output of FeatureCounts, was used for pairwise DE analyses with DESeq2 (v1.24, Love et al., 2014), to determine DEGs at each timepoint, and for time-series DE analysis with EBSeq-HMM (v1.18, Leng et al., 2015), to determine the expression dynamics of the DEGs throughout the time-course. Default settings for data normalization were used for both DESeq2 and EBSeqHMM.
Pairwise comparisons were conducted by comparing the induced and non-induced samples at days 6, 8, and 10. Additionally, samples in the same conditions were compared between timepoints. DEGs with adjusted P < 0.05 were then used in downstream pathway analyses. Time-series DE analyses were conducted separately for the NR5A1-induced and non-induced samples separately with the timepoints used as the ordering condition in EBSeqHMM. There are three possible transitions across the four ordered timepoints (days 4, 6, 8, 10) in our data, at which genes can attain one of three states: upregulation (U), downregulation (D) or stable expression (S or EE), making up 27 possible paths genes can follow in this time course (U-U-U, D-D-D, etc.). EBSeqHMM estimates the posterior probabilities (PPs) for all the possible paths and identifies genes with a non-constant expression profile over the time-course. Genes with significant DE in at least one transition between timepoints and having a PP of < 0.05 of remaining constant are identified as dynamically DEGs. The path with the highest PP (MaxPP) is assigned to each DEG. To compare the dynamically DEGs with the results from the male gonadal differentiation reported by Sepponen et al. [26] we re-analysed the male gonadal RNA-seq data with EBSeqHMM and evaluated the overlap of the dynamically DEGs between the female and male gonadal cell lines. Dynamically DEGs in our female NR5A1-induced data that were not detected as dynamically DEGs in our female non-induced data as well as the male NR5A1-induced data were identified as ‘Only in Female Induced’ DEGs (genes that show statistically significant dynamic DE profiles only in NR5A1-induced female cell line samples). Similarly, genes with statistically significant dynamic DE patterns in the NR5A1-induced male cell line samples (and not in the induced female cell line nor the non-induced male cell line samples) were identified as ‘Only in Male Induced’ DE genes. Additionally, we also identified dynamically DEGs that were detected in both cell lines (with either the same dynamic pattern or different), and those that were sex-specific having dynamically DE profiles only in one sex regardless of NR5A1 induction status.
GO enrichment analysis
GO enrichment analyses were performed using the online tool ENRICHR from the Maayan lab (ENRICHR, RRID:SCR_001575, https://maayanlab.cloud/Enrichr/).
Statistical analysis
Statistical analysis was performed with both GraphPad Prism version 9.2.0 (San Diego, CA, USA) and IBM SPSS 29 Statistics software (Chicago, IL, USA). Normal distribution of the data was assessed by using the Shapiro–Wilk normality test. Two-way ANOVA was used to assess the significance between the non-induced and induced samples across the different timepoints during differentiation. In correction for multiple comparison, Sidak’s post-hoc test was used. P values less than 0.05 were considered statistically significant. Data are shown as mean ± SEM.
Supplementary Information
Additional file 1: Supplementary figure 1. Bulk RNA-sequencing analyses showed differences between the control (-DOX-TMP) and induced (+DOX+TMP) conditions. A. Principal component analysis (PCA) comparing samples from different days of differentiation and control (CTRL, -DOX –TMP) and induced (IND, +DOX +TMP) conditions. d; day. B. Flowchart comparing the number of differentially expressed (black), coding (orange), non-coding (blue), upregulated (green) and downregulated (red) genes in induced (+DOX+TMP) and non-induced (-DOX-TMP) conditions at different timepoints in both female and male gonadal differentiation. d; day. Supplementary figure 2. Differentially expressed upregulated genes were mostly associated with (adrenal) steroidogenesis. A. RT-qPCR analysis showed that NR5A1 induction upregulated the markers HPGD, GSTA1, GSTA2, WNT6, B4GALNT2 and GNRHR. Data are reported as mean ± SEM, n=3 biological replicates. Two-way ANOVA; 0.12 (ns), 0.033 (*), 0.002 (**), < 0.001 (***). d; day. B. Bar charts showing the correlated KEGG 2021 human pathways of the female DE genes between control (CTRL, -DOX -TMP) and induced (IND, +DOX +TMP) conditions at days 6, 8 and 10 of gonadal differentiation. The bar charts show the top 10 enriched terms in the chosen library, along with their corresponding p-values. Coloured bars correspond to terms with significant p-values (< 0.05). An asterisk (*) next to a p-value indicates the term also had a significant adjusted p-value (< 0.05). Supplementary figure 3. Adrenal, testicular, and ovarian steroidogenesis markers were upregulated upon NR5A1 induction. Comparison of the expression of RNA-sequenced non-induced (CTRL, -DOX-TMP) and induced (IND, +DOX+TMP) samples. Known adrenal, testicular and ovarian steroidogenic markers were upregulated upon NR5A1 induction. Each square represents the expression of a specific gene within a technical replicate. The intensity of gene expression is indicated by a colour scale based on row z-scores (red, highest expression levels; blue, lowest expression levels). d, day.
