Abstract
Bats are exceptional among mammals for their powered flight, extended lifespans, and robust immune systems and therefore have been of particular interest in comparative genomics. Using the Oxford Nanopore Technologies long-read platform, we sequenced the genomes of two bat species with key phylogenetic positions, the Jamaican fruit bat (Artibeus jamaicensis) and the Mesoamerican mustached bat (Pteronotus mesoamericanus), and carried out a comprehensive comparative genomic analysis with a diverse collection of bats and other mammals. The high-quality, long-read genome assemblies revealed a contraction of interferon (IFN)-α at the immunity-related type I IFN locus in bats, resulting in a shift in relative IFN-ω and IFN-α copy numbers. Contradicting previous hypotheses of constitutive expression of IFN-α being a feature of the bat immune system, three bat species lost all IFN-α genes. This shift to IFN-ω could contribute to the increased viral tolerance that has made bats a common reservoir for viruses that can be transmitted to humans. Antiviral genes stimulated by type I IFNs also showed evidence of rapid evolution, including a lineage-specific duplication of IFN-induced transmembrane genes and positive selection in IFIT2. In addition, 33 tumor suppressors and 6 DNA-repair genes showed signs of positive selection, perhaps contributing to increased longevity and reduced cancer rates in bats. The robust immune systems of bats rely on both bat-wide and lineage-specific evolution in the immune gene repertoire, suggesting diverse immune strategies. Our study provides new genomic resources for bats and sheds new light on the extraordinary molecular evolution in this critically important group of mammals.
Keywords: cancer resistance, Chiroptera, comparative genomics, immunity, long reads, viral tolerance
Significance.
Bats are known for their robust immune systems and cancer resistance, but comparative genomics studies of these unique adaptations have been limited by low sample sizes and incomplete short-read genomes. The analysis of 15 bat genomes, including 8 contiguous long-read genomes, showed a shift in antiviral interferon (IFN)-α and IFN-ω gene copy numbers in bats, as well as positive selection in antiviral genes, tumor suppressors, and DNA-repair genes.
Introduction
Bats (order Chiroptera) form the second largest order of mammals and are known for a wide variety of remarkable adaptations including powered flight (Simmons et al. 2008), laryngeal echolocation (Simmons and Geisler 1998; Moss and Surlykke 2001), unusual longevity (Wilkinson and Adams 2019), and low rates of cancer (Wang et al. 2011). Bats are also hosts of diverse viruses (Calisher et al. 2006; Olival et al. 2017) and have played roles in outbreaks of emerging zoonotic viruses including Marburg virus (Amman et al. 2012), Nipah virus (Pulliam et al. 2012), and severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) (Li et al. 2005), either through direct human contact or via bridge hosts. Bats may also have played a role in the emergence of SARS-CoV-2, as closely related progenitor viruses have been detected in wild bats (Andersen et al. 2020; Holmes et al. 2021; Pekar et al. 2022). The ability of bats to tolerate viral infections may stem from unusual features of their innate immune response (Pavlovich et al. 2018). Together, these adaptations make bats a powerful system for investigating a wide variety of genotype-to-phenotype relationships, including several with implications for human health. For example, by better understanding the mechanisms of the bat immune system that allow them to tolerate viral infections (Wang et al. 2011), researchers may be better able to prevent zoonotic outbreaks (Wang et al. 2021; Becker et al. 2022). In addition, comparative genomic analyses of bats and cancer-susceptible mammals may shed new light on the causes of cancer and links between cancer and immunity (Gonzalez et al. 2018). Importantly, such studies of bats and other nonmodel organisms are highly complementary to studies based on mouse models, which are far more amenable to experimental manipulation but exhibit fewer natural adaptations relevant to human disease.
With these goals in mind, investigators have sequenced and assembled the genomes of at least 44 bat species over the past decade (supplementary table S1, Supplementary Material online). Recently, sequencing efforts in bats have been accelerated by the Bat1K global genome consortium (Teeling et al. 2018), DNA Zoo (Dudchenko et al. 2017), and Vertebrate Genome Project (Rhie et al. 2021). These new genome sequences have revealed numerous intriguing features of the immune systems of bats (Zhang et al. 2013; Escalera-Zamudio et al. 2015; Pavlovich et al. 2018; Zepeda Mendoza et al. 2018; Hawkins et al. 2019; Gorbunova et al. 2020; Jebb et al. 2020; Irving et al. 2021; Moreno Santillán et al. 2021). In particular, several genes with key roles in the innate immune system appear to have adaptively evolved in bats, including primary lines of inducible host defenses such as pathogen sensors (Escalera-Zamudio et al. 2015; Jiang et al. 2017), type I interferons (IFNs) (Zhou et al. 2016; Pavlovich et al. 2018), and antiviral genes (Fuchs et al. 2017). Specifically, bats have lost the mammalian PYHIN DNA-sensing gene family (Zhang et al. 2013; Ahn et al. 2016), they show evidence of positive selection in pathogen-sensing Toll-like receptors (TLRs) (Jiang et al. 2017), and they display copy number variation in type I IFN cytokines (Zhou et al. 2016; Pavlovich et al. 2018), which are induced by TLRs. Bat-specific modifications in immune response, tumor suppressors, DNA damage checkpoint-DNA repair pathway genes (Zhang et al. 2013), and growth hormone (Seim et al. 2013) may be associated with cancer resistance. It is thought that these adaptations in innate immunity and cancer resistance may have arisen as a result of coevolution of bats with viruses (Taylor et al. 2011; Jebb et al. 2020) and that a need for enhanced DNA repair in the face of elevated reactive oxygen species (ROS) may have been a consequence of powered flight (Zhang et al. 2013).
In this study, we augment previously existing genome sequences with new Oxford Nanopore Technologies (ONT)-based long-read assemblies for the Jamaican fruit bat (Artibeus jamaicensis) and the Mesoamerican mustached bat (Pteronotus mesoamericanus) (supplementary fig. S1, Supplementary Material online). Both species belong to Noctilionoidea, a Neotropical superfamily that comprises ∼16% of global bat diversity (www.batnames.org). Artibeus jamaicensis is one of the most common Neotropical mammals and a model species in mammalian research (Larsen et al. 2007) including work on bat immunology (Cabrera-Romo et al. 2014; Munster et al. 2016; David et al. 2022). It is a member of the family Phyllostomidae, arguably the most ecologically diverse lineage of living mammals (Fleming et al. 2020). Pteronotus mesoamericanus is a representative of the family Mormoopidae, the less well-studied sister group of the species-rich phyllostomids to which A. jamaicensis belongs (Rojas et al. 2016), making P. mesoamericanus the first long-read sequenced species in a critical outgroup for study of evolutionary changes in the phyllostomid radiation. Here, we present a comprehensive analysis of these genome sequences together with 13 previously assembled bats and other mammalian genomes. We aim to leverage the high-quality, long-read assemblies to enable accurate and complete characterization of gene duplications and losses and of genomic repeats (Vollger et al. 2019; Halo et al. 2021; Rhie et al. 2021; Blumer et al. 2022). The benefits of long-read assemblies are of particular value in studies of mammalian immunity-related genes (O’Connor and Cornwallis 2022), many of which fall in highly repetitive genomic regions including large arrays of duplicated genes (He et al. 2021). Our comparative genomic analysis of these genome sequences, which have been released as a public resource, provides several new insights into unique features of innate immune response and cancer resistance in bats.
Results
Genomic Structure of A. jamaicensis and P. mesoamericanus
Recent work in bats has generated several long-read genome assemblies (Pavlovich et al. 2018; Jebb et al. 2020; Moreno Santillán et al. 2021; Blumer et al. 2022), better enabling the study of complex regions. In this case, we were able to leverage the ONT long-read sequencing platform and an optimized flye-PEPPER-POLCA (Kolmogorov et al. 2019; Zimin and Salzberg 2020; Shafin et al. 2021) assembly and polishing strategy (see Materials and Methods) to generate reference-quality assemblies for A. jamaicensis and P. mesoamericanus with contig N50 values of 28–29 Mb (supplementary fig. S2 and supplementary table S2, Supplementary Material online) and POLCA consensus accuracy >99.99%. Using EVidenceModeler, we annotated 21,621 genes in A. jamaicensis and 21,269 genes in P. mesoamericanus. Based on the Benchmarking Sets of Universal Single-Copy Orthologs (BUSCO) protein assessment of our annotations, the gene sets in both bats are highly complete at 98.3% and 98.2%, respectively, comparable with the values of 97.4–98.3% reported for six recent PacBio-based bat assemblies (supplementary fig. S2, Supplementary Material online). Notably, all of these long-read bat assemblies have BUSCO scores approaching those of the human (99.9%) and mouse (99.9%) genomes. Orthofinder orthology detection produced 19,935 orthogroups for 15 bats and 5 outgroup mammals, of which 12,836 single-copy orthogroups were set aside for our positive selection analysis (below). Total fractions of 39.2% and 37.9% of the A. jamaicensis and P. mesoamericanus genomes consisted of repeats, respectively, with 0.4% in each genome attributed to recently active transposons including hAT, TcMariner, and piggyBac elements (supplementary fig. S2 and supplementary table S3, Supplementary Material online). We also detected nonretroviral endogenous viral elements, predominantly derived from Bornaviridae and Parvoviridae (supplementary table S4, Supplementary Material online). We provide our annotations, aligned evidence, and multiple genome alignments as a public University of California Santa Cruz (UCSC) genome browser instance (http://compgen.cshl.edu/bat).
Gene Family Expansion and Contraction Analysis
Changes in gene-family size have played an important role in shaping the immune systems of bats (Zhang et al. 2013; Ahn et al. 2016). To facilitate further analysis of gene-family expansions and contractions, we focused on our new ONT-based assemblies and the previously published long-read bat genome sequences. By comparing these bat genomes with mammalian outgroups (human, mouse, dog, pig, and horse), we identified 14 expanded gene families and 105 contracted gene families in the most recent common ancestor of bats (hereafter, the “bat MRCA”; supplementary table S5, Supplementary Material online). Thirty-nine of these 119 gene families changing size in the bat MRCA were related to immune system processes (Fisher's exact test, P = 2.1e−4), including the previously identified PYHIN gene family (PTHR12200) (Ahn et al. 2016), which was absent in all bats including the 2 newly sequenced species. There were significant differences in gene birth–death rates (λ) among Yangochiroptera, Yinpterochiroptera, and nonbat mammals (see Materials and Methods). Yangochiroptera showed the highest rate of gene birth–death, with a λ value of 0.0017 per gene per million years compared with the 0.0008 estimated for Yinpterochiroptera and 0.0005 for nonbat mammals.
The type I IFN locus is contracted in bats through losses of IFN-α but not IFN-ω
The type I IFN immune response is a critical component of the mammalian innate immune system and is responsible for activating the expression of a battery of antiviral genes following induction by pathogen-sensing components such as PYHINs, TLRs, and cGAS-STING (McNab et al. 2015). Previous comparative analyses of the type I IFN locus have shown that it is highly structurally variable in bats and other mammals (Zhou et al. 2016), with some bats, such as Pteropus alecto, showing a contraction (Zhou et al. 2016), whereas others such as Rousettus aegyptiacus (Pavlovich et al. 2018), Pteropus vampyrus, and Myotis lucifugus (Kepler et al. 2010) show evidence of expansions. However, this locus is generally large and highly duplicated across mammals (e.g., in humans, it spans ∼400 kb and contains 16 IFN genes, including 13 IFN-α genes and 1 IFN-ω gene), making it challenging to assemble and analyze.
