Abstract
A new technique for the induction of plaque formation by Chlamydia trachomatis biovar trachoma applicable to the titration of infectivity and cloning of biovar trachoma was established. Three novel strains were cloned and confirmed to be free of glycogen inclusions. The lack of glycogen accumulation correlated with the absence of a 7.5-kb plasmid, which is highly conserved in other strains of C. trachomatis. Although the growth efficiency of these plasmid-free strains was slightly lower than that of plasmid-positive strains, possession of the plasmid and glycogen accumulation were not essential for the survival of C. trachomatis.
The genus Chlamydia, the members of which are obligate intracellular bacteria, comprises four species, Chlamydia trachomatis, C. psittaci (32), C. pneumoniae (14), and C. pecorum (11). C. trachomatis is a major cause of sexually transmitted disease (STD) in developed countries (4, 21, 27). C. trachomatis has two biovars that are pathogenic for humans: biovar trachoma, which causes ocular and urogenital infections, and biovar LGV, which causes lymphogranuloma venereum. Among STDs caused by C. trachomatis strains, those caused by biovar trachoma are the most prevalent in Japan, whereas STDs caused by biovar LGV have not been documented in Japan during the past decade (24). Biovar trachoma is further divided into 18 serovars (43, 44), which are determined with monoclonal antibodies (44) and/or by restriction fragment length polymorphism analysis of PCR products from the omp-1 gene (38, 40, 45). In Japan, the dominant serovars of clinical isolates are C, D, and E, but the serotyping of isolates is not always successful, perhaps because of the coexistence of different serovars in the same clinical isolates. In addition, the coexistence of different phenotypes in the same serotype cannot be excluded. Therefore, a plaque cloning procedure is needed to obtain pure isolates for serotyping and phenotypic characterization. However, whereas many workers have succeeded in inducing plaque formation by C. psittaci and biovar LGV of C. trachomatis (1, 19, 23, 36, 37), this has not been achieved with biovar trachoma. The present study was carried out to develop a cloning technique for that biovar. By this new method, three novel strains that lacked glycogen accumulation were obtained. Although glycogen accumulation in inclusions has been regarded as a typical characteristic of C. trachomatis species (2, 9, 13), this study shows that this phenotypic property is not associated with every strain of C. trachomatis. Interestingly, these strains also lack the 7.5-kb plasmid which is otherwise highly conserved in all C. trachomatis species.
MATERIALS AND METHODS
Chlamydiae.
Reference serovars of C. trachomatis, C/TW-3/OT, D/UW-3/Cx, F/UW-6/Cx, H/UW-4/Cx, I/UW-12/Ur and L2/434/Bu, were supplied by the National Institute of Infectious Diseases (the former National Institute for Health) Japan and were maintained continuously in monolayers of the HeLa 229 or the McCoy cell line. Clinical isolates of C. trachomatis designated Ct-1555, Ct-1633, Ct-1938, Ct-1939, and Ct-1943 were randomly selected from 254 isolates and were serotyped as serotypes D, D, F, D, and H, respectively. All isolates were well adapted to the McCoy cell line.
C. psittaci Budgerigar-1 was supplied by the National Institute of Infectious Diseases Japan and was maintained in McCoy cell cultures. C. psittaci Cal 10 has been maintained in our laboratory for more than 25 years in suspension or monolayer cultures of the L929 cell line and has also been well adapted to McCoy cells. C. pneumoniae TW-183 was purchased from the Washington Research Foundation, Seattle, Wash., and was maintained by cultivation in HeLa 229 or HEp-2 cells.
Chlamydia culture.
C. trachomatis and C. psittaci were inoculated into McCoy cells, and C. pneumoniae was inoculated into HEp-2 cells. Unless otherwise noted, cell monolayers were prepared in six-well culture plates (Sumitomo Bakelite Co. Ltd., Tokyo, Japan) and were infected by centrifugation in a Beckman GP-type centrifuge (the maximum speed was limited to 2,750 rpm for cell culture plates) at 2,000 rpm (760 × g) for 60 min. The cells were then incubated at 37°C in an atmosphere of 5% CO2 in minimum essential medium (MEM; Nissui, Tokyo, Japan) containing 1 μg of cycloheximide per ml and supplemented with 10% heat-inactivated fetal calf serum (FCS; Gibco BRL Life Technologies Inc., Grand Island, N.Y.).
Plaque formation.
The following stock solutions were prepared: (i) 1.1% agarose (SeaKem ME agarose; FMC BioProducts, Rockland, Maine) in distilled water, (ii) phenol red-free 2× Eagle MEM (Nissui), (iii) 100 μg of cycloheximide per ml in phosphate-buffered saline (PBS) without Mg and Ca ions [PBS(−); Nissui], and (iv) 3% neutral red in PBS(−). All of these solutions except the agarose solution were stored at 4°C after filtration through a 0.2-μm-pore-size filter (Millipore Co., Bedford, Mass.); the agarose solution was autoclaved and was kept at room temperature. Just before overlay, the melted agarose solution was mixed with an equal volume of 2× MEM, and the mixture was maintained at 45°C. After the addition of FCS (final concentration, 10%) and cycloheximide (final concentration, 1 μg/ml), the agarose medium was overlaid on the infected cell monolayers (2 ml/well). After solidification at room temperature, 1× liquid MEM containing 10% FCS was loaded onto the agarose medium (2 ml/well) and the cultures were incubated at 37°C. The final agarose medium was prepared by adding a 1/100 volume of the neutral red stock solution to the first agarose medium and overlaying it (2 ml/well) on the first agarose medium on the appropriate days postinoculation.
Growth curve.