Acknowledgements
We would like to thank Nicholas Bolton, PhD, for revising our manuscript for the English language. We thank the following institutions for their services. Materials for generation of the activation line were kindly provided by the Biomedicum Stem Cell Centre at the University of Helsinki. Single-cell sorting to create clonal lines was performed at the Biomedicum Flow Cytometry Unit at the University of Helsinki supported by HilIFE. Library preparation, sequencing, and data analysis (FIMM-RNAseq pipeline) were performed by the FIMM Genomics NGS Sequencing unit at the University of Helsinki supported by HiLIFE and Biocentre Finland. Confocal images were taken at the Biomedicum Imaging Unit (BIU) at the University of Helsinki supported by HiLIFE.
Disclosure
The authors have nothing to disclose.
Abbreviations
- B4GALNT2
Beta-1,4-N-acetyl-galactosaminyltransferase 2
- CYP11A1
Cytochrome p450 family 11 subfamily A member 1
- CYP17A1
Cytochrome p450 family 17 subfamily A member 1
- CYP19A1
Cytochrome p450 family 19 subfamily A member 1
- DAPI
4',6-diamidino-2-phenylindole
- DE
Differential expression/differentially expressed
- DEG
Differentially expressed gene
- DOX
Doxycycline hyclate
- DSD
Differences in sex development
- EMX2
Empty spiracles homeobox 2
- FBS
Fetal bovine serum
- FOXL2
Forkhead box protein L2
- GATA4
Gata binding protein 4
- GSTA1
Glutathione S-transferase A1
- hESC
Human embryonic stem cell
- hiPSC
Human induced pluripotent stem cell
- HPGD
15-hydroxyprostaglandin dehydrogenase
- HSD3B2
Hydroxy-delta-5-steroid dehydrogenase
- HSD17B1
Hydroxysteroid 17-beta dehydrogenase 1
- hPSC
Human pluripotent stem cell
- IM
Intermediate mesoderm
- INHA
Inhibin subunit α
- KO
Knock-out
- LHX9
Lim homeobox 9
- NEAA
Non-essential amino acid
- NR5A1
Nuclear receptor subfamily 5 group A member 1
- PBS
Phosphate-buffered saline
- PCA
Principal component analysis
- POMC
Pro-opiomelanocortin
- PPIG
Peptidylprolyl isomerase G
- PS
Primitive streak
- RT-qPCR
Real time quantitative polymerase chain reaction
- ROCKi
Rho-kinase inhibitor
- RSPO1
R-spondin 1
- SF-1
Steroidogenic factor 1
- SRY
Sex-determining region on the Y chromosome
- STAR
Steroidogenic acute regulatory protein
- TMP
Trimethoprim
- WNT4
Wnt family member 4
- WNT6
Wnt family member 6
- WT1
Wilms’ tumour suppressor 1
Authors’ contributions
L.D. performed the experiments in the study with the established NR5A1 induction line, analysed and interpreted the generated data and was the principal author of this paper. K.L. assisted in the performance of the experiments, participated in the design of the study and majorly contributed to the writing of this paper. K.S. generated the NR5A1 induction line and participated in the editing and revision of the manuscript. D.Y. performed all the bioinformatics and contributed to the editing and revision of the paper. J.K. provided input on the analysis of the data. T.T. and J.T. were primarily involved in designing the study and writing of the paper. All authors reviewed the manuscript.