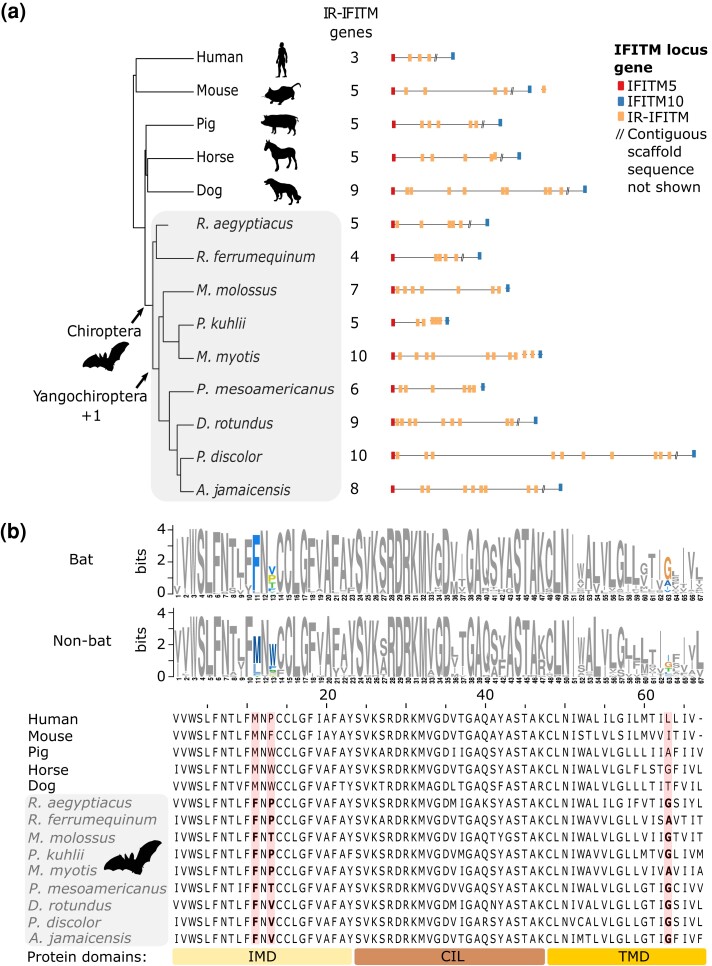
Using our expanded set of long read-based bat genomes, we found evidence of a major contraction (−9 genes; Viterbi method (De Bie et al. 2006), P = 2.7e−15) of the type I IFN gene family in the bat MRCA (fig. 1; supplementary fig. S3, Supplementary Material online). Comparison of the IFN locus in our assembly with this locus in an earlier short-read assembly of A. jamaicensis shows that the short-read assembly is both fragmented and incomplete in gene and noncoding sequence content, highlighting the value of long reads for analysis of complex loci (supplementary fig. S4, Supplementary Material online). We found that this contraction was driven specifically by loss of IFN-α genes, with gene counts of 0–4 in bats compared with 10–18 in the outgroup mammals. In contrast, IFN-ω gene counts in the bat ancestor were largely unchanged (Viterbi method, P = 0.53), ranging from three to seven in bats and zero to eight in other mammals. As a consequence, IFN-ω was 11-fold enriched relative to IFN-α in bats compared with the other mammals (Fisher's exact test, P = 4.5e-11, odds ratio = 10.80), and this enrichment was observed in every bat species (supplementary table S6, Supplementary Material online). Considering the relative ligand-binding and antiproliferative properties of IFN-ω and IFN-α (Jaks et al. 2007; Thomas et al. 2011), these changes in gene number could potentially be responsible for more potent responses to viral infections in bats relative to other mammalian orders (see Discussion).
Fig. 1.
Contraction of the type I IFN locus in bats compared with other mammals. A loss of nine genes in the bat MRCA was estimated by CAFE (Viterbi method, P = 2.7e−15). The reduction in locus size occurred together with a significant loss of IFN-α genes but not IFN-ω genes in the bat MRCA (Fisher's exact test, P = 4.5e−11, odds ratio = 10.80). The type I IFN loci in bats (gray background) are shown for long-read–based assemblies as well as a BAC-based locus assembly of P. alecto (Zhou et al. 2016) and an Illumina-based assembly of D. rotundus (Zepeda Mendoza et al. 2018). M. molossus, Molossus molossus.
Antiviral IFN-Induced Transmembrane Genes Are Expanded in Yangochiroptera Bats
An important downstream consequence of activation of type I IFNs is the expression of various antiviral IFN-stimulated genes (ISGs) (de Weerd et al. 2007). In bats, several of these genes, such as tetherin and APOBEC3, have also been shown to be under positive selection (Hayward et al. 2020) or duplicated (Jebb et al. 2020). Among antiviral ISGs, we observed an expansion of the immune-related IFN-induced transmembrane (IR-IFITM) gene family (+1 gene; Viterbi method, P = 0.018) on the branch leading to the Yangochiroptera, the suborder that includes most microbats (fig. 2). The IR-IFITMs—which have previously been reported to be under positive selection in bats (Benfield et al. 2019)—are potent broad-spectrum antiviral factors that help to prevent infection before a virus passes the lipid bilayer of the cell (Bailey et al. 2014; Desai et al. 2014). By applying a branch-site likelihood ratio test (see Materials and Methods) based on the nonsynonymous/synonymous rate ratio (dN/dS) also known as ω, we confirmed using our data that the IR-IFITMs show evidence of positive selection in the bat MRCA (P = 6.4e−3). Furthermore, we identified seven particular codon sites that show signs of episodic diversifying selection (supplementary table S7, Supplementary Material online), including three sites in the CD225 domain (codons 68, 70, and 117). Notably, codons 68 and 70 in the CD255 domain are among several sites previously shown to be critical for blocking viral infection (Benfield et al. 2019). Furthermore, the methionine-to-phenylalanine substitution at codon 68 occurs in an amphipathic helix previously shown to be essential for blocking viral infection (Chesarino et al. 2017). Together, these observations of gene duplications and positive selection in functional domains in IR-IFITMs suggest that this gene family may have played an important role in the evolution of antiviral responses in bats.
Fig. 2.
IFITM gene family expansion and positive selection associated with the bat antiviral immune response. (a) Phylogeny of bats and other mammals, showing a significant increase in gene copy number at the IFITM locus in Yangochiroptera bats based on CAFE analysis. IR-IFITM genes are shown in yellow. (b) Three codon sites in the IFITM transmembrane domains IMD and TMD in bats show evidence of positive selection (see also supplementary table S7, Supplementary Material online). The sequence logo (top) compares a 67-amino-acid region spanning these domains with orthologous regions from other mammals. The sequence alignment (middle) compares human IFITM3 with the most similar representative IR-IFITM ortholog from the other species (selected sites highlighted and shown in bold).
Expansion of PRDM9 in Phyllostomid Bats and Expansion of Heat Shock Proteins in P. mesoamericanus
A third gene family to emerge from our survey of gene expansions and contractions was PRDM9 (supplementary table S8, Supplementary Material online), which specifies the location of meiotic recombination sites (Paigen and Petkov 2018) and is known to evolve rapidly in vertebrates (Baker et al. 2017). PRDM9 may play a role in speciation (Schwartz et al. 2014, 9) and is also upregulated upon viral infection (Xie et al. 2019). We found that PRDM9 experienced a striking expansion in phyllostomid bats (+5 genes, Viterbi method P = 1.19e−05; supplementary fig. S5, Supplementary Material online), far beyond anything observed in other mammals. This observation has not, to the best of our knowledge, been made in the literature and was facilitated by our high-quality, long-read genomes. Our genomes for P. mesoamericanus and A. jamaicensis revealed that the expansion is specific to the phyllostomids and that a substantially larger expansion of PRDM9 occurred in A. jamaicensis than can be observed in the short-read assembly (supplementary table S9, Supplementary Material online). Furthermore, comparison with a recent analysis of PRDM9 in 446 vertebrates suggests that the highest PRDM9 copy number is found in phyllostomid bats (Cavassim et al. 2022). Phyllostomid bats stand out for their morphological diversity and their extensive chromosomal rearrangements (Sotero-Caio et al. 2013), raising the possibility that the expansion of PRDM9 and its effect on meiosis played a role in these traits. Intriguingly, many of the PRDM9 copies in phyllostomid bats have lost the KRAB domain (supplementary table S9, Supplementary Material online), which is thought to play an important role in recruiting the recombination machinery (Baker et al. 2017), suggesting they may have alternate functions.
Finally, we observed a major expansion in P. mesoamericanus of heat-shock proteins across multiple gene families, including heat-shock protein 70 kDa (PTHR19375, +10 genes; Viterbi method, P = 8.9e−9). Interestingly, this expansion was largely restricted to P. mesoamericanus and was not observed across other bats. Overexpression of heat-shock proteins can modulate immune responses (Tsan and Gao 2009); therefore, this duplication may have implications for immunity in P. mesoamericanus.
Positive Selection Analysis
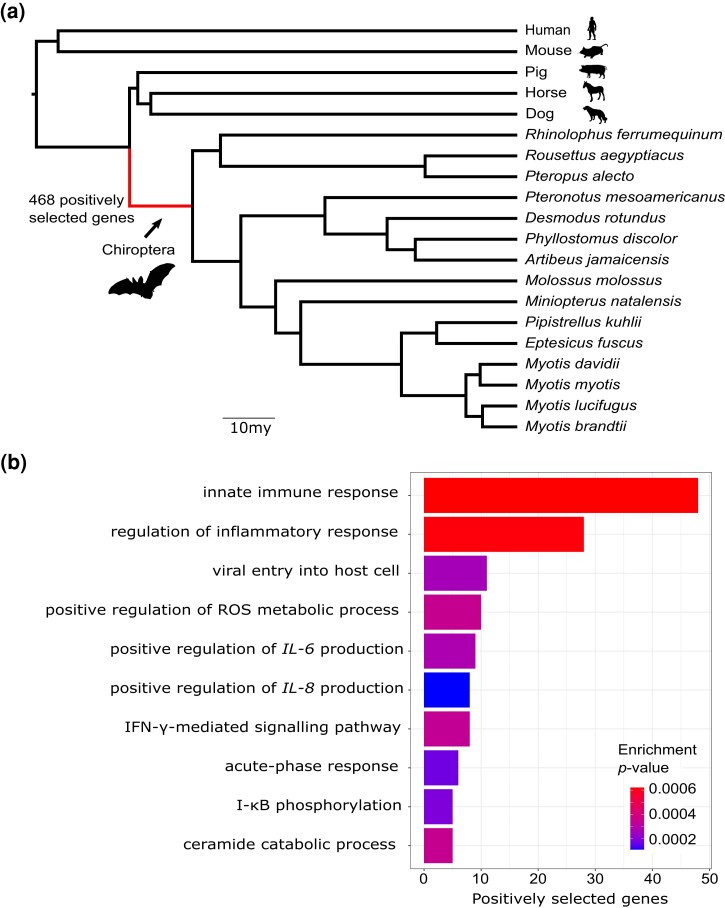
Having identified signatures of positive selection at the amino-acid level in several gene families of interest, we applied similar branch-site likelihood ratio tests (Yang and Nielsen 2002; Zhang et al. 2005; Smith et al. 2015) genome wide, focusing on 12,517 single-copy orthologs present in bats and outgroup mammals. Because we were interested in molecular traits shared by most bats, we focused on a test for positive selection on the branch leading to the bat MRCA, where we expected to have reasonably good statistical power. However, we also tested for positive selection on the lineages leading to each of the two newly sequenced bats. Observing a highly skewed distribution of nominal P-values (P > 0.98 in 86% of tests) indicative of a misspecified null model, we opted to follow a recent study in bats (Potter et al. 2021) and omit a correction for multiple comparisons across orthologs, instead adjusting only for testing on three different branches (but see supplementary table S10 and supplementary Data S1, Supplementary Material online for more conservative adjusted P-values). As the simulation study conducted by Potter et al. (2021) shows, multiple testing correction of branch-site test P-values can be overly conservative and can remove substantial numbers of true positives. Based on our testing strategy, we identified 468 positively selected genes (PSGs) with an adjusted P < 0.05 in the bat MRCA (fig. 3; supplementary table S10, Supplementary Material online). This number is roughly comparable with the 298 PSGs recently identified on the phyllostomid bat lineage using a similar filtering approach (Potter et al. 2021), but somewhat larger than the 181 PSGs (Hawkins et al. 2019) and 23 PSGs (Jebb et al. 2020) found on the bat ancestral branch in recent studies that used more stringent criteria. These PSGs were strongly enriched for immune-related functions (supplementary table S11, Supplementary Material online), including the major gene ontology (GO) biological processes “regulation of inflammatory response” (GO:0050727, P = 6.1e−4) and “innate immune response” (GO:0045087, P = 6.2e−4). In total, 125 PSGs (27% of the 468) were annotated with the parent term “immune system process” (GO:0002376). The bat MRCA branch was also enriched for PSGs involved in “positive regulation of ROS” (GO:2000379, P = 3.5e−4), possibly suggesting adaptations associated with heightened metabolic rates owing to flight (see Discussion). Moreover, we detected 6 autophagy-related PSGs, including autophagy regulator ATG9B which was previously implicated in bat longevity (Kacprzyk et al. 2021). Below, we further discuss some specific PSGs falling in two major classes: 1) pathogen sensors, cytokines, and antiviral genes and 2) DNA repair genes and tumor suppressors.
