Abstract
G protein-coupled receptors (GPCRs) play pivotal roles in regulation of cardiac function and homeostasis. To function properly, every cell needs these receptors to be stimulated only when a specific extracellular stimulus is present, and to be silenced the moment that stimulus is removed. The regulator of G protein signaling (RGS) proteins are crucial for the latter to occur at the cell membrane, where the GPCR normally resides. Perturbations in both activation and termination of G protein signaling underlie numerous heart pathologies. Although more than 30 mammalian RGS proteins have been identified, each RGS protein seems to interact only with a specific set of G protein subunits and GPCR types/subtypes in any given tissue or cell type, and this applies to the myocardium as well. A large number of studies have provided substantial evidence for the roles various RGS proteins expressed in cardiomyocytes play in cardiac physiology and heart disease pathophysiology. This review summarizes the current understanding of the functional roles of cardiac RGS proteins and their implications for the treatment of specific heart diseases, such as heart failure and atrial fibrillation. We focus on cardiac RGS4 in particular, since this isoform appears to be selectively (among the RGS protein family) upregulated in human heart failure and is also the target of ongoing drug discovery efforts for the treatment of a variety of diseases.
Keywords: arrhythmias, atrial fibrillation, cardiac myocyte, cyclic AMP, G protein-coupled receptor, G proteins, heart failure, regulator of G protein signaling, signal transduction
Introduction
G protein-coupled receptors (GPCRs) are the single largest class of pharmaceutical targets, with over 35% of the currently FDA-approved drugs directly acting on these receptors. 1 GPCRs are crucial regulators of almost every cellular physiological process, from vision to cardiovascular function and blood pressure. 2 This is largely because they always reside at the plasma membrane, thereby mediating the signal from the vast majority of extracellular stimuli that cannot pass across the cell membrane (e.g. ionized or not lipophilic enough molecules). Therefore, GPCR abnormalities result oftentimes in various pathologies, depending on the physiology of the dysfunctional receptor. Dysfunction of cardiovascular GPCRs lead to cardiovascular diseases, such as heart failure (HF), cardiomyopathies, cardiac hypertrophy, hypertension, angina, and so on.3,4 All GPCRs share a common core motif of seven largely hydrophobic α helices, each spanning the entire plasma membrane [seven transmembrane (TM)-spanning or heptahelical receptors]. 5 The heptahelical motif is essential for interaction with G proteins only upon agonist binding on the extracellular side of the receptor.6 –10 The receptor-Gα subunit interaction activates, in turn, the heterotrimeric G protein, causing guanine nucleotide exchange and the separation of Gα from the Gβγ subunits.11 –14 However, regulation of the duration of a GPCR signal is of paramount importance for cellular homeostasis and the cell utilizes various ways to terminate the GPCR signal, starting with two major processes at the level of the cell membrane. One of them operates on the receptor itself and involves GPCR phosphorylation by GPCR-kinases (GRKs), followed by arrestin binding (homologous or agonist-dependent receptor desensitization).15,16 Phosphorylation by second messenger-dependent kinases, such as protein kinase A (PKA), is also supposed to terminate G protein signaling (the so-called ‘heterologous’ or agonist-independent receptor desensitization), but whether arrestin binding follows second messenger kinase-mediated phosphorylation is still a matter of intense debate. It is more likely that second messenger-dependent kinases switch the coupling of a particular receptor to a different G protein, such as the case of PKA-induced Gs to Gi coupling switch of the β2-adrenoceptor (reviewed in Ref. 16). The other process, perhaps even more important, operates on the active G protein. The main mechanism for G protein signaling termination is guanosine triphosphate (GTP) hydrolysis to guanosine diphosphate (GDP) by the intrinsic GTPase activity of the Gα subunit. 12 As soon as GTP is converted to GDP, GDP-bound Gα subunit regains its affinity for the Gβγ subunits (switch II region loses its contacts with the guanine nucleotide and binds Gβ again) and the G protein heterotrimer reassociates, no longer being able to transduce signals (i.e. neither Gα nor Gβγ can interact with effectors now). 12