Funding
Open Access funding provided by University of Helsinki including Helsinki University Central Hospital. This project has received funding from the European Union’s Horizon 2020 research and innovation programme under Marie Sklodowska-Curie grant agreement no. 813707, Helsinki University Hospital Research Funds, the Sigrid Jusélius Foundation and the Jane and Aatos Erkko Foundation.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the NCBI Gene Expression Omnibus (GEO) repository and can be accessed through GEO Series accession number GSE236653. All material unique to this study is available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All studies were carried out according to the provisions of the Declaration of Helsinki. We have approval from the Ethics Committee of the Hospital District of Helsinki and Uusimaa for our stem cell-based research (HUS/2064/2019).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Satoh M. Histogenesis and organogenesis of the gonad in human embryos. J Anat. 1991;177:85–107. [PMC free article] [PubMed] [Google Scholar]
- 2.Rotgers E, Jørgensen A, Yao HHC. At the crossroads of fate—somatic cell lineage specification in the fetal gonad. Endocr Rev. 2018;39(5):739–759. doi: 10.1210/er.2018-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker KL, Schimmer BP, Schedl A. Genes essential for early events in gonadal development. Cell Mol Life Sci. 1999;55(6):831–8. doi: 10.1007/s000180050337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makiyan Z. Studies of gonadal sex differentiation. Organogenesis. 2016;12(1):42–51. doi: 10.1080/15476278.2016.1145318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koopman P, Münsterberg A, Capel B, Vivian N, Lovell-Badge R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990;348(6300):450–452. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- 6.Gomes NL, Chetty T, Jorgensen A, Mitchell RT. Disorders of sex development—novel regulators, impacts on fertility, and options for fertility preservation. Int J Mol Sci. 2020;21(7):2282. doi: 10.3390/ijms21072282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vainio S, Heikkilä M, Kispert A, Chin N, McMahon AP. Female development in mammals is regulated by Wnt-4 signalling. Nature. 1999;397(6718):405–409. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- 8.Kim Y, Kobayashi A, Sekido R, DiNapoli L, Brennan J, Chaboissier MC, et al. Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol. 2006;4(6):e187. doi: 10.1371/journal.pbio.0040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biason-Lauber A, Chaboissier MC. Ovarian development and disease: the known and the unexpected. Semin Cell Dev Biol. 2015;45:59–67. doi: 10.1016/j.semcdb.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 10.Rey R, Josso N, Racine C. Sexual Differentiation. In: Feingold KR, Anawalt B, Boyce A, Chrousos G, de Herder WW, Dhatariya K, editors. Endotext. South Dartmouth (MA): MDText.com, Inc.; 2000. Available from: http://www.ncbi.nlm.nih.gov/books/NBK279001/. Cited 2021 Jun 23.