Fig. 3.
Positive selection scan on the bat ancestral branch. (a) Maximum likelihood phylogeny based on codon-site partitioned analysis of 3,632 gene alignments, with the bat ancestral branch indicated. (b) TopGO hierarchical GO enrichment analysis of the PSGs against the background set of tested genes, suggesting strong enrichment of innate immunity genes. The ten most significant GO terms are shown, eight of which are related to innate immunity including genes involved in interleukin regulation and IFN pathways.
Pathogen Sensors, Interleukins, and Antiviral Genes in Bats Are Rapidly Evolving
The 468 PSGs on the bat ancestral branch included several genes that encode proteins with pathogen-sensing roles. For example, the TLR-encoding genes TLR7 and TLR8, are included in our set (supplementary table S10, Supplementary Material online), and the related TLR9 (which shows reduced activation in bats [Banerjee et al. 2017]) was on the threshold of statistical significance (P = 0.05). These genes have been identified in previous scans for positive selection (Escalera-Zamudio et al. 2015; Jiang et al. 2017). Another previously identified PSG in our set is the IFN stimulator STING (Xie et al. 2018). The TLRs and STING, as well as the PSG NLRP3 (Zhang et al. 2013; Ahn et al. 2019), all play important roles in inflammation and are considered therapeutic targets for inflammatory disease (Hennessy et al. 2010; Fitzgerald and Kagan 2020; Gouravani et al. 2020; Decout et al. 2021). Positive selection in these genes may play a role in dampening downstream responses to pathogens. Interestingly, the location of a bat-specific substitution in codon 358 of STING that was linked to dampened IFN activation (Xie et al. 2018) was identified in our scan as one of ten positively selected sites (supplementary table S7, Supplementary Material online). In addition to these previously identified PSGs, we identified TLR2 to be under positive selection. Unlike the nucleic-acid sensing TLRs 7, 8, and 9, TLR2 recognizes lipoproteins of pathogens such as bacteria and enveloped viruses (Oliveira-Nascimento et al. 2012). TLR2 signaling also induces the inflammatory cytokine TNF-α in infections with viruses like SARS-CoV-2, and blocking TLR2 protects against pathogenesis caused by the “cytokine storm” (Zheng et al. 2021). We additionally found evidence of positive selection in the key TLR regulator UNC93B1 (Kim et al. 2008), which is regulated by type I IFNs (Panchanathan et al. 2013). UNC93B1 is thought to regulate nucleic-acid sensing TLRs such as TLR7 and TLR9 upstream of the process of TLR trafficking from the endoplasmic reticulum to endolysosomes (Lee et al. 2013; Pelka et al. 2018).
Another group prominently represented in our PSGs are genes encoding the interleukins, a collection of cytokines with diverse functions in immunity and inflammation (Garlanda et al. 2013; Veldhoen 2017). In bats, earlier work identified reduced expression of interleukin-8 (Banerjee et al. 2017), positive selection of several interleukins and interleukin receptors (Moreno Santillán et al. 2021), and loss of interleukin-36γ (Jebb et al. 2020), suggesting that rapid evolution of interleukins may have contributed to unique molecular adaptations in bat immunity. We found that 5 interleukin-related GO categories were significantly enriched for PSGs in bats (supplementary table S11, Supplementary Material online), including “interleukin-1β production” (GO:0032611, P = 0.00075) and “positive regulation of interleukin-6 production” (GO:0032755, P = 0.00028). In addition, we identified the pleiotropic cytokine-encoding genes interleukin-6 and interleukin-15 as PSGs as well as the genes encoding several interleukin-associated receptors (supplementary table S10, Supplementary Material online). Interleukin-6, which encodes one of the most important cytokines during infection (Velazquez-Salinas et al. 2019), showed 6 sites predicted to be positively selected in bats and a high ratio of nonsynonymous substitutions per nonsynonymous site to the number of synonymous substitutions per synonymous site on the bat MRCA branch (dN/dS = 2.72). Inflammatory cytokine-encoding genes such as interleukin-6 and interleukin-15 that were under positive selection in the bat MRCA could be additional contributors to dampened inflammation in bats.
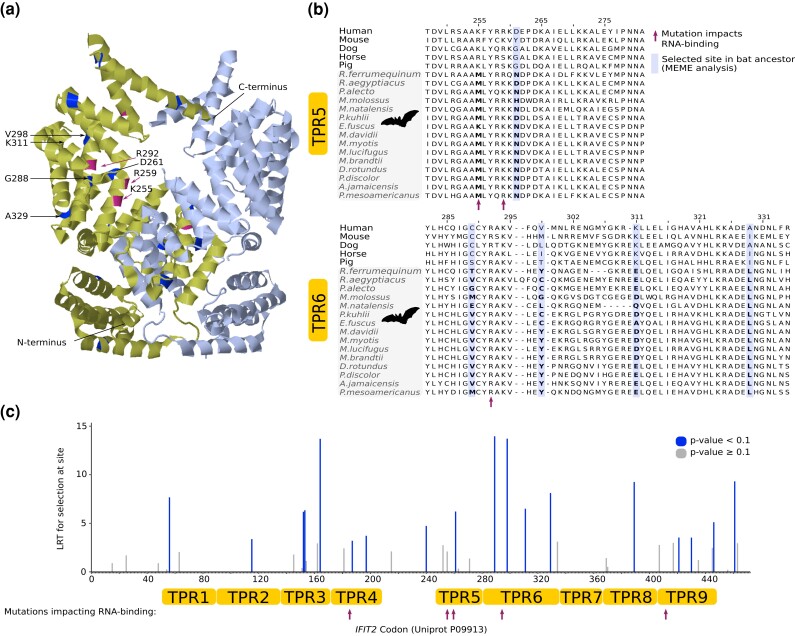
Our PSGs also included several antiviral ISGs, which, as noted above, are activated by type I IFNs (de Weerd et al. 2007) and in several cases (including APOBEC3 and Mx) have been shown to be duplicated (Jebb et al. 2020) and/or under positive selection (Fuchs et al. 2017; Hayward et al. 2018) in bats. For example, we detected positive selection in PARP9, whose gene product interacts with DTX3L and STAT1 to enhance IFN responsiveness (Zhang et al. 2015; Xing et al. 2021). We also found strong evidence of positive selection, affecting 18 codons, in IFIT2, whose product inhibits replication of a broad range of RNA and DNA viruses (Diamond and Farzan 2013; Fensterl and Sen 2014) (fig. 4). Ten of the positively selected sites overlap or are physically close to sites known to impact RNA-binding in the TPR4, TPR5, TPR6, and TPR9 motifs (Yang et al. 2012), and a bat-specific lysine-to-methionine substitution occurs at codon 255 which is involved in RNA-binding (Yang et al. 2012) (fig. 4). Positive selection of IFIT2 in bats may therefore alter, and possibly enhance, expression of numerous antiviral response genes. Taken together, our results provide novel evidence for rapid evolution in the innate immune system of bats (fig. 5).
Fig. 4.
The antiviral gene IFIT2 is positively selected in RNA-binding regions in bats. (a) 3D structure of the IFIT2 protein (PDB:4G1T) showing sites selected in the bat ancestor and sites known to be involved in RNA-binding function of the protein. (b) Amino acid alignment of the tetratricopeptide repeat (TPR) regions TPR5 and TPR6 (based on UniProt annotation for P09913) showing sites selected in the bat ancestor (highlighted and bold) and a fixed bat lysine-to-methionine substitution in codon 255 which is located in a region involved in RNA-binding (indicated with arrow). (c) MEME analysis of IFIT2 codon sites showing 18 sites selected in bats based on a likelihood ratio test.
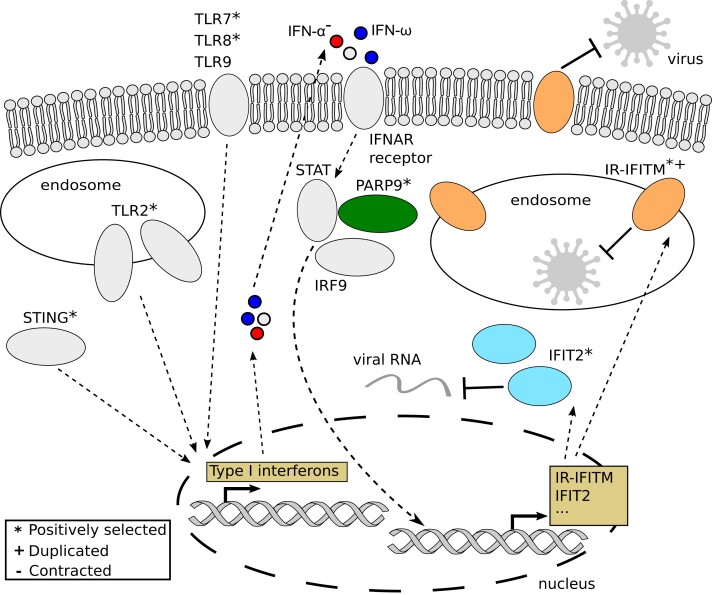
Fig. 5.