A one-step growth curve for each strain was made as reported previously (28). Briefly, McCoy cells were infected by centrifugation with appropriately diluted inocula, incubated at 37°C, and then harvested at appropriate times postinoculation. After sonication and brief centrifugation, each supernatant of the cell homogenate was inoculated onto a McCoy cell monolayer on a coverslip (diameter, 14 mm) set in a 24-well cell culture plate (Sumitomo Bakelite Co. Ltd.), and infection was carried out under the conditions described above. Two coverslip cultures for each sample were stained with fluorescein-conjugated monoclonal antibody (MAb) directed against the genus-specific antigen (Chlamydia FA Seiken; Denka Seiken, Tokyo, Japan), and the chlamydial inclusions were then counted. The infectivity (numbers of inclusion-forming units per milliliter) of each sample was calculated from the mean number of inclusions per coverslip.
Quantitation of glycogen.
The glycogen in the infected cells was quantified by the anthrone reaction (16). Infected cells from two wells of a six-well culture plate were carefully suspended in PBS(−) with a rubber policeman, transferred to a glass centrifuge tube with a tight cap, and then centrifuged at 300 × g for 10 min. After the supernatant was removed, the pellet was stored at −80°C until use. The pellet was then solubilized in 1 ml of 50% KOH at 100°C for 3 h and cooled at room temperature. Glycogen was then precipitated by the addition of 3 ml of distilled water and 8 ml of ethanol. After washing twice with ice-cold 60% ethanol, the precipitated glycogen was dried in a vacuum desiccator overnight. The precipitate was dissolved in 1 ml of distilled water, and then 5 ml of anthrone solution prepared as described by Hanson and Phillips (16) was added. After thorough mixing, the reaction mixture was allowed to stand for 10 min at 100°C. The color that developed was measured in a spectrophotometer at 625 nm.
Purification of EBs.
The elementary body (EB) suspension for extraction of chromosomal and plasmid DNAs was prepared as follows. McCoy cells with well-developed inclusions were collected at 72 h postinoculation and were sonicated to facilitate the release of chlamydial organisms from the host cells. After cell debris was removed by centrifugation at 900 × g for 10 min, the supernatant was layered onto a 25% (wt/vol) sucrose cushion in 30 mM Tris-HCl buffer (pH 7.2, and centrifuged at 8,000 × g for 60 min at 4°C. The pellet obtained was suspended in 0.2 M Tris-HCl buffer (pH 7.2) containing 10 mM MgCl2 and was then exposed continuously to DNase (20 μg/ml), RNase (20 μg/ml), and trypsin (100 μg/ml) at 37°C for 60 min each. The suspension was then centrifuged again by sucrose-cushioning centrifugation, and the resulting pellet was resuspended in sucrose-phosphate-glutamate (SPG) buffer and stored at −80°C until it was required for DNA extraction.
For PCR and ligase chain reaction (LCR), a highly purified EB suspension was prepared. The crude EB suspension was layered onto a two-layer cushion; the bottom layer consisted of a 50% (wt/vol) sucrose solution and the top layer consisted of 30% (vol/vol) Urografin (3.5-diacetamido-2,4,6-triisobenzoic acid; Schering AG, Berlin, Germany) in 30 mM Tris-HCl buffer (pH 7.2). It was then centrifuged at 8,000 × g for 60 min. The pellet and turbid bottom layer were suspended in the SPG buffer, and the mixture was then centrifuged at 12,000 × g for 30 min. After resuspension of the pellet in SPG buffer, the suspension was layered onto a continuous Urografin gradient column (40 to 52% [vol/vol]) and centrifuged at 8,000 × g for 60 min. Next, the EB band in the gradient column was recovered and was diluted with SPG buffer, and the EBs were precipitated by centrifugation at 12,000 × g for 30 min. The EBs obtained were then suspended in 30 mM Tris-HCl buffer and stored at −80°C. Finally, the number of EBs in the highly purified suspension was counted as described previously (29).
Preparation and restriction analysis of plasmid and chromosomal DNAs.
Plasmid DNA was extracted with the QIAprep Spin Miniprep kit (QIAGEN, Hilden, Germany). On the basis of the results reported by Palmer and Falkow (34) and Peterson et al. (35), EcoRI was used for restriction fragment analysis. Chromosomal DNA was extracted by the method of Fukushi and Hirai (12). Briefly, the EBs were treated with proteinase K (200 μg/ml) in the presence of 5% 2-mercaptoethanol, 20 mM EDTA, and 0.5% sodium-N-laurylsarcosine at 45°C for 60 min and were then extracted with a mixture of phenol-chloroform-isoamyl alcohol (25:24:1). This was followed by treatment with DNase-free RNase A (1 μg/ml) at 37°C for 30 min and precipitation with ethanol. The DNA solution was dialyzed against 10 mM Tris-HCl buffer (pH 8.0) containing 1 mM EDTA. BamHI, EcoRI, HindIII, KpnI, PvuII, and SalI were used for restriction fragment length polymorphism analysis. Digestion and electrophoresis of the plasmid and chromosomal DNAs were done by the methods of Sambrook et al. (39).
PCR and LCR.
Both the PCR test kit (AMPLICOR Chlamydia trachomatis; Roche Diagnostic Systems, Branchburg, N.J.) (25) and the LCR test kit (Abbott Laboratories, North Chicago, Ill.) (8) are designed to detect the 7.5-kb plasmid DNA commonly contained in C. trachomatis. We used them to determine if the cloned strains possessed the 7.5-kb plasmid by assaying highly purified EB suspensions as reported previously (30, 31).
Fluorescence and light microscopies.