- 11.Lin YT, Capel B. Cell fate commitment during mammalian sex determination. Curr Opin Genet Dev. 2015;32:144–152. doi: 10.1016/j.gde.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biason-Lauber A. WNT4, RSPO1, and FOXL2 in sex development. Semin Reprod Med. 2012;30(5):387–395. doi: 10.1055/s-0032-1324722. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, Workman S, Wilson MJ. The molecular pathways underlying early gonadal development. J Mol Endocrinol. 2019;62(1):R47–64. doi: 10.1530/JME-17-0314. [DOI] [PubMed] [Google Scholar]
- 14.Rice DA, Mouw AR, Bogerd AM, Parker KL. A shared promoter element regulates the expression of three steridogenic enzymes. Mol Endocrinol. 1991;5(10):1552–1561. doi: 10.1210/mend-5-10-1552. [DOI] [PubMed] [Google Scholar]
- 15.Lala DS, Rice DA, Parker KL. Steroidogenic factor I, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor I. Mol Endocrinol. 1992;6(8):1249–1258. doi: 10.1210/mend.6.8.1406703. [DOI] [PubMed] [Google Scholar]
- 16.Morohashi K, Honda S, Inomata Y, Handa H, Omura T. A common trans-acting factor, Ad4-binding protein, to the promoters of steroidogenic P-450s. J Biol Chem. 1992;267(25):17913–17919. [PubMed] [Google Scholar]
- 17.Parker KL, Schimmer BP. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr Rev. 1997;18(3):361–377. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- 18.Lin L, Achermann JC. Steroidogenic factor-1 (SF-1, Ad4BP, NR5A1) and disorders of testis development. Sex Dev. 2008;2(4–5):200–209. doi: 10.1159/000152036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schimmer BP, White PC. Minireview: steroidogenic factor 1: its roles in differentiation, development, and disease. Mol Endocrinol. 2010;24(7):1322–37. doi: 10.1210/me.2009-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadovsky Y, Crawford PA. Developmental and physiologic roles of the nuclear receptor steroidogenic factor-I in the reproductive system. Reprod Sci. 1998;5(1):6–12. doi: 10.1016/s1071-5576(97)00096-8. [DOI] [PubMed] [Google Scholar]
- 21.Zhao L, Bakke M, Hanley NA, Majdic G, Stallings NR, Jeyasuria P, et al. Tissue-specific knockouts of steroidogenic factor 1. Mol Cell Endocrinol. 2004;215(1):89–94. doi: 10.1016/j.mce.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 22.Jeyasuria P, Ikeda Y, Jamin SP, Zhao L, de Rooij DG, Themmen APN, et al. Cell-specific knockout of Steroidogenic factor 1 reveals its essential roles in gonadal function. Mol Endocrinol. 2004;18(7):1610–1619. doi: 10.1210/me.2003-0404. [DOI] [PubMed] [Google Scholar]
- 23.Zhao H, Li Z, Cooney AJ. Orphan nuclear receptor function in the ovary. Front Biosci. 2007;12:3398. Available from: https://pubmed.ncbi.nlm.nih.gov/17485308/. Cited 2022 Oct 20 . [DOI] [PubMed]
- 24.Werner R, Mönig I, Lünstedt R, Wünsch L, Thorns C, Reiz B, et al. New NR5A1 mutations and phenotypic variations of gonadal dysgenesis. PLoS One. 2017;12(5):e0176720. doi: 10.1371/journal.pone.0176720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Khairi R, Achermann JC. Steroidogenic factor-1 and human disease. Semin Reprod Med. 2012;30(5):374–381. doi: 10.1055/s-0032-1324720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sepponen K, Lundin K, Yohannes DA, Vuoristo S, Balboa D, Poutanen M, et al. Steroidogenic factor 1 (NR5A1) induces multiple transcriptional changes during differentiation of human gonadal-like cells. Differentiation. 2022;S0301–4681(22):00062–00067. doi: 10.1016/j.diff.2022.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Sepponen K, Lundin K, Knuus K, Väyrynen P, Raivio T, Tapanainen JS, et al. The role of sequential BMP signaling in directing human embryonic stem cells to bipotential gonadal cells. J Clin Endocrinol Metab. 2017;102(11):4303–4314. doi: 10.1210/jc.2017-01469. [DOI] [PubMed] [Google Scholar]
- 28.Mamsen LS, Ernst EH, Borup R, Larsen A, Olesen RH, Ernst E, et al. Temporal expression pattern of genes during the period of sex differentiation in human embryonic gonads. Sci Rep. 2017;7(1):15961. doi: 10.1038/s41598-017-15931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.del Valle I, Buonocore F, Duncan AJ, Lin L, Barenco M, Parnaik R, et al. A genomic atlas of human adrenal and gonad development. Wellcome Open Res. 2017;2:25. doi: 10.12688/wellcomeopenres.11253.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lecluze E, Rolland AD, Filis P, Evrard B, Leverrier-Penna S, Maamar MB, et al. Dynamics of the transcriptional landscape during human fetal testis and ovary development. Hum Reprod. 2020;35(5):1099–1119. doi: 10.1093/humrep/deaa041. [DOI] [PubMed] [Google Scholar]
- 31.Gallo-Payet N. 60 YEARS OF POMC: adrenal and extra-adrenal functions of ACTH. J Mol Endocrinol. 2016;56(4):T135–156. doi: 10.1530/JME-15-0257. [DOI] [PubMed] [Google Scholar]
- 32.Lang K, Sturmer A, Adam P, Fassnacht M, Quinkler M, Morcos M, et al. Chemokine receptor expression in the adrenal cortex and in adrenocortical tumours. Endocr Abstr. 2009;20. Available from: https://www.endocrine-abstracts.org/ea/0020/ea0020p20. Cited 2023 Feb 2.