Unlike the monomeric Ras-like G proteins, all 16 human Gα subunits of heterotrimeric G proteins, that is, the two members of the Gs, the eight members of the Gi/o, the four members of the Gq/11, and the two members of the G12 family, possess intrinsic GTPase activity.12,17 Nonetheless, rates of GTP hydrolysis vary considerably for the various Gα subunits, with certain isoforms (Gαq, Gαz) being extremely slow at converting GTP to GDP.12,18,19 Importantly, the GTP hydrolysis rates for all heterotrimeric G protein Gα subunits measured in vitro appear slow and probably incompatible with in vivo functions.20 –22 This is why the cell utilizes ‘regulator of G protein signaling (RGS)’ domain-containing proteins, a ~120-amino acid-long domain that can bind the Gα subunit and dramatically accelerate GTP hydrolysis.20 –30 The proteins that contain this RGS domain, first discovered in yeast and in the nematode worm Caenorhabditis elegans, are called RGS proteins.20 –30 GTP hydrolysis is enormously (up to 2000 times higher) accelerated by RGS proteins, and both the amplitude and duration of Gα and free Gβγ subunit signaling are markedly reduced.12,31 Every protein with a functional RGS domain is categorized into a subfamily, designated by a letter (A–F) and the name of a representative member of that particular subfamily (next to the letter ‘R’).31 –35 For instance, the A/RZ subfamily is named after the representative RGSZ protein member. 36 Some RGS proteins, for example, RGS4 or RGS2, also interfere with the interaction of active (GTP-bound) Gα subunits with downstream effectors. 26 By acting as GTPase-activating proteins (GAPs) on Gα subunits, RGS proteins also accelerate free Gβγ signaling termination, since the heterotrimer reassembles.26,27 It was initially thought that there might be a specific RGS protein for each of the 16 different Gα subunits but we now know that this could not have been further from the truth. 27 Not only do the RGS proteins outnumber the Gα subunits, but also several of them can act upon more than one Gα type/family (e.g. RGS4 inactivates both Gαi/o and Gαq/11 subunits). Furthermore, Gαs is not a substrate for any RGS protein, and it is still an open question whether Gα12/13 are. However, it seems that most (if not all) RGS proteins inactivate G proteins in a cell type- and GPCR-specific manner, that is, they do not inactivate their Gα subunit substrates at all times or under any circumstances. 27 The identity of the receptor that has stimulated the G protein seems to play a crucial role in whether that G protein serves as a substrate for the RGS protein. For example, RGS4 inactivates angiotensin II type 1 receptor (AT1R)-stimulated Gαq but not gonadotropin-releasing hormone receptor-stimulated Gαq subunits.37,38 This is extremely important to consider because it bestows RGS protein functions with exceptional receptor-G protein signaling pathway specificity that can be exploited for therapeutic purposes.
In the present review, we first discuss the current literature on the regulation of cardiac GPCRs by RGS proteins in the context of heart physiology but also of heart disease, followed by a closer look at cardiac RGS4, which has been documented to be implicated in HF and atrial fibrillation (AFib). Our review focuses exclusively on the cardiac effects of the B/R4 family of RGS proteins (RGS1–5, RGS8, RGS13, RGS16, RGS18, and RGS21), the smallest mammalian RGS protein family members that function primarily (if not exclusively) as G protein GAPs, that is, are bona fide RGS proteins. Other proteins that contain RGS homology domains but serve other primary functions (e.g. GRKs, which are serine/threonine kinases), as well as a thorough discussion of the biology and physiology of RGS proteins in tissues outside the heart, are beyond the scope of the present review.
Cardiac RGS proteins and regulation of GPCR signaling pathways
RGS1, RGS2, and RGS3 are expressed in both cardiac myocytes and fibroblasts. RGS2 is also robustly expressed in both vascular smooth muscle and endothelial cells.32,39 RGS4 is highly expressed in the brain, heart, and adrenal glands.24,30,31 RGS5 displays mainly vascular expression.34,40,41 RGS8, RGS13, and RGS18 are mainly expressed in immune cells although RGS18 is also present in platelets. RGS16 and RGS21 are expressed in the heart.34,42 –44 RGS3 exists in multiple isoforms, 34 of which the PDZ-containing one is expressed in cardiac atria and both its long and short isoforms are abundant in the ventricles. 37 In human aortic smooth muscle cells, RGS3 regulates sphingosine 1-phosphate receptor, endothelin-1 (ET-1) receptor, and AT1R signaling. 37 Cardiac-specific overexpression of RGS3 blocks maladaptive hypertrophy and fibrosis and improves cardiac function. 45 RGS3 is also upregulated in spontaneously hypertensive heart failure (SHHF) rat hearts. 46 However, in a SHHF rat model that developed congestive HF over time, RGS3 was found downregulated in the myocardium. 46 Consistent with these findings, RGS3 appears elevated in myocardial samples from human end-stage HF patients, suggesting a role in human chronic and advanced HF. 47 Nevertheless, the specific GPCRs or signaling mechanisms affected by the RGS3 expression changes in human HF are unknown, so it is unclear at present whether these RGS3 changes are causative or not.