Schema of cellular innate immunity processes associated with genes rapidly evolving in bats. Proteins shown in color are the most significant innate immunity proteins highlighted in this study. Pathogen sensing pathways involving TLRs and cGAS-STING can induce expression of type I IFNs including IFN-α and IFN-ω. IFN-α genes were lost in the bat ancestor, potentially giving a greater role to IFN-ω. The type I IFNs trigger the induction of ISGs via pathways including STAT, which interacts with the positively selected PARP9. The ISGs include the positively selected IFIT2 gene and the immune-related (IR) IFITM genes duplicated in Yangochiroptera bats, with both IFIT2 and IR-IFITM genes playing prominent roles in antiviral defenses. The overall schema is based on reviews of type I IFNs, PARP9, IFIT, and IFITM proteins (Diamond and Farzan 2013; Zhu and Zheng 2021) and studies on IFITM interactions with RNA viruses (Brass et al. 2009; Foster et al. 2016).
DNA Repair Genes and Tumor Suppressors Are Positively Selected in Bats
Enhanced DNA repair has been proposed as a mechanism for longevity and cancer resistance in various mammals including bats (Seim et al. 2013; Tollis et al. 2017). We identified six DNA repair-related PSGs and 46 PSGs that were “cancer-related” (supplementary table S10, Supplementary Material online), meaning they were included in either the Tumor Suppressor Database (Zhao et al. 2016) or the Catalogue Of Somatic Mutations In Cancer (Forbes et al. 2011). Notably, cancer-related genes were enriched more than 2-fold among PSGs on the bat ancestral branch relative to a set of mammalian branches (Fisher's exact test: P = 9e−3, odds ratio = 2.2). Among DNA repair genes, we detected evidence of positive selection in the tumor suppressor-encoding PALB2 and in four DNA polymerase-encoding genes (POLA1, POLD1, POLK, and POLM). PALB2 is a crucial component of the BRCA complex and is required for homologous recombination repair (Sy et al. 2009; Deveryshetty et al. 2019; Belotserkovskaya et al. 2020). In bats, it shows three sites under selection as well as seven bat-specific coding indels, including a 21-nucleotide deletion in a RAD51/BRCA1-interacting region (Uniprot annotation: Q86YC2). Despite previous evidence in a long-lived bat (Zhang et al. 2013), we did not find a signal for selection in BRCA2, although it does contain 14 bat-specific indels. Similarly, we did not identify the DNA repair genes RAD50 and KU80 (Zhang et al. 2013) or the key tumor suppressor gene TP53 as PSGs, but we did find four sites (codons 35, 38, 54, 97) in TP53 that are potentially selected in bats as well as a previously described bat-specific indel in the nuclear localization signal domain (Zhang et al. 2013) (supplementary fig. S6, Supplementary Material online).
Although TP53 did not appear among our PSGs, we did identify genes encoding two other tumor suppressors that interact with it (table 1): BCL-2 interacting killer (BIK) and large tumor suppressor kinase (LATS2). Both genes showed highly significant signals of selection in bats in our data set but have not been identified in tests of other mammalian branches (Nielsen et al. 2005; Kosiol et al. 2008) or in earlier studies in bats (Hawkins et al. 2019; Jebb et al. 2020). LATS2 is a kinase that modulates the functions of tumor suppressors such as TP53 as well as canonical growth-related Hippo signaling effectors YAP/TAZ (Furth and Aylon 2017). We found that LATS2 is predominantly under negative selection (bat ancestor ω = 0.23, outgroup mammals ω = 0.11) but nevertheless contains 13 nonsynonymous substitutions in bats as well as seven bat-specific microindels in its coding region (supplementary fig. S7 and supplementary Data S2, Supplementary Material online). Four of the substitutions fall within known functional domains of the protein, with a codon 134 glutamic-acid-to-aspartic-acid substitution in the LATS conserved domain 1 (LCD1) predicted to have an effect on the protein based on SNPeffect (Reumers et al. 2005). Previous experimental work in mice has shown that the LCD1 domain is critical for tumor suppressor activity of LATS2 (Yu et al. 2015). The second tumor suppressor, BIK, belongs to the proapoptotic BH3-only family of proteins that are upregulated in response to various stress signals and act as antagonists of prosurvival proteins (Huang and Strasser 2000; Zou et al. 2002). BIK is regulated by TP53 (Hur et al. 2006; López et al. 2017) and contributes to the apoptotic response induced by chemotherapy treatments (Zou et al. 2002; Real et al. 2006). The rapid molecular evolution of LATS2 and BIK suggests that these genes may contribute to bat molecular adaptations in tumor suppression.
Table 1.
Positively Selected Genes Involved in Cancer (P < 1.0e−3 with At Least One Site Selected in the Bat MRCA) that Showed the Strongest Statistical Significance. The P-Values Shown (aBSREL P-Value) are Derived from the Branch-Site Likelihood Ratio Test and Adjusted for Multiple Comparisons Across Branches but not Across Genes (see Text). Values of ω > 10 can Occur Due to Limited Signal and Imply Uncertain Estimates. Sites with Evidence of Positive Selection in the Bat MRCA were Identified Using Site-Wise MEME Analysis with a Significance Threshold of P < 0.1
| Symbol | Name | aBSREL P-Value | Sites under Selection | Bat Branch ω | Outgroup Branch ω |
|---|---|---|---|---|---|
| CDH1 | Cadherin-1 | 2.0e−6 | 3 | 0.17 | 0.29 |
| CAT | Catalase | 7.0e−6 | 1 | 0.38 | >10 |
| BIK | BCL2 interacting killer | 3.7e−5 | 1 | 1.92 | 0.89 |
| PALB2 | Partner and localizer of BRCA2 | 1.8e−4 | 3 | 0.67 | 0.43 |
| LATS2 | Large tumor suppressor kinase 2 | 2.5e−4 | 10 | 0.23 | 0.11 |
| SLC39A4 | Solute carrier family 39 member 4 | 3.8e−4 | 3 | 0.55 | 0.42 |
| SPARCL1 | SPARC like 1 | 4.0e−4 | 4 | 0.88 | 0.95 |
| PLA2G7 | Phospholipase A2 group VII | 8.5e−4 | 5 | >10 | 1.70 |
| GALR1 | Galanin receptor 1 | 8.7e−4 | 1 | 0.10 | 0.08 |
| CD79A | CD79a molecule | 9.0e−4 | 1 | >10 | 0.37 |
| MYO1A | Myosin IA | 9.8e−4 | 7 | 0.94 | 0.14 |
The two cancer-related genes displaying the strongest signals of positive selection, cadherin-1 (CDH1) and catalase (CAT) (table 1), also play roles in tumor development. CDH1 belongs to a family of transmembrane glycoproteins that mediate cell–cell adhesion and regulate cell growth, making them therapeutic targets for preventing tumor progression (Yu et al. 2019). CAT is an antioxidant enzyme that can protect from ROS-induced stress (Glorieux and Calderon 2017) and plays a role in TP53-mediated ROS regulation in response to DNA damage (Kang et al. 2013, 53). Although these functions make CAT and CDH1 intriguing examples of genes that have potentially evolved to enhance bat cancer resistance, previous studies have also found a signal for selection in CAT (Enard et al. 2016; Slodkowicz and Goldman 2020) and CDH1 (Enard et al. 2016; Vicens and Posada 2018; Slodkowicz and Goldman 2020) in other mammals. Experimental validation of the bat-specific mutations detected here will thus be important to demonstrate their potential adaptive value.
Discussion
High-quality and complete genome sequences are indispensable for revealing patterns of genomic variation within and between species. In this study, we used long-read sequencing to assemble the genomes of the bats A. jamaicensis and P. mesoamericanus and analyzed them together with 13 additional bat genomes to provide insights into the unique molecular evolution of bats. We found evidence of positive selection and structural variation in three key components of the bat innate immune system, including pathogen sensing, type I IFN cytokine signaling, and IFN-stimulated antiviral genes (see schema in fig. 5). Our results highlight how bat immune systems rely on both bat-wide and lineage-specific evolution in the immune gene repertoire, suggesting an interplay of diverse immune strategies with core molecular adaptations that occurred in the bat ancestor such as the loss of the PYHIN inflammatory gene family. In particular, the loss of IFN-α genes and the potentially increased reliance of bats on IFN-ω may play a role in their tolerance of viral infections. The expansion of antiviral IR-IFITM genes and PRDM9 as well as APOBEC3 (Jebb et al. 2020) and MHC-I (Moreno Santillán et al. 2021) class genes in major bat clades may have further contributed to lineage-specific evolution. Our findings underline the rapid evolution of the bat innate immune system, which has also been shown to have undergone additional gene losses in pathways related to defensins, natural killer signaling, and IFNs (Ahn et al. 2016; Jebb et al. 2020; Moreno Santillán et al. 2021). Thus, a combination of expansions of antiviral genes and losses of proinflammatory genes may contribute to the dampened inflammatory response and viral tolerance in bats. We also found evidence of positive selection in the bat MRCA in 46 cancer-related genes, suggesting a possible link to the unusually low incidence of cancer in bats.
Perhaps our most striking finding, building on earlier comparative genomic studies of the complex type I IFN locus (Zhou et al. 2016; Pavlovich et al. 2018), is that nine type I IFNs were lost in the bat MRCA. Notably, we found that bats have lost most or—in the case of Pipistrellus kuhlii, Myotis myotis, and P. mesoamericanus—all of their IFN-α genes, making their type I IFN locus particularly distinct among mammals. The overall contraction of the type I IFN locus in bats has been hypothesized to allow a smaller number of constitutively expressed IFN-α genes to perform the functions of the 13 IFN-α genes in humans (Zhou et al. 2016). The constitutive expression and rapid evolution of bat IFNs may also partly result from dampened inflammation caused by evolutionary changes to inflammosome genes such as AIM2, caspase-1 and IL-1β (Goh et al. 2020), because inflammosomes may negatively regulate type I IFN sensors (Yu et al. 2018). Indeed, the regulation of type I IFNs in bats also appears to have evolved adaptively, as positive selection of the key transcription factor IRF3 in bats was previously shown to enhance antiviral responses via type I IFN activation (Banerjee et al. 2020). However, our results suggest that constitutive expression of IFN-α is not common to all bats, in line with expression analyses in R. aegyptiacus (Pavlovich et al. 2018). We hypothesize that by relying on the potentially more potent IFN-ω rather than IFN-α, bats may further enhance their antiviral responses. Although further work will be needed to demonstrate a functional shift to IFN-ω, the lack of any IFN-α genes in three bat species strongly suggests a shift has occurred at least in these cases. It is possible that an enhanced antiviral response owing to IFN-ω helps to compensate for an overall dampened inflammatory response in bats. If these properties of IFN-ω can be established, they may open the door to new therapeutic uses of IFN-ω (Zhao et al. 2009; Li et al. 2017).