To simultaneously confirm the C. trachomatis species and examine the inclusion morphology, the infected McCoy cell monolayers on coverslips (diameter, 14 mm) were fixed with ethanol or acetone and stained directly with a fluorescein-conjugated MAb to the species-specific C. trachomatis antigen (MicroTrak; Syva Co., Palo Alto, Calif.). Otherwise, a fluorescein-conjugated MAb to the genus-specific antigen (Chlamydia FA Seiken; Denka Seiken) was used to stain the inclusions of C. trachomatis, C. pneumoniae, and C. psittaci. To determine the level of accumulation of glycogen in the C. trachomatis inclusions, infected McCoy cells were air dried at 40 h postinoculation and were then fixed with methanol for 20 min at room temperature, after which iodine staining was carried out by a method described previously (26).
Electron microscopy.
Infected cells were pelleted by brief centrifugation (300 × g for 3 min) and doubly fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.3) for 90 min followed by 2.5% OsO4 fixation in the same buffer for 90 min. Thin sections were prepared by a previously reported method (26) and were stained with both uranyl acetate and lead citrate solutions. Plasmid DNA specimens were prepared by the protein surface spreading method described by Griffith (15). Briefly, the sample suspension was mixed with 0.5 M ammonium acetate (pH 8.0) containing 100 μg of cytochrome c (Sigma Chemical Co., St. Louis, Mo.) per ml, and then the mixture was slowly spread down a glass slide onto a 0.01 M ammonium acetate solution. The sample-containing protein film was picked up with a carbon-coated grid and was dehydrated in 90% ethanol. To enhance the plasmid DNA contrast, the grid was shadowcast with a platinum-palladium alloy at an angle of 1:8 on a rotating stage. Both specimens, thin sections and shadow-cast plasmid specimens, were examined with a JEM 2000 EX or a Hitachi H-500 electron microscope at accelerating voltages of 80 and 75 kV, respectively.
RESULTS
Plaque formation.
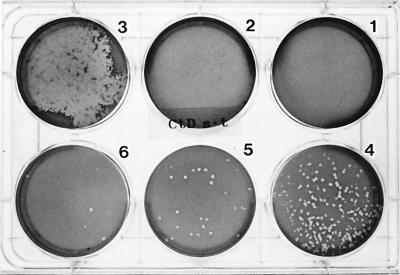
Figure 1 shows plaques of serovar D that formed on day 13 postinoculation. Plaques were clearly seen in the culture wells infected with inocula at dilutions of 10−6 and 10−7 whereas no plaques were seen in the wells infected with inocula diluted to 10−2 and 10−3. In the well infected with an inoculum at a dilution of 10−4, the plaques were extensively fused. Thus, it is clear that the absence of plaques in the first two wells resulted from plaque fusion. All of the serovars and clinical isolates so far tested could form plaques. To form clear plaques, serovar C required 10 days, serovars D, F, H, and L2 required 13 days, and serovar I and the clinical isolates needed 14 days. C. psittaci Cal 10 and Budgerigar-1, on the other hand, formed clear plaques on day 7 postinoculation, and these increased in size, ranging from 4 to 5 mm in diameter by day 13 postinoculation. Consequently, plaque fusion frequently occurred even at infectious doses lower than those for the C. trachomatis strains (data not shown).
FIG. 1.
Plaques formed by C. trachomatis serovar D on day 13 postinoculation. The numbers labeled on the wells from 1 to 6 indicate inocula diluted to 10−2, 10−3, 10−4, 10−5, 10−6, and 10−7, respectively. The plate was photographed from the bottom.
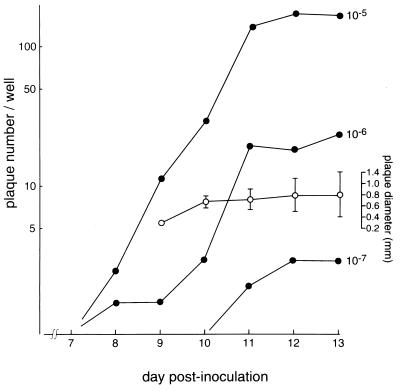
Figure 2 shows the time course of plaque formation and plaque size. Small plaques were first seen on day 8 postinoculation with the inocula at dilutions of 10−5 and 10−6 and reached a maximum level on days 12 and 13 for all inocula. The plaque diameter was measured with a magnifying glass with graduations of 0.1 mm. The mean diameter of well-isolated plaques was approximately 0.8 mm on days 10 to 13, but the range of diameters increased each day. A linear correlation between the plaque numbers and inoculum dilution was clearly seen on days 11 to 13 postinoculation.
FIG. 2.
Time course of plaque formation and plaque diameter in McCoy cell monolayers infected with C. trachomatis serovar D. Each closed circle indicates the mean plaque number calculated from the total number in six wells for each inoculum. The open circles indicate the mean diameters of 18 plaques on day 9, 30 plaques on day 10, and 60 plaques on days 11, 12, and 13 postinoculation.
Cloning of novel strains.