Cardiac RGS4 is most abundant in the sinoatrial (SA) and atrioventricular (AV) nodal regions, as well as throughout the atria.48,49 It is also expressed in aorta and in ventricles.37,46,47 Its functions in the heart are discussed in detail in the following sections below. RGS2 plays a critical role in vascular tone regulation but has been shown to affect cardiac compensation to pressure overload. 50 It also appears to be involved in the counter-regulatory effects of atrial natriuretic factor against AT1R-induced hypertrophy. 51 Notably, RGS2 is the only RGS protein reported to date to directly oppose Gs protein signaling, albeit not by acting as a GAP for Gαs but rather by interacting with adenylyl cyclase (the effector of Gαs) and inhibiting it.52 –54 No RGS protein acting as Gαs-GAP has been reported to date. 36 RGS5 has also been reported to participate in cardioprotection against pressure overload, although no specific receptors were examined in that study. 55 RGS5 or RGS2 knockouts lead to worsened pressure overload-induced cardiac fibrosis in mice.50,55 Gq/11-coupled receptors AT1R endothelin type A receptor (ETAR) are major profibrotic mediators in human cardiac fibroblasts.56 –58 RGS2 opposes AT1R signaling-dependent cell proliferation and collagen synthesis in ventricular fibroblasts. 59 However, cardiomyocyte-residing RGS2, acting in a paracrine fashion, may have contributed to these anti-fibrotic effects of cardiac RGS2.
RGS13 is one of the two RGS proteins (the other one being RGS2) that typically localizes in the cell nucleus. 60 Indeed, upon cyclic 3′,5′-adenosine monophosphate (cAMP) synthesis and cAMP-dependent protein kinase (PKA) activation, RGS13 translocates to the nucleus and interacts with the PKA-phosphorylated transcription factor cAMP response element-binding (CREB) protein, inhibiting gene transcription downstream of CREB. 61 However, this may not occur in the heart, given the very low RGS13 expression in the myocardium. 31 RGS16 is present in both cardiac myocytes and fibroblasts31,62,63 and is one of the very few RGS proteins identified to date that act as Gα12/13-GAPs. 64 Bacterial lipopolysaccharide (LPS) endotoxin impairs cardiac contractility and precipitates acute septic HF. 65 Treatment of cardiomyocytes with LPS or ET-1 upregulates RGS16 transcriptionally, lowering phospholipase C (PLC)-β activation by ET-1-activated ETARs in cardiac myocytes.32,66
Therapeutic potential of cardiac RGS4
Cardiac RGS4 and HF
Exogenous overexpression of RGS4 in cardiomyocytes attenuates ETAR signaling through PLCβ activation, thereby reducing contractility but also hypertrophy.67 –69 Indeed, RGS4 overexpression in murine cardiac myocytes inhibits compensation for aortic banding-induced afterload increase. 68 Cardiac RGS4-overexpressing mice also suffered from increased postoperative mortality following aortic banding. 68 This could have occurred because of reduced Gq signaling-dependent adaptive hypertrophic/inotropic responses. 67 Surprisingly however, positive inotropy in response to dobutamine was preserved in the RGS4-overexpressing mice, 68 so β-adrenergic-dependent contractility was intact. Perhaps the excess mortality happened because of RGS4-mediated blockade of Gi/o protein signaling, which is essential for anti-apoptosis in the myocardium.70,71 Importantly, RGS4 overexpression ameliorated cardiac hypertrophy in the survivors by inhibiting the ‘fetal’ gene program activation induced by the Gq protein/calcium signaling pathway. 68 The salutary effects of RGS4 in Gq-dependent hypertrophy induction were also observed in transgenic mice overexpressing both RGS4 and Gαq in the same hearts. 69 Thus, RGS4 was established more than 20 years ago as cardioprotective against hypertrophic signals and increased afterload courtesy of its Gq signaling inhibition. Corroborating this role for RGS4 is the fact that it is found upregulated in an experimental rat model of cardiac hypertrophy as well. 46
Importantly, cardiac RGS4 was found upregulated in advanced human HF in two different populations.47,72 In a German study, RGS4 was found selectively upregulated, that is, the only 1 out of 10 RGS proteins examined, at both the mRNA and protein levels, in human dilated or ischemic cardiomyopathy-related end-stage HF. 72 In the English study, RGS4 mRNA and protein levels were increased in both end-stage and acute human HF. 47 Additionally, RGS4 dampened PLC activity in human left ventricular membranes, along with terminating ETAR-dependent Gq/PLC/Ca2+ signaling. 72 In conclusion, cardiac RGS4 appears to be cardioprotective and its upregulation in the failing human heart may very well serve as a compensatory mechanism in the face of excessive hypertrophic and maladaptive (metabolically demanding) Gq/PLC/Ca2+ signaling by certain cardiac GPCRs.