In addition, our findings suggest that, compared with other parts of the mammalian phylogeny, the lineage leading to bats is enriched for positively selected cancer-related genes. Rapid evolution of DNA repair genes as well as tumor suppressors has been proposed as a mechanism for cancer resistance in other long-lived mammals such as whales (Tejada-Martinez et al. 2021). Intriguingly, it was previously hypothesized that selection of cancer resistance components such DNA repair genes in bats resulted from a need to reduce the negative effects of ROS generated as a consequence of flight (Zhang et al. 2013). This suggestion appears consistent with our findings of positively selected ROS regulators and DNA repair genes. The evolution of cancer resistance in bats may also be associated with adaptations in the bat immune system. There is substantial overlap of genes related to cancer and the immune system (de Fonseca et al. 2010), with immune-related genes being known to play a role in cancer surveillance (Ostrand-Rosenberg 2008) and tumor suppression (Li et al. 2011; Siegrist et al. 2011; Alteber et al. 2018). Specific examples include immune-related genes that are rapidly evolving in bats. For example, ROS plays an important role in NLRP3 inflammasome activation (Moossavi et al. 2018; Yang et al. 2019), IFIT2 plays a role in apoptosis and cancer progression (Pidugu et al. 2019), and IFNs contribute to antitumor activity (Aricò et al. 2019; Lu et al. 2019). Positive selection in such genes may be driven by fitness trade-offs between roles in immunity and cancer (de Fonseca et al. 2010). For example, inhibition of inflammation can promote longevity (Youm et al. 2013; Marín-Aguilar et al. 2020) and suppress tumor growth (Lee et al. 2019; Tengesdal et al. 2021). Comparative analyses of gene expression across mammals and experimental validation may help resolve the different roles of ROS regulation, DNA repair, inflammation, and immunity in cancer resistance. We anticipate that our evolutionary findings and the novel genomic resources we have made available (including a genome browser) will encourage and facilitate further genomic research in bats, particularly as models that can lead to new strategies for addressing major challenges to human health such as infectious diseases and cancer.
Materials and Methods
Sample Background and Acquisition
Fresh liver samples from a single individual of A. jamaicensis (AMNH.Mammals.279493, male) and one P. mesoamericanus (AMNH.Mammals.279536, male) were collected by N.B.S. in April 2017 at the Lamanai Archaeological Reserve in Orange Walk District, Belize (17.75117°N, 88.65446°W). Sampling followed best practices for humane capture, handling, and euthanasia of live mammals outlined by the American Society of Mammalogists (Sikes 2016). All work was conducted with permission of the Belize Forest Department under permit number WL/2/1/17 (19) with Institutional Animal Care and Use Committee (IACUC) approval from the American Museum of Natural History (AMNHIACUC-20170403) and University of Georgia (A2014 04-016-Y3-A5). Bats were captured in ground-level mist nets and placed in individual cloth bags for transport to the Lamanai Field Research Center. After identification, the bats were euthanized using isoflurane, and the liver was removed immediately after death. Samples were placed in multiple individual 2 ml cryotubes and flash-frozen by placement in a liquid nitrogen dry shipper. The cold chain was maintained through shipment to the AMNH, storage in the AMNH Ambrose Monell Cryo Collection, and subsequent sample processing and transfers.
Genome Sequencing and Assembly
Approximately 40 mg of liver tissue from each bat was received at Cold Spring Harbor Laboratory (CSHL) and stored at −8 °C. The liver samples were crushed with a micro pestle and mixed with 10 ml of TLB buffer and 50 μl of RNase A immediately before use. After 1-h incubation at 37°C, 50 μl of proteinase K was added, followed by incubation at 5 °C for 3 h with hourly inversion mixing. After addition of 10 ml of a phenol chloroform/isoamyl alcohol mixture, each sample was rocked for 10 min and then centrifuged at 4500 RPM for 10 min. The top aqueous layer was retained and an equal volume of chloroform/isoamyl alcohol was added, and rocking and centrifugation were repeated as above. The top layer was transferred to a fresh tube with 4 ml of 5 M ammonium acetate. Following the addition of 30 ml of ice cold 100% ethanol, the sample was rocked for 10 min. The visible DNA was then extracted with a glass pipet and placed in a 1.5 ml tube. The sample was washed once with 100% ethanol and centrifuged for 5 min at 10,000 RPM. The ethanol was removed, and any remaining ethanol was evaporated on a 37 °C heat block for 10 min. The final DNA was resuspended in 10 mM Tris HCl pH 8.5 and stored overnight at 4 °C.
DNA was then sheared to ∼50–75 kb using a Diagnode Megarupter following manufacturer's recommendations. DNA was further enriched for long fragments via the Circulomics small read eliminator XL kit, which iteratively degrades short fragments. DNA was prepared for Nanopore sequencing using the ONT 1D sequencing by ligation kit (SQK-LSK109). Briefly, 1–1.5 μg of fragmented DNA was repaired with the NEB FFPE repair kit, followed by end repair and A-tailing with the NEB Ultra II end-prep kit. After an Ampure clean-up step, prepared fragments were ligated to ONT specific adapters via the NEB blunt/TA master mix kit. The library underwent a final clean-up and was loaded onto a PromethION PRO0002 flowcell per manufacturer's instructions. The flowcells were sequenced with standard parameters for 3 days. Basecalling was performed in real time with Guppy 3.2 and reads were later re-basecalled with Guppy 4 to improve quality. Nanopore reads were filtered for minimum length of 10 kb and minimum 85% accuracy using filtLong 0.2.0 (http://github.com/rrwick/Filtlong). The resulting coverage was 96.11× for A. jamaicensis (read N50 of 25.47 kb and maximum read length of 815.21 kb) and 99.19× for P. mesoamericanus (read N50 of 26.01 kb and maximum read length of 1.68 Mb).
Illumina short read libraries were prepared from the same tissue as above with the Illumina TruSeq DNA kit, targeting a 550 bp insert size with polymerase chain reaction enrichment. Libraries were sequenced at the New York Genome Center, on a NovaSeq S4 flowcell in a paired-end 150 bp format to ∼30× genome coverage. Short reads were used only for polishing, not assembly.
Reads were assembled using flye 2.8.3 (Kolmogorov et al. 2019) with “–nano-raw –no-alt-contigs” flags, after evaluation of additional assemblies generated with wtbg2 2.5 (Ruan and Li 2020), NextDenovo 2.2 (Hu et al. 2023), and Shasta 0.7.0 (Shafin et al. 2020) (supplementary table S12, Supplementary Material online). Assembly was followed by one round of long-read polishing using minimap 2.17 (Li 2018) with default parameters for alignment and PEPPER 0.1 (Shafin et al. 2021) with the “PromethION_r941_guppy305_HAC_human.pkl” model for polishing. Next, bwa-mem 0.7.17 (Li 2013) was used to align the Illumina short-read data to the long-read polished assembly with default parameters, and one round of short-read polishing was carried out with POLCA from (Zimin and Salzberg 2020) from MaSuRCA 3.4.0 using default parameters. To compare assembly contiguity, error rates, and completeness, assemblies were then assessed using Merqury 1.0 (Rhie et al. 2020), as well as with BUSCO 4.0.5 (Simão et al. 2015) for mammals (odb9). Additionally, we used python scripts to compute the cumulative sum of contigs length (N [X] length) versus the cumulative sum of N (X)% of the total genome. Further assembly statistics were calculated using BBTools 38.86 (http://sourceforge.net/projects/bbmap/). Duplicated haplotypes in the assemblies were purged using purge_dups 1.2.3 (Guan et al. 2020).
The assemblies were aligned to human, mouse, and pig assemblies as well as to four bat assemblies (M. myotis, Phyllostomus discolor, Desmodus rotundus, and Rhinolophus ferrumequinum) using Cactus 1.0.0 (Armstrong et al. 2020). Bat-specific small indels (<1 kb) were called relative to the human reference from the multiple alignment using a custom python script. Indels were called in alignment blocks with at least ten bases and seven species aligned including at least two nonbat mammals. Only fixed indels present in all bats or all bats but one were retained. Bat-specific insertions and deletions relative to the human reference were distinguished based on the sequence of the pig outgroup.
Gene Annotation
Public RNA-seq data from SRA were downloaded for A. jamaicensis and Pteronotus parnelli (supplementary table S13, Supplementary Material online), and proteins for human (GCF_000001405.39), mouse (GCF_000001635.26), and seven bat species (Myotis brandtii, GCF_000412655.1; Myotis davidii, GCF_000327345.1; M. lucifugus, GCF_000147115.1; P. discolor, GCF_004126475.1; P. alecto, GCF_000325575.1; Rhinolophus ferumequinum, GCF_004115265.1; and R. aegyptiacus, GCF_001466805.2) were downloaded from RefSeq. RNA-seq reads were aligned to the new reference genomes using HISAT 2.2.0 (Kim et al. 2019) with the parameters “–no-mixed –no-discordant –downstream-transcriptome-assembly”. Transcripts were assembled using StringTie 2.1.1 (Kovaka et al. 2019). To reduce potential loss of transcripts due to limitations of short-read alignment, transcriptomes were also de novo assembled with Trinity 2.9.1 (Grabherr et al. 2011) using default parameters. PASA 2.4.1 (Haas et al. 2003) was used to generate the final set of transcripts based on alignment with GMAP (Wu and Watanabe 2005) and BLAT (Kent 2002) using minimum thresholds of 90% of transcript length aligned at 90% identity. GeMoMa was used to project gene annotations from six bat assemblies (Jebb et al. 2020) to the genomes of A. jamaicensis and P. mesoamericanus with default parameters. Transdecoder with default parameters was used to predict coding sequences within the assembled transcripts (Haas et al. 2013). We used the transcripts and proteins sequences as evidence for the MAKER3 annotation pipeline (Cantarel et al. 2008) with the ab initio gene predictors SNAP 2006.07.28 (Korf 2004) and Augustus 3.3.3 (Stanke et al. 2006). GlimmerHMM (Majoros et al. 2004) and GeneMark 4.68 (Lomsadze et al. 2005) were used to generate further ab initio gene predictions. Final annotations were generated with EVidenceModeler 1.1.1 (Haas et al. 2008), weighting ab initio and MAKER predictions as 1, protein alignment evidence as 2, transcript evidence as 5, and PASA evidence as 10. To reduce the number of potentially missing genes caused by lack of protein and RNA-seq evidence, we used intact TOGA 1.0.0 (Kirilenko et al. 2023) gene projections from the recently assembled A. jamaicensis (GCF_014825515.1) to add gene annotations that did not intersect the Evidence Modeler annotations. The completeness of the final predicted protein set was assessed using BUSCO.
Repeat Analysis
Repeat masking was carried out using an iterative masking and de novo repeat detection approach. After masking repeats with RepeatMasker 4.0.9 (Tarailo-Graovac and Chen 2009) and the combined RepBase-20181026 and Dfam-3.1 repeat databases, novel repeats were detected in the masked genomes with RepeatModeler 2.0.1 using default parameters. The consensus de novo repeats longer than 100 bp were then concatenated with a vertebrate repeat library including novel bat repeats (Jebb et al. 2020) and clustered with CD-HIT 4.81 (Fu et al. 2012). All clustered novel sequences with >80% sequence similarity across >80% of the length of the clustered sequences were excluded (Wicker et al. 2007). Novel repeats were then aligned to the nt database using BLAST+ 2.7.1, and all repeats matching annotated transcripts were removed. A further BLAST analysis was used to exclude repeats with fewer than 10 alignments to the reference genome from which they were derived. Transposable elements were classified and assigned families using TEclass (Abrusán et al. 2009) and DeepTE (Yan et al. 2020). The consensus de novo repeats were then concatenated with the vertebrate repeat library, and a final masking of the genome was carried out with RepeatMasker using the “sensitive” setting. The recently diverged repeat landscape was analyzed using the RepeatMasker script calcDivergenceFromAlign.pl with correction of substitution rates based on the Kimura 2-Parameter model. Transposons were considered recently diverged at 7% divergence from the consensus (Jebb et al. 2020), which approximates an insertion <30 Ma, assuming a mammalian substitution rate of 2.2 × 10−9 (Kumar and Subramanian 2002). To allow comparison of the repeat landscape within Noctilionoidea bat clade, the same repeat masking approach was carried out for several closely related bats (D. rotundus, P. discolor, Sturnira hondurensis, and M. myotis).