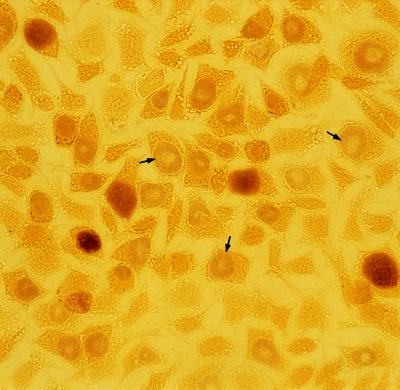
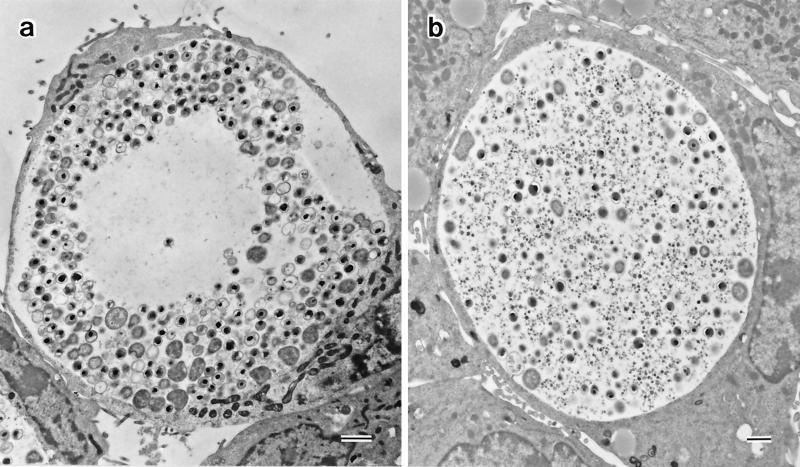
A number of single plaques were removed with agar-well punchers (diameter, 2 mm) commonly used to make holes in the Ouchterlony immunodiffusion test. Each agarose plug was sonicated and centrifuged at 300 × g for 5 min, and the supernatant was inoculated onto McCoy cell monolayers on a coverslip as described above. After incubation for 2 days, the McCoy cells were examined by iodine staining and immunostaining. To obtain a pure strain, this cloning procedure was repeated three times for each strain. From serovars D and F and a clinical isolate, we obtained three novel strains with inclusions that did not stain with iodine. Therefore, these strains were designated glycogen-negative strains D-9-3, F4.12t, and Ct-1943-3.1, respectively. In addition, every glycogen-positive strain always produced approximately 1% glycogen-negative inclusions among the glycogen-positive inclusions even after three successive clonings. To date, we have been unable to isolate any strain which produces 100% glycogen-positive inclusions. Probably, during every replication in a host cell, 1% of the C. trachomatis population loses the glycogen accumulation phenotype. Conversely, the clone producing 100% glycogen-negative inclusions was easily obtained by selecting a glycogen-negative plaque. Nevertheless, we chose D-12N, F4.5N, and Ct-1943-3N as glycogen-positive strains corresponding to the D-9-3, F4.12t, and Ct-1943-3.1 strains, respectively, and examined their biological characteristics. To compare the inclusions from glycogen-negative and glycogen-positive strains in the same microscopic field, the Ct-1943-3.1 and Ct-1943-3N strains were mixed at a ratio of 5:1 and were inoculated into McCoy cell monolayers (Fig. 3). There were two types of differently stained inclusions: a densely stained one and a faintly stained one, indicating that the former was of Ct-1943-3N origin and that the latter was of Ct-1943-3.1 origin. The Ct-1943-3.1 inclusion frequently showed a clear zone at the center, a morphology resembling a target-like bull’s eye. This unique morphology was readily recognizable under a phase-contrast microscope at 30 h postinoculation. It was also observed in the other glycogen-negative strains, strains D-9-3 and F4.12t. The same morphological features were revealed by immunostaining with both genus-specific and species-specific MAbs (data not shown), and the same results were also obtained by electron microscopy (Fig. 4). Chlamydial bodies, such as EBs, intermediate forms, and reticulate bodies, were excluded from the large, translucent area, corresponding to the clear zone observed by iodine staining and immunostaining (Fig. 4a). Many empty vesicles, which appeared to be derived from intermediate forms and EBs because they were smaller in size than reticulate bodies, were noted. Compression of the host nucleus was regularly observed. It should be noted that no glycogen particles were encountered in the glycogen-negative inclusions examined, whereas inclusions of the D-12N, F4.5N, and Ct-1943-3N glycogen-positive strains normally accumulated glycogen (Fig. 4b).
FIG. 3.
Inclusions of the Ct-1943-3.1 (glycogen-negative) and Ct-1943-3N (glycogen-positive) C. trachomatis strains in McCoy cells stained with iodine. Arrows indicate the inclusions in strain Ct-1943-3.1; these inclusions are distinct from the normally stained inclusions of strain Ct-1943-3N in terms of stainability and morphology.
FIG. 4.
Electron micrographs of C. trachomatis Ct-1943-3.1 (a) and Ct-1943-3N (b) inclusions at 40 h postinoculation. The center of the Ct-1943-3.1 inclusion consists of a large, translucent area from which chlamydial bodies are excluded; no glycogen particles are seen throughout the inclusion vacuole. In contrast, chlamydial bodies and glycogen particles are scattered rather homogeneously in the Ct-1943-3N inclusion (b). Compression of the nucleus to the periphery is seen in both host cells. Bars, 1 μm.
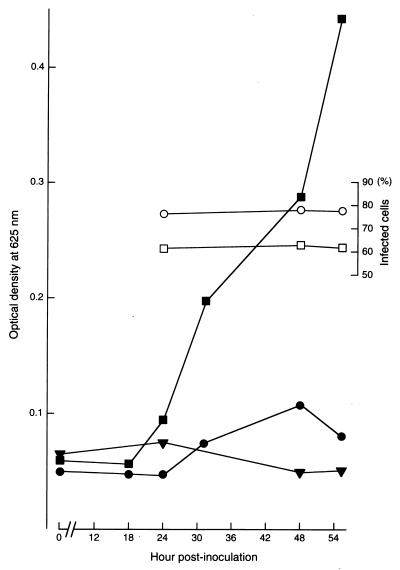
To confirm biochemically the absence of glycogen accumulation, McCoy cells infected with glycogen-positive or glycogen-negative strains were examined in a timed sequential manner by use of the anthrone reaction (Fig. 5). The results showed that the magnitude of the reaction in McCoy cells infected with the glycogen-negative strain was just faintly higher than that in noninfected McCoy cells. In contrast, the reaction for a glycogen-positive strain rapidly increased beginning at 18 h postinoculation, even though the number of infected cells was fewer than that for the cells infected with the glycogen-negative strain. The same results were obtained with the D-9-3 and Ct-1943-3.1 strains. The serotypes of the glycogen-negative strains did not differ from those of the parent glycogen-positive strains. The glycogen-negative strains have been stably maintained in our laboratory to date.