Consistent with this, we recently uncovered that RGS4 also opposes the Gi/o protein signaling of the short-chain free fatty acid receptor (FFAR)-3 in cultured cardiomyocytes. 73 FFAR3 is activated mainly by gut microbial metabolites propionate and butyrate, but also by other free fatty acids with a shorter than six carbon atoms-long chain.74,75 Like the other three human FFARs (FFAR1, FFAR2, FFAR4), FFAR3 is a Gi/o-coupled GPCR that promotes inflammation through interleukin (IL)-6 and IL-1β induction, transforming growth factor (TGF)-β-dependent fibrosis, and increased norepinephrine release (via Gi/o-derived free Gβγ-activated PLCβ activation and subsequent Ca2+ signaling).76 –79 RGS4 was found to be essential for the blockade of cardiac FFAR3-mediated inflammation and fibrosis, as well as for neuronal FFAR3-dependent sympatholysis that preserved cardiac βAR reserve (cardiomyocyte β-adrenergic receptor (AR) membrane density). 73 These findings suggest a protective role for cardiac RGS4 in reverse remodeling and in mitigation of sympathetic nervous system hyperactivity induced by gut microbiota-derived nutrient metabolites, such as propionic and butyric acids.73,77 Of note, ketone bodies like β-hydroxybutyrate have been reported to antagonize FFAR3,76,80 so it appears that RGS4 can mimic (at least some of) the beneficial actions of ketone bodies in the heart.
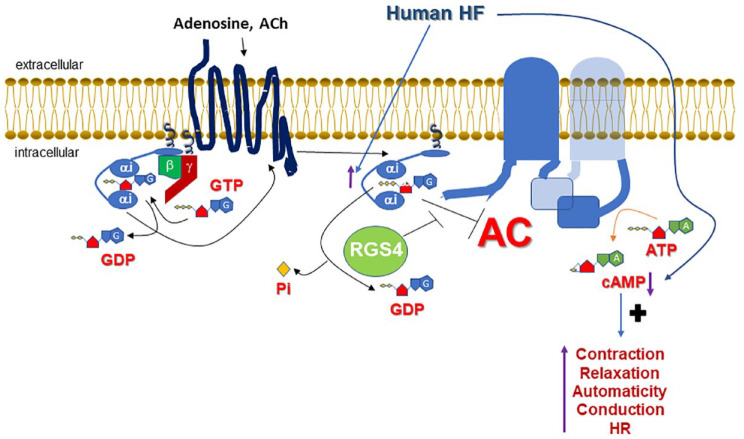
Another signaling mechanism that could potentially endow RGS4 with therapeutic benefit potential in human HF is the positive regulation of cardiac cAMP levels it may exert courtesy of its GAP activity at Gαi subunits (Figure 1). As has been suggested for RGS4 in pancreatic beta cells and other tissues,81,82 termination of Gαi subunit signaling by RGS4 would relieve adenylyl cyclase from Gαi inhibition, thereby indirectly promoting cAMP synthesis (and PKA activation) by Gs-coupled GPCRs, such as the cardiac βARs (Figure 1). The fact that the response of the RGS4-overexpressing mice to dobutamine post-aortic banding was normal also argues in favor of this scenario. 68 This mechanism might be particularly important in the setting of human HF, given that Gαi (but not Gαs or Gαq) is known to be selectively upregulated in the failing human heart, regardless of the type of failure (acute or chronic end-stage) or etiology (ischemic or dilated cardiomyopathy)83 –88 (Figure 1). This Gαi upregulation is driven by norepinephrine overstimulation of cardiac β1ARs, which transcriptionally upregulate Gαi via the Gs protein/cAMP/PKA signaling axis, and thus probably serves as a feedback, counter-regulatory mechanism against catecholaminergic overdrive of the failing heart.84,85 However, increased Gαi activity means that basal and hormone-activated adenylyl cyclase activities are suppressed, leading to chronically low cAMP levels in the failing human heart (Figure 1). Indeed, several lines of evidence point to the fact that cAMP levels are low and cAMP synthesis is deficient in the failing human heart.89 –92 Although this might initially serve as an adaptive response of the failing myocardium to protect itself from excessive norepinephrine stimulation (the developing sympathetic nervous system overdrive), low cAMP levels can become maladaptive over time, because cAMP is essential not only for the contractile (systolic) function