Mining of Endogenous Viral Elements
To scan for endogenous viral elements that might provide evidence of past infections, we aligned the complete RVDB-prot viral protein database (Bigot et al. 2019) to our genomes using BLAST with an e-value threshold of 1e−5. Alignments intersecting coding sequence and those shorter than 100aa were excluded. Nonretroviral matches were aligned to the NR protein database using blastx with an e-value threshold of 1e−5 and TaxonKit 0.6.0 (Shen and Xiong 2019) was used to retain sequences with a best match to a viral lineage.
Identification of Orthologous Gene Groups
We analyzed bat genes for lineage-specific signals of positive selection and gene duplications based on clustered ortholog groups. Coding sequences for the seven bats used for annotation and six outgroup mammals (human, mouse, dog, pig, and horse) were downloaded from RefSeq. Annotated open reading frames that did not show a nucleotide number that was a multiple of three or that contained internal stop codons were discarded. The longest isoform for each gene was retained and translated to an amino acid sequence using Biopython (Cock et al. 2009). All proteins were clustered with the proteins of our bats using OrthoFinder 2.3.11 (Emms and Kelly 2019). A set of single copy orthologs was extracted from the OrthoFinder results, retaining orthologs with at least 12 bat species and three mammalian outgroups. Genes were then aligned using PRANK v.170427 (Löytynoja 2014) with the species tree provided to guide the alignment. Cancer-associated ortholog clusters were identified based on the databases TSG (Zhao et al. 2016) and COSMIC (Forbes et al. 2011).
Gene Family Expansion Analysis
Alignments of 3,632 single copy genes that were present in all species were concatenated into a single alignment, which was divided into three partitions corresponding to codon positions. A maximum-likelihood phylogeny was inferred using RAxML 8.2.12 (Stamatakis 2014) rapid bootstrapping under the GTR + G + I model with 100 bootstraps. A dated phylogeny was then generated using MCMCtree in PAML 4.9 (Zhang et al. 2005). Node ages were calibrated based on TimeTree (Kumar et al. 2017) ages for Euarchontoglires, bats, Yangochiroptera, and Yinpterochiroptera. Convergence was assessed based on analysis of two replicate runs with tracer (Rambaut et al. 2018). The dated phylogeny was used to calculate gene family expansions and contractions with CAFE 5 (De Bie et al. 2006) based on a P-value threshold of 0.05. CAFE was executed using the base model with an error model (1.7% of gene families were estimated to have an error in gene size) and three λ values (gene birth–death rates) for the lineages Yinpterochiroptera, Yangochiroptera, and nonbat mammals. Assessment using likelihood ratio tests showed that two λ values (one for bats and one for nonbats) were a significantly better fit than one (P < 0.01) and three λ values were a significantly better fit than two (P < 0.01). Orthofinder orthogroups were collapsed into 7,405 PANTHER gene families based on BiomaRt 2.46.3 (Durinck et al. 2005) data. For each of 19,935 orthogroups, a representative gene was selected to obtain a PANTHER assignment. For 2,466 orthogroups, where no human or mouse gene was represented, we selected a gene for sequence-based annotation using interproscan 5.50 (Blum et al. 2021) and eggNOG 2.1.3 (Cantalapiedra et al. 2021). As CAFE assumes all genes were present in the common ancestor of all analyzed species, orthogroups represented in less than two bats and two outgroup mammals were excluded. Final copy numbers and genomic coordinates for IFITM (supplementary table S14, Supplementary Material online) and type I IFN (supplementary table S15, Supplementary Material online) genes were visualized using genoPlotR 0.8.11 (Guy et al. 2010). For the analysis of the PRDM9 expansion in phyllostomids, recently generated short-read assemblies of S. hondurensis (GCA_014824575.2) and A. jamaicensis (GCA_014825515.1) were included in the phylogenetic analysis (supplementary fig. S5, Supplementary Material online) but not the CAFE analysis. Sequences and RAxML phylogenies are provided for IFITM, IFN, and PRDM9 orthogroups (supplementary Data S3, Supplementary Material online). Silhouettes of species for our figures were obtained from PhyloPic (https://www.phylopic.org/).
Positive Selection Analysis
We aimed to determine whether each gene was positively selected in one or more of three groups: the bat MRCA, P. mesoamericanus, and A. jamaicensis. We detected positive selection using the adaptive branch-site random effects model (aBSREL) method implemented in HyPhy 2.5.12 (Kosakovsky Pond et al. 2019). A likelihood ratio test was used to determine whether a lineage-specific group of codons in the alignment is experiencing significant positive selection. Unlike the branch site tests implemented in PAML (Zhang et al. 2005), aBSREL allows rate variation among the background branches, which can reduce false positive errors (Smith et al. 2015). In addition, we also used PAML to apply Test 2, a branch site test of positive selection. This test compares the alternative model where some branches are under positive selection and thus exhibit sites with ω > 1 with the corresponding null model where ω is fixed as 1. We computed P-values according to a χ2 distribution with one degree of freedom. Although accounting for nonadaptive processes when inferring selection in population-level analyses can be critical (Johri et al. 2022, 2023), branch site tests are well-powered to discriminate positive selection from negative or relaxed selection with low false positive rates in phylogenetic-scale analyses (Anisimova et al. 2002; Zhang et al. 2005; Yang and dos Reis 2011; Smith et al. 2015).
The P-values calculated by PAML and HyPhy were adjusted for multiple testing of three branches per gene using the Benjamini–Hochberg method for controlling false discovery rate (FDR) implemented in base R. We also corrected for multiple testing of all genes using FDR (supplementary table S10, Supplementary Material online). We used the maximum-likelihood species tree inferred using 3,048 orthologs (see Materials and Methods section “Gene Family Expansion Analysis”) to provide the topology for the positive selection scans. In this tree, bats are the sister group to Fereungulata (Cetartiodactyla, Perissodactyla, Carnivora, and Pholidota) (Jebb et al. 2020). Relationships within Fereungulata remain challenging to resolve (Doronina et al. 2017; Jebb et al. 2020), and here, we followed the TimeTree topology that places Perissodactyla + Carnivora as sister to Cetartiodactyla. For comparison with a previous scan for positive selection on all branches of a mammalian phylogeny (including human, chimpanzee, macaque, mouse, rat, and dog), we used the set of 8,594 genes that were tested for positive selection using PAML in both this study and in (Kosiol et al. 2008). MEME (Murrell et al. 2012) analysis was used to identify sites potentially under selection in the bat MRCA with a significance threshold of P < 0.1. Alignment of genes positively selected in the bat MRCA is provided in supplementary Data S4, Supplementary Material online.
Gene Ontology Enrichment
GO, Reactome, and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations for genes were obtained via BiomaRt. A total of 18,452 orthogroups were annotated with at least one feature and 17,840 were assigned GO features. We carried out enrichment analysis on groups of genes that were positively selected or that showed gene family expansions. Enrichment analysis was performed using topGO 2.42.0, with the elim algorithm and Fisher's exact test (P < 0.01). All genes tested for selection were used as the background. The elim algorithm is a conservative approach that processes the GO graph from the bottom up, to eliminate higher order terms that would otherwise appear enriched due to correlation with more specific terms. GO terms with fewer than ten genes annotated were excluded.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
This research was supported, in part, by US National Institutes of Health (NIH) grants P30-CA045508 (to D. Tuveson and colleagues) and R35-GM127070 (to Ad.S.) and by the Simons Center for Quantitative Biology. Computational work was performed with assistance from the NIH Grant S10OD028632-01. We would further like to acknowledge funding support from the CSHL/Northwell Health Affiliation for purchase of the ONT PromethION sequencer used in this study. We thank the CSHL Cancer Center and Director David Tuveson for support as well as use of the Sequencing Technologies and Analysis Shared Resource. The genome browser was set up with help from Ritika Ramani. W.R.M. is the Davis Family Professor of Human Genetics at CSHL. D.J.B. was supported by the National Science Foundation (BII 2213854). We also thank Semir Beyaz for helpful comments on the manuscript. Finally, we thank Brock Fenton, Neil Duncan, the staff at the Lamanai Field Research Center, and many other colleagues who assisted with the fieldwork necessary for this study. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or other funding sources.
Contributor Information
Armin Scheben, Simons Center for Quantitative Biology, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA.
Olivia Mendivil Ramos, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA.
Melissa Kramer, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA.
Sara Goodwin, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA.
Sara Oppenheim, American Museum of Natural History, Institute for Comparative Genomics, New York, New York, USA.
Daniel J Becker, School of Biological Sciences, University of Oklahoma, Norman, Oklahoma, USA.
Michael C Schatz, Simons Center for Quantitative Biology, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA; Departments of Computer Science and Biology, Johns Hopkins University, Baltimore, Maryland, USA.
Nancy B Simmons, Department of Mammalogy, Division of Vertebrate Zoology, American Museum of Natural History, New York, New York, USA.
Adam Siepel, Simons Center for Quantitative Biology, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA.
W Richard McCombie, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA.
Author Contributions
Ad.S. and W.R.M. conceived the study. N.B.S. and S.O. provided DNA samples. S.G. provided sequencing resources. Ar.S. carried out the analysis, with assistance from O.M.R. and M.K. D.J.B. helped interpret the endogenous virus data. M.C.S advised on the methodology. Ar.S. drafted the manuscript, and all authors reviewed and edited the final manuscript.
Data Availability
The raw sequencing data and final genome assemblies generated in this study have been submitted to the NCBI BioProject database under accession number PRJNA751559. Gene annotations can be downloaded or viewed as UCSC genome browser tracks from our laboratory website (http://compgen.cshl.edu/bat/). The data underlying this article are available in figshare at https://doi.org/10.6084/m9.figshare.15223014.v3 (Scheben 2021). Code is available on GitHub at https://github.com/CshlSiepelLab/bat_genome_analysis.