FIG. 5.
Time course of glycogen accumulation in McCoy cells infected with the glycogen-positive strain F4.5N (square) and the glycogen-negative strain F4.12t (circle), as assayed by the anthrone reaction. Each open symbol indicates the percentage of infected cells marked with the same closed symbol showing the indicated reaction magnitude. Closed triangles indicate the results for uninfected McCoy cells.
Growth curve.
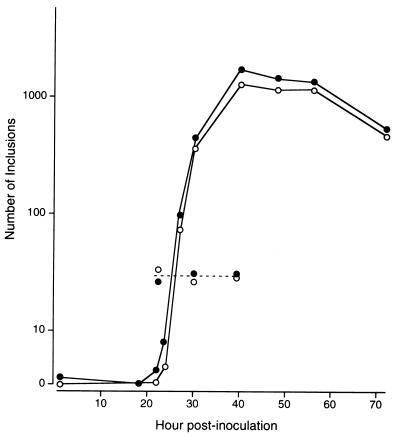
Glycogen-negative and glycogen-positive strains of the same titer were inoculated into cells, and the growth rates were compared (Fig. 6). Infectious EB progeny of glycogen-positive strain F4.5N appeared abruptly at 22 h and reached a maximum level at 40 h postinoculation. A similar curve was obtained for glycogen-negative strain F4.12t. However, the EB progeny of F4.12t appeared 3 h after those of F4.5N, and the titer of the former was always slightly lower than that of the latter. The same results were obtained when other glycogen-negative and glycogen-positive strains were compared (data not shown). These results indicated that the growth efficiency of the glycogen-negative strains is slightly lower than that of the glycogen-positive strains under the culture conditions used.
FIG. 6.
One-step growth curves of glycogen-positive strain F4.5N (closed circles) and glycogen-negative strain F4.12t (open circles) in McCoy cells. The numbers of inclusions in more than 1,500 cells at 20, 30, and 40 h postinoculation were determined, and then the infectious center (broken line) in each well was calculated.
Plasmid and chromosomal DNAs.
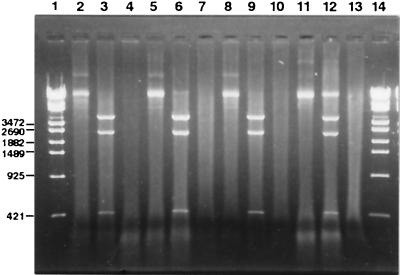
The electrophoresis of plasmid DNA extracted from approximately 108 EBs of the glycogen-positive and glycogen-negative strains is illustrated in Fig. 7. No band was detected in the lanes loaded with extracts from the glycogen-negative strains, whereas a band of 7.5 kb for an intact plasmid and three fragments of 4.6, 2.5, and 0.4 kb resulting from EcoRI digestion were clearly seen in the lanes with the glycogen-positive strains. The EcoRI fragment patterns agreed well with those reported by Palmer and Falkow (34). These results strongly indicated the absence of the 7.5-kb common plasmid in the glycogen-negative strains. To confirm these results, assays with 103 EBs of each strain were done with both PCR and LCR test kits, which were capable of detecting two or more EBs per assay under the experimental conditions used (30, 31). No positive reaction with the glycogen-negative strains was obtained with either test kit. The results were also confirmed by electron microscopy with 108 EBs of each strain. Circular plasmid DNA molecules of about 2 μm in length were seen in the extracts of the glycogen-positive strains, whereas no plasmid molecules have been encountered in the samples prepared from the glycogen-negative strains so far examined. When restriction fingerprints of the chromosomal DNAs were examined, no difference was found between the glycogen-positive and glycogen-negative strains (data not shown).
FIG. 7.
Agarose gel electrophoresis of 7.5-kb plasmid fractions prepared from glycogen-positive and glycogen-negative strains. Lanes 1 and 14, DNA molecular mass markers (λ-EcoT14I digest; Takara, Tokyo, Japan). The numbers of base pairs for representative bands are indicated in the left margin. Lanes 2, 5, 8, and 11, plasmid fractions extracted from glycogen-positive strains D-12N, F4.5N, Ct-1943-3N, and L2/434/Bu, respectively; lanes 3, 6, 9, and 12, fractions after digestion with EcoRI of lanes 2, 5, 8, and 11, respectively; lanes 4, 7, and 10, extracts of strains D.9-3, F4.12t, and Ct-1943-3.1, respectively; lane 13, fraction prepared from C. pneumoniae TW-183. Each fraction was prepared from approximately 108 EBs.