of the heart but also for its relaxation (diastolic) function.86,93 In addition to inotropy, automaticity, and dromotropy, cAMP increases lusitropy of the myocardium, as well. This is mainly achieved by a combination of PKA-dependent phosphorylations that primarily activate sarco(endo)plasmic reticulum calcium adenosine triphosphatase (SERCA) in the sarcoplasmic reticulum (SR) (via phospholamban phosphorylation) to remove Ca2+ from the cytoplasm back into the SR, 94 the sodium pump in the plasma membrane (via phospholemman phosphorylation) to drive sodium/calcium exchanger-mediated Ca2+ extrusion out of the cardiomyocyte, 95 and even accelerate actomyosin filament relaxation (via myosin-binding protein-C3 phosphorylation).96,97 All these actions combined reverse the intracellular free [Ca2+] elevation induced by cAMP during contraction and allow for the myocardium to relax and fill with blood during diastole.98,99 It is thus quite plausible that RGS4 is selectively (among all RGS proteins expressed in the human heart) upregulated in the failing human heart as a compensatory mechanism for the myocardium in an effort to counterbalance the Gαi upregulation and maintain some basic level of adenylyl cyclase activity and cAMP synthesis necessary for proper cardiomyocyte homeostasis (Figure 1). In fact, one of the first articles reporting the Gαi upregulation in human HF, by Böhm and colleagues in 1990, concluded with the quote: ‘Inactivation of Giα could be a potential target for the medical treatment of chronic heart failure’. 83 The RGS proteins discovered a few years later, specifically RGS4, could fill this role perfectly. Nevertheless, this RGS4 upregulation is evidently insufficient to increase cAMP levels in the failing human heart to a substantial extent, given that the cAMP levels measured in advanced human HF are still low.89,90 Thus, RGS4 upregulation alone does not suffice to halt (let alone reverse) the progressive deterioration of cardiac function in humans with chronic HF. Finally, it is worth noting that the fact that Gαi is elevated in the failing human heart means that interventions such as GRK2 inhibition, aimed at increasing βAR-elicited Gs protein signaling that is depressed in human HF due to elevated GRK2-dependent desensitization,100,101 would probably be ineffective at sufficiently improving cAMP levels and, consequently, cardiac function.
Figure 1.
Role of cardiomyocyte RGS4 in the context of human HF. Basal and hormone induced (e.g. by adenosine A1 and A3 or M2 muscarinic cholinergic receptors) Gαi activity is elevated, so cAMP levels are low in human HF. RGS4, by accelerating GTP hydrolysis on Gαi, functionally opposes/terminates Gαi actions, thereby (indirectly) promoting AC activation and cAMP synthesis. cAMP exerts multiple effects in the heart crucial for cardiomyocyte function, such as contraction followed by relaxation, automaticity, and positive chronotropy and dromotropy (conduction). Thus, RGS4 can potentially reverse part of the molecular abnormalities present in the failing human myocardium.
A, adenine; AC, adenylyl cyclase; ATP, adenosine triphosphate; cAMP, 3′,5′-adenosine monophosphate; G, guanine; HF, heart failure; P, phosphorylation; Pi, inorganic phosphate; RGS, regulator of G protein signaling. See text for more details and all other molecular acronym descriptions.