- 33.Doghman M, Arhatte M, Thibout H, Rodrigues G, De Moura J, Grosso S, et al. Nephroblastoma overexpressed/cysteine-rich protein 61/connective tissue growth factor/nephroblastoma overexpressed gene-3 (NOV/CCN3), a selective adrenocortical cell proapoptotic factor, is down-regulated in childhood adrenocortical tumors. J Clin Endocrinol Metab. 2007;92(8):3253–3260. doi: 10.1210/jc.2007-0342. [DOI] [PubMed] [Google Scholar]
- 34.Côté M, Guillon G, Payet MD, Gallo-Payet N. Expression and regulation of adenylyl cyclase isoforms in the human adrenal gland. J Clin Endocrinol Metab. 2001;86(9):4495–4503. doi: 10.1210/jcem.86.9.7837. [DOI] [PubMed] [Google Scholar]
- 35.Cava C, Bertoli G, Castiglioni I. Portrait of tissue-specific coexpression networks of noncoding RNAs (miRNA and lncRNA) and mRNAs in normal tissues. Comput Math Methods Med. 2019;2019:9029351. doi: 10.1155/2019/9029351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abel MH, Baban D, Lee S, Charlton HM, O’Shaughnessy PJ. Effects of FSH on testicular mRNA transcript levels in the hypogonadal mouse. J Mol Endocrinol. 2009;42(4):291–303. doi: 10.1677/JME-08-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perera EM, Martin H, Seeherunvong T, Kos L, Hughes IA, Hawkins JR, et al. Tescalcin, a novel gene encoding a putative EF-Hand Ca2+-binding protein, Col9a3, and renin are expressed in the mouse testis during the early stages of gonadal differentiation**This work was supported in part by the Department of Pediatrics at the University of Miami School of Medicine. Endocrinology. 2001;142(1):455–63. doi: 10.1210/endo.142.1.7882. [DOI] [PubMed] [Google Scholar]
- 38.Ai A, Tang Z, Liu Y, Yu S, Li B, Huang H, et al. Characterization and identification of human immortalized granulosa cells derived from ovarian follicular fluid. Exp Ther Med. 2019;18(3):2167–2177. doi: 10.3892/etm.2019.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller WL, Auchus RJ. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr Rev. 2011;32(1):81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallinelli A, Garuti G, Matteo ML, Genazzani AR, Facchinetti F. Genetics: expression of pro-opiomelanocortin gene in human ovarian tissue. Hum Reprod. 1995;10(5):1085–9. doi: 10.1093/oxfordjournals.humrep.a136099. [DOI] [PubMed] [Google Scholar]
- 41.Cheung LWT, Wong AST. Gonadotropin-releasing hormone: GnRH receptor signaling in extrapituitary tissues. FEBS J. 2008;275(22):5479–5495. doi: 10.1111/j.1742-4658.2008.06677.x. [DOI] [PubMed] [Google Scholar]
- 42.Maeda K, Kitawaki J, Yokota K, Noguchi T, Urabe M, Yamamoto T, et al. [Effects of gonadotropin-releasing hormone and its analogue (buserelin) on aromatase in cultured human granulosa cells] Nihon Sanka Fujinka Gakkai Zasshi. 1996;48(2):89–95. [PubMed] [Google Scholar]
- 43.Moreau F, Mittre H, Benhaim A, Bois C, Bertherat J, Carreau S, et al. Aromatase expression in the normal human adult adrenal and in adrenocortical tumors: biochemical, immunohistochemical, and molecular studies. Eur J Endocrinol. 2009;160(1):93–99. doi: 10.1530/EJE-08-0215. [DOI] [PubMed] [Google Scholar]
- 44.Luisi S, Florio P, Reis FM, Petraglia F. Inhibins in female and male reproductive physiology: role in gametogenesis, conception, implantation and early pregnancy. Hum Reprod Update. 2005;11(2):123–135. doi: 10.1093/humupd/dmh057. [DOI] [PubMed] [Google Scholar]