DISCUSSION
Banks et al. (1) suggested that plaque formation by chlamydiae might depend on the strains’ growth rates, and consequently, for slowly growing strains such as biovar trachoma long-term maintenance of the cell monolayer in agar medium might be required for plaque formation. In the present study, two points were crucial to the success of plaque formation by biovar trachoma. (i) The liquid culture medium had to be layered onto a solid agarose medium and changed at 4- to 5-day intervals; this treatment may refresh the cells and support chlamydial growth until plaque formation occurs. (ii) The agarose concentration was intentionally reduced to 0.5%, although 1% is the usual concentration for plaque formation by lytic viruses. The reduced agarose concentration may serve to keep the cells in good condition. The quality of the agarose was also important because purified agar was unsuitable for long-term maintenance of the cells. It is also likely that the agarose overlay might create appropriate conditions for the attachment of progeny EBs to host cells by electrostatic interaction (17) and successive adhesion of EBs to host cell receptors (3), followed by endocytosis (20). Interestingly, cycloheximide in the liquid culture medium did not improve the efficiency of chlamydial replication. It was effective only in the first agarose medium (data not shown).
As shown in Fig. 2, a linear correlation between the inoculum dilution and plaque number was noted. This indicates that titration of chlamydial inoculum is possible by this technique. However, plaque formation required approximately 2 weeks. This requirement may limit the application of this technique to the titration of EBs, since the infectious titer of EBs can be determined by inclusion counting on day 2 or 3 postinoculation. However, plaque formation by biovar trachoma was very useful for obtaining pure strains. This method enabled us to isolate and characterize three novel glycogen-negative strains, strains D-9-3, F4.12t, and Ct-1943-3. Their inclusions were characterized by a target-like bull’s eye morphology that appeared with the formation of a central translucent area that began to be clearly seen at about 30 h postinoculation. This area expanded continuously until the host cell lysed. Chlamydial organisms were excluded from this central area and were pushed to the periphery of the inclusion vacuole. Many envelope-like structures, which might be derived from disintegrated organisms, were seen among intact organisms. Inclusions were round or oval in shape and compressed the host nuclei to the periphery at the late stages of infection, as observed in infections with the parent strains. It is therefore very likely that glycogen accumulation is not essential for the growth of C. trachomatis, although the presence of glycogen is one of the characteristics that differentiates C. trachomatis from the other three chlamydial species (32).
The 7.5-kb plasmid is believed to encode genes essential for survival of the organisms, since it has been strictly conserved in the species C. trachomatis (5, 34, 41). However, as shown in the present study, this plasmid was not essential for the growth of C. trachomatis. Peterson et al. (35) also isolated a plasmid-free chlamydia of serovar L2 and reported that the isolate did not contain any detectable plasmid sequence integrated into its chromosome. From these results, it is reasonable to conclude that plasmid-free organisms can be maintained as stable strains. The present study demonstrated a close correlation between the presence of a plasmid and the accumulation of glycogen. Furthermore, the glycogen-positive strains always produced a small population of glycogen-negative inclusions, even after repeated cloning purifications. On the basis of these findings, it can be speculated that the plasmid-free strains possibly arose through omission of plasmids from the plasmid-possessing organisms during replication in host cells.
The entire nucleotide sequence of the 7.5-kb plasmid has been determined (6, 18, 41), and it has nine open reading frames (ORFs) which can encode for polypeptides of 10 kDa or longer (41). A 28-kDa protein, the product of ORF 3, has been determined to be a potential immunogen of the outer membranes of C. trachomatis strains that infect humans (7), but the biological function of the plasmid is still unknown. Moulder (33) stated that the glycogen synthetase in cells infected with C. trachomatis biovar LGV was of the bacterial type. Using the DNASIS homology search program (Hitachi Software Engineering Co. Ltd., Tokyo, Japan), we compared every amino acid sequence translated from each ORF of the 7.5-kb plasmid (6) with those of enzymes involved in bacterial glycogen synthesis (22, 42) but found no significant homology. Thus, it is likely that the glycogen accumulation by C. trachomatis in infected cells is indirectly regulated by an unknown factor(s) encoded by the plasmid.
The existence of stable plasmid-free C. trachomatis may cause undesirable problems with diagnosis with PCR and LCR test kits, because the primers in these kits are designed to detect the plasmid. In fact, Farencena et al. (10) recently reported on a clinical isolate of biovar trachoma that lacked the plasmid. Such isolates cannot be detected with current PCR and LCR kits.
In the present study, the growth characteristics of the plasmid-free strains were almost the same as those of the plasmid-positive parent strains in tissue culture systems. However, there is a possibility that the gene products of the 7.5-kb plasmid may be essential for infection of animal organs but not for chlamydial growth in tissue culture systems. Consequently, the pathogenicity of the plasmid-free strains in experimental animals is an important subject for further investigation.
ACKNOWLEDGMENTS
We thank P. B. Wyrick of the Department of Microbiology and Immunology, University of North Carolina School of Medicine, Chapel Hill, N.C., and David H. Waterbury, Kawasaki Medical School, Kurashiki, Japan, for critical reviews of the manuscript.