Cardiac RGS4 and AFib
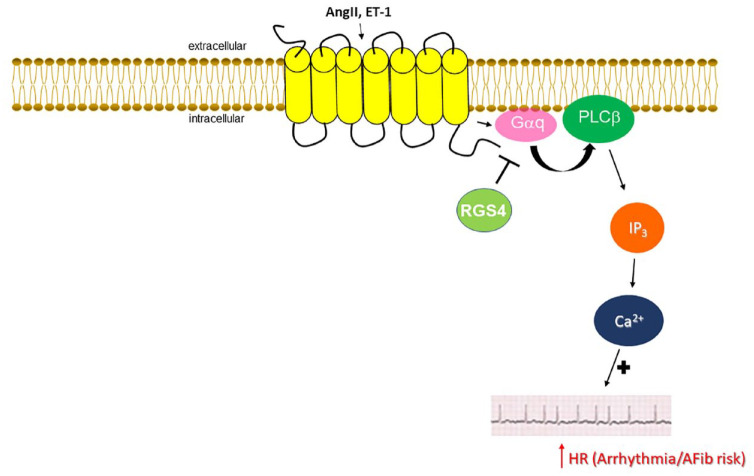
Apart from its putative roles in regulation of cardiac inotropy and lusitropy, RGS4 has been documented to play a crucial role in cardiac chronotropy regulation. 48 Cholinergic regulation of heart rate (HR) is mainly mediated by the Gi/o-coupled M2 muscarinic cholinergic receptor (mAChR).89,102 The underlying mechanism for acetylcholine (ACh)-induced bradycardia is activation of Gi/o-derived free Gβγ subunits, which help open atrial G protein-coupled inwardly rectifying K+ (GIRK) channels, resulting in ACh-induced potassium hyperpolarizing currents (IKACh).27,48,102 M2 mAChR-stimulated, as well as adenosine receptor-stimulated, Gαi-dependent inhibition of adenylyl cyclase also contributes to cholinergic (and adenosinergic) slowing of HR since cAMP is essential for the operation of hyperpolarization-activated cyclic Nucleotide-gated (HCN)-4 channels, responsible for the generation of the pacemaker ‘funny’ current (If) in SA nodal pacemaker cells.103,104 cAMP also enhances depolarizing Ca2+ influx currents in AV nodal cells (via PKA-mediated phosphorylation and opening of L-type calcium channels and of ryanodine receptor 2 channels), which is responsible for propagation of electrical conduction throughout the atria, AV node, and over to the ventricles (Purkinje fibers and Hiss bundle).26,27,94,105 In other words, cAMP lowering reduces automaticity and induces negative dromotropy in the heart. RGS4 and RGS6 have long been known to function as key regulators of cholinergic control of HR.106 –108 RGS4 or RGS6 genetic deletion results in severe bradycardia from vagal stimulation in vivo.106 –108 However, RGS6 may use a different mechanism for slowing HR, since, unlike RGS4, RGS6 can directly interact with Gβ5 via its Gγ-like domain and form a complex that suppresses IKACh. 106 In fact, the role of RGS4 in negative regulation of normal, basal IKACh currents in the SA node has been challenged by several studies.109,110 Indeed, it appears that, under normal basal vagal tone conditions, RGS6 and RGS10 are mainly responsible for IKACh desensitization.109,111 In conditions that enhance vagal tone, however, RGS4 takes over and suppresses the excess IKACh currents that promote AFib development secondary to physical exercise or other AFib-precipitating stimuli.108,110 Further supporting a cardioprotective role for RGS4 against AFib pathogenesis is the fact that RGS4 is essential for suppression of pro-arrhythmogenic Ca2+ signaling by Gq/11 protein-coupled receptors, primarily the endothelin ETA and angiotensin II AT1 receptors, in the heart 112 (Figure 2). Indeed, RGS4 knockout atrial myocytes developed AFib more frequently and exhibited higher endothelin-dependent Ca2+ spark frequencies than controls. 112 Thus, RGS4 protects against AFib induced by uncontrolled Gq/11-PLCβ/inositol trisphosphate (IP3)/Ca2+ signaling, causing abnormal beats/electrical events. 112 Finally, RGS4 has been shown to suppress PLC activity (and subsequent Ca2+ signaling), both basally and upon ET-1 stimulation, in human cardiomyocyte membranes. 72 In conclusion, RGS4 appears essential for suppression of excessive Ca2+ and excessive cholinergic IKACh signaling in human atria, both of which are arrhythmogenic and can lead to AFib development (Figure 2). This strongly suggests that pharmacological interventions to enhance cardiac RGS4 expression and/or activity might have significant therapeutic value in AFib treatment and prevention, especially since RGS4 does not seem to negatively affect normal vagal HR regulation, which would be arrhythmogenic on its own and also appears to be protective against pathological cardiac hypertrophy. 113
Figure 2.
Role of (atrial) cardiomyocyte RGS4 in the context of human AFib. RGS4 terminates Gq protein signaling induced by AngII and ET-1 receptors, thereby attenuating pro-arrhythmic calcium signaling and reducing risk of AFib development.