- 45.Hayes FJ, Hall JE, Boepple PA, Crowley WF., Jr Differential control of gonadotropin secretion in the human: endocrine role of inhibin1. J Clin Endocrinol Metab. 1998;83(6):1835–1841. doi: 10.1210/jcem.83.6.4884. [DOI] [PubMed] [Google Scholar]
- 46.Smith OE, Morin F, Roussel V, Bertucci M, Boyer A, Murphy BD. The role of steroidogenic factor 1 (SF-1) in somatic cell function of the testes and ovaries of mature mice. Reproduction. 2022;1(aop). Available from: https://rep.bioscientifica.com/view/journals/rep/aop/rep-22-0049/rep-22-0049.xml. Cited 2022 Oct 20. [DOI] [PubMed]
- 47.Stévant I, Kühne F, Greenfield A, Chaboissier MC, Dermitzakis ET, Nef S. Dissecting cell lineage specification and sex fate determination in gonadal somatic cells using single-cell transcriptomics. Cell Rep. 2019;26(12):3272–3283e3. doi: 10.1016/j.celrep.2019.02.069. [DOI] [PubMed] [Google Scholar]
- 48.Balboa D, Weltner J, Eurola S, Trokovic R, Wartiovaara K, Otonkoski T. Conditionally stabilized dCas9 activator for controlling gene expression in human cell reprogramming and differentiation. Stem Cell Reports. 2015;5(3):448–59. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary figure 1. Bulk RNA-sequencing analyses showed differences between the control (-DOX-TMP) and induced (+DOX+TMP) conditions. A. Principal component analysis (PCA) comparing samples from different days of differentiation and control (CTRL, -DOX –TMP) and induced (IND, +DOX +TMP) conditions. d; day. B. Flowchart comparing the number of differentially expressed (black), coding (orange), non-coding (blue), upregulated (green) and downregulated (red) genes in induced (+DOX+TMP) and non-induced (-DOX-TMP) conditions at different timepoints in both female and male gonadal differentiation. d; day. Supplementary figure 2. Differentially expressed upregulated genes were mostly associated with (adrenal) steroidogenesis. A. RT-qPCR analysis showed that NR5A1 induction upregulated the markers HPGD, GSTA1, GSTA2, WNT6, B4GALNT2 and GNRHR. Data are reported as mean ± SEM, n=3 biological replicates. Two-way ANOVA; 0.12 (ns), 0.033 (*), 0.002 (**), < 0.001 (***). d; day. B. Bar charts showing the correlated KEGG 2021 human pathways of the female DE genes between control (CTRL, -DOX -TMP) and induced (IND, +DOX +TMP) conditions at days 6, 8 and 10 of gonadal differentiation. The bar charts show the top 10 enriched terms in the chosen library, along with their corresponding p-values. Coloured bars correspond to terms with significant p-values (< 0.05). An asterisk (*) next to a p-value indicates the term also had a significant adjusted p-value (< 0.05). Supplementary figure 3. Adrenal, testicular, and ovarian steroidogenesis markers were upregulated upon NR5A1 induction. Comparison of the expression of RNA-sequenced non-induced (CTRL, -DOX-TMP) and induced (IND, +DOX+TMP) samples. Known adrenal, testicular and ovarian steroidogenic markers were upregulated upon NR5A1 induction. Each square represents the expression of a specific gene within a technical replicate. The intensity of gene expression is indicated by a colour scale based on row z-scores (red, highest expression levels; blue, lowest expression levels). d, day.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the NCBI Gene Expression Omnibus (GEO) repository and can be accessed through GEO Series accession number GSE236653. All material unique to this study is available from the corresponding author on reasonable request.