This work was supported by project research grants (grants 5-503, 8-404, and 9-505) from the Kawasaki Medical School and a Grant-in-Aid for Scientific Research (grant 02454102) from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Banks J, Eddie B, Schachter J, Meyer K F. Plaque formation by Chlamydia in L cells. Infect Immun. 1970;1:259–262. doi: 10.1128/iai.1.3.259-262.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker Y, Mashiab P, Bernkopf H. Biochemical changes in FL cell cultures infected with a trachoma agent. Nature. 1962;193:271–272. doi: 10.1038/193271a0. [DOI] [PubMed] [Google Scholar]
- 3.Byrne G I, Moulder J W. Parasite-specified phagocytosis of Chlamydia psittaci and Chlamydia trachomatis by L and HeLa cells. Infect Immun. 1978;19:598–606. doi: 10.1128/iai.19.2.598-606.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cates W, Jr, Wasserheit J N. Genital chlamydial infections: epidemiology and reproductive sequelae. Am J Obstet Gynecol. 1991;164:1771–1781. doi: 10.1016/0002-9378(91)90559-a. [DOI] [PubMed] [Google Scholar]
- 5.Comanducci M, Ricci S, Ratti G. The structure of a plasmid of Chlamydia trachomatis believed to be required for growth within mammalian cells. Mol Microbiol. 1988;2:531–538. doi: 10.1111/j.1365-2958.1988.tb00060.x. [DOI] [PubMed] [Google Scholar]
- 6.Comanducci M, Ricci S, Cevenini R, Ratti G. Diversity of the chlamydial common plasmid in biovars with different pathogenicity. Plasmid. 1990;23:149–154. doi: 10.1016/0147-619x(90)90034-a. [DOI] [PubMed] [Google Scholar]
- 7.Comanducci M, Manetti R, Bini L, Santucci A, Pallini V, Cevenini R, Sueur J-M, Orfila J, Ratti G. Humoral immune response to plasmid protein pgp3 in patients with Chlamydia trachomatis infection. Infect Immun. 1994;62:5491–5497. doi: 10.1128/iai.62.12.5491-5497.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dille B J, Butzen C C, Birkenmeyer L G. Amplification of Chlamydia trachomatis DNA by ligase chain reaction. J Clin Microbiol. 1993;31:729–731. doi: 10.1128/jcm.31.3.729-731.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan V S, Jenkin H M. Glycogen metabolism in chlamydia-infected HeLa cells. J Bacteriol. 1970;104:608–609. doi: 10.1128/jb.104.1.608-609.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farencena A, Comanducci M, Donati M, Ratti G, Cevenini R. Characterization of a new isolate of Chlamydia trachomatis which lacks the common plasmid and has properties of biovar trachoma. Infect Immun. 1997;65:2965–2969. doi: 10.1128/iai.65.7.2965-2969.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukushi H, Hirai K. Proposal of Chlamydia pecorum strain derived from ruminants. Int J Syst Bacteriol. 1992;42:306–308. doi: 10.1099/00207713-42-2-306. [DOI] [PubMed] [Google Scholar]
- 12.Fukushi H, Hirai K. Genetic diversity of avian and mammalian Chlamydia psittaci strains and relation to host origin. J Bacteriol. 1989;171:2850–2855. doi: 10.1128/jb.171.5.2850-2855.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon F B, Quan A L. Occurrence of glycogen in inclusions of the psittacosis-lymphogranuloma venereum-trachoma agents. J Infect Dis. 1965;115:186–191. doi: 10.1093/infdis/115.2.186. [DOI] [PubMed] [Google Scholar]
- 14.Grayston J T, Kuo C-C, Campbell L A, Wang S-P. Chlamydia pneumoniae sp. nov. for chlamydia sp. strain TWAR. Int J Syst Bacteriol. 1989;39:88–90. [Google Scholar]
- 15.Griffith J D. Electron microscopic visulization of DNA in association with cellular components. In: Prescott D M, editor. Methods in cell biology. VII. New York, N.Y: Academic Press, Inc.; 1973. pp. 129–146. [DOI] [PubMed] [Google Scholar]
- 16.Hanson R S, Phillips J A. Chemical composition. In: Gerhandt P, et al., editors. Manual of methods for general bacteriology. Washington, D.C: American Society for Microbiology; 1981. pp. 328–364. [Google Scholar]
- 17.Hatch I P, Vance D W, Jr, Al-Hossainy E. Attachment of Chlamydia psittaci to formaldehyde-fixed and unfixed L cells. J Gen Microbiol. 1981;125:273–283. doi: 10.1099/00221287-125-2-273. [DOI] [PubMed] [Google Scholar]
- 18.Hatt C, Ward M E, Clarks I N. Analysis of the entire nucleotide sequence of the cryptic plasmid of Chlamydia trachomatis serovar L1. Evidence for involvement in DNA replication. Nucleic Acids Res. 1988;16:4053–4067. doi: 10.1093/nar/16.9.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higashi N, Tamura A. A plaque assay for meningopneumonitis virus in monolayers of strain L cells. Virology. 1960;12:578–588. doi: 10.1016/0042-6822(60)90180-x. [DOI] [PubMed] [Google Scholar]
- 20.Hodinka R L, Davis C H, Choong J, Wyrick P B. Ultrastructural study of endocytosis of Chlamydia trachomatis by McCoy cells. Infect Immun. 1988;56:1456–1463. doi: 10.1128/iai.56.6.1456-1463.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Judson F N. Assessing the number of genital and chlamydial infections in the United States. J Reprod Med. 1985;30:269–272. [PubMed] [Google Scholar]
- 22.Kiel J A, Boels J M, Beldman G, Venema G. Glycogen in Bacillus subtilis: molecular characterization of an operon encoding enzyme involved in glycogen biosynthesis and degradation. Mol Microbiol. 1994;11:203–218. doi: 10.1111/j.1365-2958.1994.tb00301.x. [DOI] [PubMed] [Google Scholar]