ACh, acetylcholine; AFib, atrial fibrillation; AngII, angiotensin II; ET-1, endothelin-1; HR, heart rate; IP3, inositol 1′,4′,5′-trisphosphate; RGS, regulator of G protein signaling. See text for more details and all other molecular acronym descriptions.
Conclusions and future perspectives
A lot of progress has been made over the past 20 years in elucidating the signaling actions and biological effects of RGS proteins in the heart, as in other organs and organ systems. RGS proteins could be attractive therapeutic targets in diseases of the heart, the kidneys, the central nervous system, but also in oncology and other disease areas. The major question that needs to be answered for each disease and each RGS protein is whether its inhibition or potentiation is therapeutically desirable, which, of course, depends on each individual tissue type and disease setting in question. RGS protein inhibition generally enhances GPCR signaling, which can theoretically be beneficial for reducing dosage (and side effects) of other drugs that act as GPCR agonists (e.g. β2-adrenergic agonists in asthma). Moreover, by blocking activation of certain effectors by certain G proteins (e.g. RGS2-mediated blockade of adenylyl cyclase activation, RGS4-mediated blockade of PLCβ) RGS protein inhibitors fine-tune GPCR signaling in response to GPCR agonist drugs. On the flip side, RGS protein stimulation can be desirable in many pathological conditions characterized by aberrant G protein signaling and low RGS protein activity or expression.
Although a considerable amount of work still needs to be done to fully elucidate its function in the heart and in other organs, RGS4 already emerges as a potential therapeutic target in both human AFib and HF. Coupled with its potential in treatment of kidney injury/disease, 114 cancer,115,116 asthma, 117 and diabetes, 82 not to mention its already substantiated potential as a genetic risk factor for psychiatric disorders, 118 development of a pharmacological ‘magic bullet’ based on RGS4 activity augmentation in the future will not be surprising.
Interestingly, a number of small molecule inhibitors for RGS4 have been developed over the past 10–15 years, 40 largely for the purpose of delineating the effects of this protein in vivo, that is, as an alternative to RGS4 knockouts (see O’Brien et al. 40 for an excellent recent review on the chemistry and pharmacology of RGS protein-targeting compounds). Indeed, in vivo experiments with the RGS4 small molecule inhibitor CCG-50014 confirmed the crucial role RGS4 plays in modulating analgesia, including opioid receptor-mediated pain relief, which was significantly enhanced by coadministration of this RGS4 inhibitor. 40 However, while RGS4 inhibition may be therapeutically advantageous in analgesia or in brain disorders, the findings from cardiovascular studies discussed above strongly argue for RGS4 potentiation being advantageous in HF and AFib. Unfortunately, design and development of RGS protein enhancers is inherently more difficult than that of RGS protein inhibitors. Besides, a Gαi or a Gαq inhibitor can (in theory at least) do the same job as an RGS4 enhancer. An interesting, alternative approach toward augmentation of RGS4 expression/activity could be protein stabilization, that is, inhibition of RGS4 proteasomal degradation via ubiquitination. 119 Indeed, pharmacological inhibition of the N-end rule pathway that degrades several R4 RGS proteins including RGS440,119 with the neurostimulant agent para-chloroamphetamine has been shown to increase RGS4 protein stability/levels. 120 Pharmacological augmentation of RGS4 levels/activity is thus feasible.
Admittedly, our present review has several limitations, such as relying on in vitro and animal model studies; focusing exclusively on the myocardium and on cardiomyocytes without taking into account the complex interplay between cardiac myocytes, fibroblasts, and endothelial cells, as well as, of course, between the heart, blood vessels, and neurons that innervate the myocardium; and, finally, focusing specifically on RGS4, while it is almost certain that RGS4 works in concert with other RGS proteins and G protein-interacting partners to produce its biological effects in the heart and in other tissues/organs. Nevertheless, we attempted herein to document a case for the beneficial effects of RGS4, and hence, for its pharmacological potentiation being potentially therapeutic, specifically in human HF and AFib. The arrival of better isoform-specific small organic molecules and of other molecular tools that modulate activity or expression or subcellular localization of RGS proteins in the near future will be instrumental in defining the appropriate place of each individual RGS protein, RGS4 included, on the map of targets for the current and future therapeutic arsenals for cardiac hypertrophy, HF, AFib, arrhythmias, hypertension, and other cardiovascular diseases.
Acknowledgments
None.