Literature cited
- Abrusán G, Grundmann N, DeMester L, Makalowski W. 2009. TEclass—a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 25:1329–1330. [DOI] [PubMed] [Google Scholar]
- Ahn M, et al. 2019. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat Microbiol. 4:789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn M, Cui J, Irving AT, Wang L-F. 2016. Unique loss of the PYHIN gene family in bats amongst mammals: implications for inflammasome sensing. Sci Rep. 6:21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteber Z, et al. 2018. The anti-inflammatory IFITM genes ameliorate colitis and partially protect from tumorigenesis by changing immunity and microbiota. Immunol Cell Biol. 96:284–297. [DOI] [PubMed] [Google Scholar]
- Amman BR, et al. 2012. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. 8:e1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. 2020. The proximal origin of SARS-CoV-2. Nat Med. 26:450–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z. 2002. Accuracy and power of Bayes prediction of amino acid sites under positive selection. Mol Biol Evol. 19:950–958. [DOI] [PubMed] [Google Scholar]
- Aricò E, Castiello L, Capone I, Gabriele L, Belardelli F. 2019. Type I interferons and cancer: an evolving story demanding novel clinical applications. Cancers (Basel). 11:1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J, et al. 2020. Progressive cactus is a multiple-genome aligner for the thousand-genome era. Nature 587:246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CC, Zhong G, Huang I-C, Farzan M. 2014. IFITM-family proteins: the cell's first line of antiviral defense. Annu Rev Virol. 1:261–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker Z, et al. 2017. Repeated losses of PRDM9-directed recombination despite the conservation of PRDM9 across vertebrates. Elife 6:e24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, et al. 2020. Positive selection of a serine residue in bat IRF3 confers enhanced antiviral protection. iScience 23:100958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Rapin N, Bollinger T, Misra V. 2017. Lack of inflammatory gene expression in bats: a unique role for a transcription repressor. Sci Rep. 7:2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker DJ, et al. 2022. Serum proteomics identifies immune pathways and candidate biomarkers of coronavirus infection in wild vampire bats. Front Virol. 2. doi: 10.3389/fviro.2022.862961 [DOI] [Google Scholar]
- Belotserkovskaya R, et al. 2020. PALB2 chromatin recruitment restores homologous recombination in BRCA1-deficient cells depleted of 53BP1. Nat Commun. 11:819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfield CT, et al. 2019. Bat IFITM3 restriction depends on S-palmitoylation and a polymorphic site within the CD225 domain. Life Sci Alliance. 3:e201900542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot T, Temmam S, Pérot P, Eloit M. 2019. RVDB-prot, a reference viral protein database and its HMM profiles. F1000Res 8:530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M, et al. 2021. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49:D344–D354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer M, et al. 2022. Gene losses in the common vampire bat illuminate molecular adaptations to blood feeding. Sci Adv. 8:eabm6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, et al. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, west Nile virus, and dengue virus. Cell 139:1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Romo S, et al. 2014. Experimental inoculation of Artibeus jamaicensis bats with dengue virus serotypes 1 or 4 showed no evidence of sustained replication. Am J Trop Med Hyg. 91:1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. 2006. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 19:531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol. 38:5825–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, et al. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18:188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavassim MIA, et al. 2022. PRDM9 Losses in vertebrates are coupled to those of paralogs ZCWPW1 and ZCWPW2. Proc Natl Acad Sci U S A. 119:e2114401119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarino NM, et al. 2017. IFITM3 Requires an amphipathic helix for antiviral activity. EMBO Rep. 18:1740–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJA, et al. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca RR, Kosiol C, Vinař T, Siepel A, Nielsen R. 2010. Positive selection on apoptosis related genes. FEBS Lett. 584:469–476. [DOI] [PubMed] [Google Scholar]
- [dataset] Scheben A. 2021. Long-read sequencing reveals rapid evolution of immunity- and cancer-related genes in bats. figshare. Dataset. doi: 10.6084/m9.figshare.15223014.v2. [DOI]
- David Q, Schountz T, Schwemmle M, Ciminski K. 2022. Different but not unique: deciphering the immunity of the Jamaican fruit bat by studying its viriome. Viruses 14:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW. 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22:1269–1271. [DOI] [PubMed] [Google Scholar]
- Decout A, Katz JD, Venkatraman S, Ablasser A. 2021. The cGAS–STING pathway as a therapeutic target in inflammatory diseases. Nat Rev Immunol 21:548–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai TM, et al. 2014. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 10:e1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveryshetty J, et al. 2019. Novel RNA and DNA strand exchange activity of the PALB2 DNA binding domain and its critical role for DNA repair in cells. Elife 8:e44063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Weerd NA, Samarajiwa SA, Hertzog PJ. 2007. Type I interferon receptors: biochemistry and biological functions. J Biol Chem. 282:20053–20057. [DOI] [PubMed] [Google Scholar]
- Diamond MS, Farzan M. 2013. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 13:46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina L, et al. 2017. Speciation network in Laurasiatheria: retrophylogenomic signals. Genome Res. 27:997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, et al. 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356:92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, et al. 2005. Biomart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21:3439–3440. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. Orthofinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard D, Cai L, Gwennap C, Petrov DA. 2016. Viruses are a dominant driver of protein adaptation in mammals. Elife 5:e12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalera-Zamudio M, et al. 2015. The evolution of bat nucleic acid-sensing Toll-like receptors. Mol Ecol. 24:5899–5909. [DOI] [PubMed] [Google Scholar]
- Fensterl V, Sen GC. 2014. Interferon-induced IFIT proteins: their role in viral pathogenesis. J Virol. 89:2462–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, Kagan JC. 2020. Toll-like receptors and the control of immunity. Cell 180:1044–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming TH, Dávalos LM, Mello MAR. 2020. Phyllostomid bats: a unique mammalian radiation. Chicago, IL: University of Chicago Press. [Google Scholar]
- Forbes SA, et al. 2011. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 39:D945–D950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TL, et al. 2016. Resistance of transmitted founder HIV-1 to IFITM-mediated restriction. Cell Host Microbe. 20:429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J, et al. 2017. Evolution and antiviral specificities of interferon-induced Mx proteins of bats against ebola, influenza, and other RNA viruses. J Virol. 91:e00361-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth N, Aylon Y. 2017. The LATS1 and LATS2 tumor suppressors: beyond the Hippo pathway. Cell Death Differ. 24:1488–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlanda C, Dinarello CA, Mantovani A. 2013. The interleukin-1 family: back to the future. Immunity 39:1003–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorieux C, Calderon PB. 2017. Catalase, a remarkable enzyme: targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biol Chem. 398:1095–1108. [DOI] [PubMed] [Google Scholar]
- Goh G, et al. 2020. Complementary regulation of caspase-1 and IL-1β reveals additional mechanisms of dampened inflammation in bats. Proc Natl Acad Sci U S A. 117:28939–28949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez H, Hagerling C, Werb Z. 2018. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 32:1267–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Kennedy BK. 2020. The world goes bats: living longer and tolerating viruses. Cell Metab. 32:31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouravani M, et al. 2020. The NLRP3 inflammasome: a therapeutic target for inflammation-associated cancers. Expert Rev Clin Immunol. 16:175–187. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, et al. 2020. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics 36:2896–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L, Roat Kultima J, Andersson SG. 2010. Genoplotr: comparative gene and genome visualization in R. Bioinformatics 26:2334–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31:5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halo JV, et al. 2021. Long-read assembly of a Great Dane genome highlights the contribution of GC-rich sequence and mobile elements to canine genomes. Proc Natl Acad Sci. 118:e2016274118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JA, et al. 2019. A metaanalysis of bat phylogenetics and positive selection based on genomes and transcriptomes from 18 species. Proc Natl Acad Sci. 116:11351–11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward JA, et al. 2018. Differential evolution of antiretroviral restriction factors in pteropid bats as revealed by APOBEC3 gene complexity. Mol Biol Evol. 35:1626–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward JA et al. 2020. Unique evolution of antiviral tetherin in bats. bioRxiv. 2020.04.08.031203. doi: 10.1101/2020.04.08.031203.
- He K, Minias P, Dunn PO. 2021. Long-read genome assemblies reveal extraordinary variation in the number and structure of MHC loci in birds. Genome Biol Evol. 13:evaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy EJ, Parker AE, O’Neill LAJ. 2010. Targeting Toll-like receptors: emerging therapeutics? Nat Rev Drug Discov. 9:293–307. [DOI] [PubMed] [Google Scholar]
- Holmes EC, et al. 2021. The origins of SARS-CoV-2: a critical review. Cell 184:4848–4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J et al. 2023. An efficient error correction and accurate assembly tool for noisy long reads. 2023.03.09.531669. doi: 10.1101/2023.03.09.531669.
- Huang DC, Strasser A. 2000. BH3-Only proteins-essential initiators of apoptotic cell death. Cell 103:839–842. [DOI] [PubMed] [Google Scholar]
- Hur J, et al. 2006. Regulation of expression of BIK proapoptotic protein in human breast cancer cells: p53-dependent induction of BIK mRNA by fulvestrant and proteasomal degradation of BIK protein. Cancer Res. 66:10153–10161. [DOI] [PubMed] [Google Scholar]
- Irving AT, Ahn M, Goh G, Anderson DE, Wang L-F. 2021. Lessons from the host defences of bats, a unique viral reservoir. Nature 589:363–370. [DOI] [PubMed] [Google Scholar]
- Jaks E, Gavutis M, Uzé G, Martal J, Piehler J. 2007. Differential receptor subunit affinities of type I interferons govern differential signal activation. J Mol Biol. 366:525–539. [DOI] [PubMed] [Google Scholar]
- Jebb D, et al. 2020. Six reference-quality genomes reveal evolution of bat adaptations. Nature 583:578–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, et al. 2017. Selective evolution of Toll-like receptors 3, 7, 8, and 9 in bats. Immunogenetics 69:271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri P, et al. 2022. Recommendations for improving statistical inference in population genomics. PLOS Biol 20:e3001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri P, Pfeifer SP, Jensen JD. 2023. Developing an evolutionary baseline model for humans: jointly inferring purifying selection with population history. Mol. Biol. Evol 40:msad100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacprzyk J, et al. 2021. Evolution of mammalian longevity: age-related increase in autophagy in bats compared to other mammals. Aging (Albany NY). 13:7998–8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MY, et al. 2013. The critical role of catalase in prooxidant and antioxidant function of p53. Cell Death Differ. 20:117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB, et al. 2010. Chiropteran types I and II interferon genes inferred from genome sequencing traces by a statistical gene-family assembler. BMC Genomics. 11:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-M, Brinkmann MM, Paquet M-E, Ploegh HL. 2008. UNC93B1 Delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 452:234–238. [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilenko BM, et al. 2023. Integrating gene annotation with orthology inference at scale. Science 380:eabn3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37:540–546. [DOI] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics. 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, et al. 2019. Hyphy 2.5—a customizable platform for evolutionary hypothesis testing using phylogenies. Mol Biol Evol. 37:295–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosiol C, et al. 2008. Patterns of positive selection in six mammalian genomes. PLoS Genet. 4:e1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S, et al. 2019. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. 2017. Timetree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34:1812–1819. [DOI] [PubMed] [Google Scholar]
- Kumar S, Subramanian S. 2002. Mutation rates in mammalian genomes. Proc Natl Acad Sci. 99:803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PA, et al. 2007. Phylogenetics and phylogeography of the Artibeus jamaicensis complex based on cytochrome-b DNA sequences. J Mammal. 88:712–727. [Google Scholar]
- Lee BL, et al. 2013. UNC93B1 Mediates differential trafficking of endosomal TLRs. Elife 2:e00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HE, et al. 2019. Inhibition of NLRP3 inflammasome in tumor microenvironment leads to suppression of metastatic potential of cancer cells. Sci Rep. 9:12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, et al. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679. [DOI] [PubMed] [Google Scholar]
- Li D, et al. 2011. KLF4-mediated negative regulation of IFITM3 expression plays a critical role in colon cancer pathogenesis. Clin Cancer Res. 17:3558–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv. doi: 10.48550/arXiv.1303.3997.