- 23.Kozikowski E H, Hahon N. Plaque formation by psittacosis virus. J Bacteriol. 1964;88:533–534. doi: 10.1128/jb.88.2.533-534.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumamoto Y, Sato T, Hirose T, Nishimura M, Koroku M, Murakami S, Saito I, Kojima H, Okazaki T, Kawamura N, Hisazumi H, Kawada Y, Maeda S, Kamidono S, Ohmori H, Kumazawa J, Ohi Y, Fujimoto M, Katagiri S, Takahashi K, Matsuda S, Takada M, Sugase M, Noguchi M, Okada H, Nakano H, Motomura R. Epidemiologic study of urogenital Chlamydia trachomatis infection. Jpn Arch Sex Transm Dis. 1994;5:32–42. doi: 10.1097/00007435-199411000-00006. . (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 25.Loeffelholz M J, Lewinski C A, Silver S R, Purohit A P, Herman S A, Buonagurio D A, Dragon E A. Detection of Chlamydia trachomatis in endocervical specimens by polymerase chain reaction. J Clin Microbiol. 1992;30:2847–2851. doi: 10.1128/jcm.30.11.2847-2851.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto A, Bessho H, Uehira K, Suda T. Morphological studies of the association of mitochondria with chlamydial inclusions and the fusion of chlamydial inclusions. J Electron Microsc. 1991;40:356–363. [PubMed] [Google Scholar]
- 27.McGregor J A, French J I. Chlamydia trachomatis infection in pregnancy. Am J Obstet Gynecol. 1991;164:1782–1789. doi: 10.1016/0002-9378(91)90560-e. [DOI] [PubMed] [Google Scholar]
- 28.Miyashita N, Kubota Y, Kimura M, Nakajima M, Niki Y, Soejima R, Matsumoto A. Characterization of a Chlamydia pneumoniae isolated from a 57-year-old man. Microbiol Immunol. 1994;38:857–864. doi: 10.1111/j.1348-0421.1994.tb02138.x. [DOI] [PubMed] [Google Scholar]
- 29.Miyashita N, Matsumoto A. Establishment of a particle counting method for purified elementary bodies of chlamydiae and evaluation of sensitivities of the IDEIA Chlamydia kit and DNA probe by using the purified elementary bodies. J Clin Microbiol. 1992;30:2911–2916. doi: 10.1128/jcm.30.11.2911-2916.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyashita N, Iijima Y, Matsumoto A. Evaluation of the sensitivity and specificity of polymerase chain reaction test kit AMPLICOR Chlamydia trachomatis. Microbiol Immunol. 1994;38:81–85. doi: 10.1111/j.1348-0421.1994.tb01747.x. [DOI] [PubMed] [Google Scholar]
- 31.Miyashita N, Matsumoto A, Niki Y, Matsushima T. Evaluation of the sensitivity and specificity of a ligase chain reaction test kit for the detection of Chlamydia trachomatis. J Clin Pathol. 1996;49:1–3. doi: 10.1136/jcp.49.6.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moulder J W, Hatch T T, Kuo C-C, Schachter J, Storz J. Order II. Chlamydiales Storz and Page 1971, 334AL. In: Krieg N R, editor. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 729–739. [Google Scholar]
- 33.Moulder J W. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer L, Falkow S. A common plasmid of Chlamydia trachomatis. Plasmid. 1986;16:52–62. doi: 10.1016/0147-619x(86)90079-x. [DOI] [PubMed] [Google Scholar]
- 35.Peterson E M, Markoff B A, Schachter J, De La Maza L M. The 7.5-kb plasmid present in Chlamydia trachomatis is not essential for the growth of this microorganism. Plasmid. 1990;23:144–148. doi: 10.1016/0147-619x(90)90033-9. [DOI] [PubMed] [Google Scholar]
- 36.Piraino F, Abel C. Plaque assay for psittacosis virus in monolayers of chick embryo fibroblasts. J Bacteriol. 1964;87:1503–1511. doi: 10.1128/jb.87.6.1503-1511.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piraino F. Plaque formation in chick embryo fibroblast cells by Chlamydia isolated from avian and mammalian sources. J Bacteriol. 1969;98:475–480. doi: 10.1128/jb.98.2.475-480.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez P, Vekris A, de Barbeyrac B, Dutilh B, Bonnet J, Bebear C. Typing of Chlamydia trachomatis by restriction endonuclease analysis of the amplified major outer membrane protein gene. J Clin Microbiol. 1991;29:1132–1136. doi: 10.1128/jcm.29.6.1132-1136.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning; a laboratory manual. 2nd ed. 1989. pp. 5.28–5.32. and p. 9.32–9.33. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 40.Sayada C, Denamur E, Xerri B, Orfila J, Elion J. Rapid genotyping of the Chlamydia trachomatis major outer membrane protein by the polymerase chain reaction. FEMS Microbiol Lett. 1991;83:73–78. doi: 10.1016/0378-1097(91)90447-i. [DOI] [PubMed] [Google Scholar]
- 41.Sriprakash K S, Macavoy E S. Characterization and sequence of a plasmid from the trachoma biovars of Chlamydia trachomatis. Plasmid. 1987;18:205–214. doi: 10.1016/0147-619x(87)90063-1. [DOI] [PubMed] [Google Scholar]
- 42.Uttaro A D, Ugalde R A. A chromosomal cluster of genes encoding ADP-glucose synthetase, glycogen synthase and phosphoglucomutase in Agrobacterium tumefaciens. Gene. 1994;150:117–122. doi: 10.1016/0378-1119(94)90869-9. [DOI] [PubMed] [Google Scholar]
- 43.Wang S-P, Grayston J T. Three new serovars of Chlamydia trachomatis: Da, Ia and L2a. J Infect Dis. 1991;163:403–405. doi: 10.1093/infdis/163.2.403. [DOI] [PubMed] [Google Scholar]
- 44.Wang S-P, Kuo C-C, Barnes R C, Stephens R S, Grayston J T. Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J Infect Dis. 1985;152:791–800. doi: 10.1093/infdis/152.4.791. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida H, Kishi Y, Shiga S, Inouye S, Hagiwara T. Serotyping of Chlamydia trachomatis by polymerase chain reaction. Jpn Arch Sex Transm Dis. 1995;6:40–45. . (In Japanese.) [Google Scholar]