- Li S, et al. 2017. Interferon-omega: current status in clinical applications. Int Immunopharmacol. 52:253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33:6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López I, et al. 2017. p53-mediated suppression of BiP triggers BIK-induced apoptosis during prolonged endoplasmic reticulum stress. Cell Death Differ. 24:1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A. 2014. Phylogeny-aware alignment with PRANK. Methods Mol Biol. 1079:155–170. [DOI] [PubMed] [Google Scholar]
- Lu C, et al. 2019. Type I interferon suppresses tumor growth through activating the STAT3-granzyme B pathway in tumor-infiltrating cytotoxic T lymphocytes. J Immunother Cancer. 7:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoros WH, Pertea M, Salzberg SL. 2004. Tigrscan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20:2878–2879. [DOI] [PubMed] [Google Scholar]
- Marín-Aguilar F, et al. 2020. NLRP3 Inflammasome suppression improves longevity and prevents cardiac aging in male mice. Aging Cell. 19:e13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. 2015. Type I interferons in infectious disease. Nat Rev Immunol. 15:87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moossavi M, Parsamanesh N, Bahrami A, Atkin SL, Sahebkar A. 2018. Role of the NLRP3 inflammasome in cancer. Mol Cancer. 17:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Santillán DD, et al. 2021. Large-scale genome sampling reveals unique immunity and metabolic adaptations in bats. Mol Ecol. 30:6449–6467. [DOI] [PubMed] [Google Scholar]
- Moss CF, Surlykke A. 2001. Auditory scene analysis by echolocation in bats. J Acoust Soc Am. 110:2207–2226. [DOI] [PubMed] [Google Scholar]
- Munster VJ, et al. 2016. Replication and shedding of MERS-CoV in Jamaican fruit bats (artibeus jamaicensis). Sci Rep. 6:21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, et al. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8:e1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, et al. 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLOS Biol. 3:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor EA, Cornwallis CK. 2022. Immunity and lifespan: answering long-standing questions with comparative genomics. Trends Genet. 38:650–661. [DOI] [PubMed] [Google Scholar]
- Olival KJ, et al. 2017. Host and viral traits predict zoonotic spillover from mammals. Nature 546:646–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Nascimento L, Massari P, Wetzler LM. 2012. The role of TLR2 in infection and immunity. Front Immunol. 3:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrand-Rosenberg S. 2008. Immune surveillance: a balance between protumor and antitumor immunity. Curr Opin Genet Dev. 18:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Petkov PM. 2018. PRDM9 and its role in genetic recombination. Trends Genet. 34:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan R, Liu H, Choubey D. 2013. Expression of murine Unc93b1 is up-regulated by interferon and estrogen signaling: implications for sex bias in the development of autoimmunity. Int Immunol. 25:521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovich SS, et al. 2018. The Egyptian rousette genome reveals unexpected features of bat antiviral immunity. Cell 173:1098–1110.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekar JE, et al. 2022. The molecular epidemiology of multiple zoonotic origins of SARS-CoV-2. Science 377:960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka K, et al. 2018. The chaperone UNC93B1 regulates Toll-like receptor stability independently of endosomal TLR transport. Immunity 48:911–922.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidugu VK, Pidugu HB, Wu M-M, Liu C-J, Lee T-C. 2019. Emerging functions of human IFIT proteins in cancer. Front Mol Biosci. 6:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter JHT, et al. 2021. Dietary diversification and specialization in Neotropical bats facilitated by early molecular evolution. Mol Biol Evol. 38:3864–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam JRC, et al. 2012. Agricultural intensification, priming for persistence and the emergence of Nipah virus: a lethal bat-borne zoonosis. J R Soc Interface. 9:89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst Biol. 67:901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real PJ, et al. 2006. Transcriptional activation of the proapoptotic BIK gene by E2F proteins in cancer cells. FEBS Lett. 580:5905–5909. [DOI] [PubMed] [Google Scholar]
- Reumers J, et al. 2005. SNPeffect: a database mapping molecular phenotypic effects of human non-synonymous coding SNPs. Nucleic Acids Res. 33:D527–D532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, et al. 2021. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM. 2020. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas D, Warsi OM, Dávalos LM. 2016. Bats (Chiroptera: Noctilionoidea) challenge a recent origin of extant Neotropical diversity. Syst Biol. 65:432–448. [DOI] [PubMed] [Google Scholar]
- Ruan J, Li H. 2020. Fast and accurate long-read assembly with wtdbg2. Nat Methods. 17:155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JJ, Roach DJ, Thomas JH, Shendure J. 2014. Primate evolution of the recombination regulator PRDM9. Nat Commun. 5:4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seim I, et al. 2013. Genome analysis reveals insights into physiology and longevity of the Brandt's Bat Myotis brandtii. Nat Commun. 4:2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafin K, et al. 2020. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat Biotechnol. 38:1044–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafin K, et al. 2021. Haplotype-aware variant calling with PEPPER-Margin-DeepVariant enables high accuracy in nanopore long-reads. Nat Methods. 18:1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Xiong J. 2019. TaxonKit: a cross-platform and efficient NCBI taxonomy toolkit. bioRxiv. 513523. doi: 10.1101/513523.
- Siegrist F, Ebeling M, Certa U. 2011. The small interferon-induced transmembrane genes and proteins. J Interferon Cytokine Res. 31:183–197. [DOI] [PubMed] [Google Scholar]
- Sikes RS. 2016. 2016 guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. J Mammal. 97:663–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Simmons NB, Geisler JH. 1998. Phylogenetic relationships of Icaronycteris, Archaeonycteris, Hassianycteris, and Palaeochiropteryx to extant bat lineages, with comments on the evolution of echolocation and foraging strategies in Microchiroptera. Bulletin of the AMNH ; no. 235. Relationships of Eocene bats. Available from: http://digitallibrary.amnh.org/handle/2246/1629 (cited 2020 Sep 7).
- Simmons NB, Seymour KL, Habersetzer J, Gunnell GF. 2008. Primitive early eocene bat from Wyoming and the evolution of flight and echolocation. Nature 451:818–821. [DOI] [PubMed] [Google Scholar]
- Slodkowicz G, Goldman N. 2020. Integrated structural and evolutionary analysis reveals common mechanisms underlying adaptive evolution in mammals. Proc Natl Acad Sci U S A. 117:5977–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, et al. 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol. 32:1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotero-Caio CG, et al. 2013. Chromosomal evolution among leaf-nosed nectarivorous bats—evidence from cross-species chromosome painting (Phyllostomidae. Chiroptera). BMC Evol Biol. 13:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, et al. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34:W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy SMH, Huen MSY, Chen J. 2009. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 106:7155–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinforma. Chapter 4:4.10.1–4.10.14. [DOI] [PubMed] [Google Scholar]
- Taylor DJ, Dittmar K, Ballinger MJ, Bruenn JA. 2011. Evolutionary maintenance of filovirus-like genes in bat genomes. BMC Evol Biol. 11:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling EC, et al. 2018. Bat biology, genomes, and the Bat1k project: to generate chromosome-level genomes for all living bat species. Annu Rev Anim Biosci. 6:23–46. [DOI] [PubMed] [Google Scholar]
- Tejada-Martinez D, de Magalhães JP, Opazo JC. 2021. Positive selection and gene duplications in tumour suppressor genes reveal clues about how cetaceans resist cancer. Proc Biol Sci. 288:20202592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tengesdal IW, et al. 2021. Targeting tumor-derived NLRP3 reduces melanoma progression by limiting MDSCs expansion. Proc Natl Acad Sci. 118:e2000915118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, et al. 2011. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell 146:621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Schiffman JD, Boddy AM. 2017. Evolution of cancer suppression as revealed by mammalian comparative genomics. Curr Opin Genet Dev. 42:40–47. [DOI] [PubMed] [Google Scholar]
- Tsan M-F, Gao B. 2009. Heat shock proteins and immune system. J Leukoc Biol. 85:905–910. [DOI] [PubMed] [Google Scholar]
- Velazquez-Salinas L, Verdugo-Rodriguez A, Rodriguez LL, Borca MV. 2019. The role of interleukin 6 during viral infections. Front Microbiol. 10:1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M. 2017. Interleukin 17 is a chief orchestrator of immunity. Nat Immunol. 18:612–621. [DOI] [PubMed] [Google Scholar]
- Vicens A, Posada D. 2018. Selective pressures on human cancer genes along the evolution of mammals. Genes (Basel). 9:582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollger MR, et al. 2019. Long-read sequence and assembly of segmental duplications. Nat Methods. 16:88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L-F, Gamage AM, Chan WOY, Hiller M, Teeling EC. 2021. Decoding bat immunity: the need for a coordinated research approach. Nat Rev Immunol. 21:269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L-F, Walker PJ, Poon LLM. 2011. Mass extinctions, biodiversity and mitochondrial function: are bats ‘special’ as reservoirs for emerging viruses? Curr Opin Virol. 1:649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 8:973–982. [DOI] [PubMed] [Google Scholar]
- Wilkinson GS, Adams DM. 2019. Recurrent evolution of extreme longevity in bats. Biol Lett. 15:20180860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21:1859–1875. [DOI] [PubMed] [Google Scholar]
- Xie J, et al. 2018. Dampened STING-dependent interferon activation in bats. Cell Host Microbe. 23:297–301.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Liu P-S, Percipalle P. 2019. Analysis of global transcriptome change in mouse embryonic fibroblasts after dsDNA and dsRNA viral mimic stimulation. Front Immunol. 10:836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, et al. 2021. Identification of poly(ADP-ribose) polymerase 9 (PARP9) as a noncanonical sensor for RNA virus in dendritic cells. Nat Commun. 12:2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Bombarely A, Li S. 2020. DeepTE: a computational method for de novo classification of transposons with convolutional neural network. Bioinformatics 36:4269–4275. [DOI] [PubMed] [Google Scholar]
- Yang Z, et al. 2012. Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Res. 22:1328–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, dos Reis M. 2011. Statistical properties of the branch-site test of positive selection. Mol Biol Evol. 28:1217–1228. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 19:908–917. [DOI] [PubMed] [Google Scholar]
- Yang Y, Wang H, Kouadir M, Song H, Shi F. 2019. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis. 10:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youm Y-H, et al. 2013. Canonical NLRP3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab. 18:519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, et al. 2018. Inflammasome activation negatively regulates MyD88-IRF7 type I IFN signaling and anti-malaria immunity. Nat Commun. 9:4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Bachman J, Lai Z-C. 2015. Mutation analysis of large tumor suppressor genes LATS1 and LATS2 supports a tumor suppressor role in human cancer. Protein Cell. 6:6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Yang L, Li T, Zhang Y. 2019. Cadherin signaling in cancer: its functions and role as a therapeutic target. Front Oncol. 9:989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepeda Mendoza ML, et al. 2018. Hologenomic adaptations underlying the evolution of sanguivory in the common vampire bat. Nat Ecol Evol. 2:659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, et al. 2013. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 339:456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. 2015. PARP9-DTX3L ubiquitin ligase targets host histone H2BJ and viral 3C protease to enhance interferon signaling and control viral infection. Nat Immunol. 16:1215–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22:2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhao X, et al. 2009. Characterization and virus-induced expression profiles of the porcine interferon-omega multigene family. J Interferon Cytokine Res. 29:687–693. [DOI] [PubMed] [Google Scholar]
- Zhao M, Kim P, Mitra R, Zhao J, Zhao Z. 2016. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 44:D1023–D1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, et al. 2021. TLR2 Senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat Immunol. 22:829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, et al. 2016. Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc Natl Acad Sci U S A. 113:2696–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Zheng C. 2021. When PARPs meet antiviral innate immunity. Trends Microbiol. 29:776–778. [DOI] [PubMed] [Google Scholar]
- Zimin AV, Salzberg SL. 2020. The genome polishing tool POLCA makes fast and accurate corrections in genome assemblies. PLoS Comput Biol. 16:e1007981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, et al. 2002. Systemic tumor suppression by the proapoptotic gene BIK. Cancer Res. 62:8–12. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data and final genome assemblies generated in this study have been submitted to the NCBI BioProject database under accession number PRJNA751559. Gene annotations can be downloaded or viewed as UCSC genome browser tracks from our laboratory website (http://compgen.cshl.edu/bat/). The data underlying this article are available in figshare at https://doi.org/10.6084/m9.figshare.15223014.v3 (Scheben 2021). Code is available on GitHub at https://github.com/CshlSiepelLab/bat_genome_analysis.