Footnotes
ORCID iD: Anastasios Lymperopoulos  https://orcid.org/0000-0001-9817-6319
https://orcid.org/0000-0001-9817-6319
Contributor Information
Giselle Del Calvo, Laboratory for the Study of Neurohormonal Control of the Circulation, Department of Pharmaceutical Sciences, Barry and Judy Silverman College of Pharmacy, Nova Southeastern University, Fort Lauderdale, FL, USA.
Teresa Baggio Lopez, Laboratory for the Study of Neurohormonal Control of the Circulation, Department of Pharmaceutical Sciences, Barry and Judy Silverman College of Pharmacy, Nova Southeastern University, Fort Lauderdale, FL, USA.
Anastasios Lymperopoulos, Laboratory for the Study of Neurohormonal Control of the Circulation, Department of Pharmaceutical Sciences, Barry and Judy Silverman College of Pharmacy, Nova Southeastern University, 3200 South University Drive, HPD (Terry) Building/Room 1350, Fort Lauderdale, FL 33328-2018, USA.
Declarations
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Author contributions: Giselle Del Calvo: Investigation; Writing – original draft.
Teresa Baggio Lopez: Investigation; Writing – original draft.
Anastasios Lymperopoulos: Conceptualization; Investigation; Project administration; Resources; Supervision; Writing – original draft; Writing – review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: AL is supported by a grant from the National Institutes of Health (NIH)/National Heart, Lung, and Blood Institute (NHLBI) (R01 #HL155718-01).
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Availability of data and materials: Not applicable.
References
- 1. Insel PA, Sriram K, Gorr MW, et al. GPCRomics: an Approach to discover GPCR drug targets. Trends Pharmacol Sci 2019; 40: 378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sriram K, Insel PA. G protein-coupled receptors as targets for approved drugs: how many targets and how many drugs? Mol Pharmacol 2018; 93: 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li J, Ge Y, Huang JX, et al. Heterotrimeric G proteins as therapeutic targets in drug discovery. J Med Chem 2020; 63: 5013–5030. [DOI] [PubMed] [Google Scholar]
- 4. Hauser AS, Attwood MM, Rask-Andersen M, et al. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov 2017; 16: 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weis WI, Kobilka BK. The molecular basis of G protein-coupled receptor activation. Annu Rev Biochem 2018; 87: 897–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Venkatakrishnan AJ, Deupi X, Lebon G, et al. Molecular signatures of G-protein-coupled receptors. Nature 2013; 494: 185–194. [DOI] [PubMed] [Google Scholar]
- 7. Huang CC, Tesmer JJG. Recognition in the face of diversity: interactions of heterotrimeric G proteins and G protein-coupled receptor (GPCR) kinases with activated GPCRs. J Biol Chem 2011; 286: 7715–7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rasmussen SG, DeVree BT, Zou Y, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 2011; 477: 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chung KY, Rasmussen SGF, Liu T, et al. Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature 2011; 477: 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dror RO, Mildorf TJ, Hilger D, et al. Signal transduction. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science 2015; 348: 1361–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem 1994; 140: 1–22. [DOI] [PubMed] [Google Scholar]
- 12. Sprang SR. Invited review: Activation of G proteins by GTP and the mechanism of Gα-catalyzed GTP hydrolysis. Biopolymers 2016; 105: 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Knight KM, Ghosh S, Campbell SL, et al. A universal allosteric mechanism for G protein activation. Mol Cell 2021; 81: 1384–1396.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DeVree BT, Mahoney JP, Vélez-Ruiz GA, et al. Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature 2016; 535: 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Desimine VL, McCrink KA, Parker BM, et al. Biased agonism/antagonism of cardiovascular GPCRs for heart failure therapy. Int Rev Cell Mol Biol 2018; 339: 41–61. [DOI] [PubMed] [Google Scholar]
- 16. Ferguson SS. Evolving concepts in G protein-coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol Rev 2001; 53: 1–24. [PubMed] [Google Scholar]
- 17. Van Dop C, Tsubokawa M, Bourne HR, et al. Amino acid sequence of retinal transducin at the site ADP-ribosylated by cholera toxin. J Biol Chem 1984; 259: 696–698. [PubMed] [Google Scholar]
- 18. Berstein G, Blank JL, Jhon DY, et al. Phospholipase C-beta 1 is a GTPase-activating protein for Gq/11, its physiologic regulator. Cell 1992; 70: 411–418. [DOI] [PubMed] [Google Scholar]
- 19. Casey PJ, Fong HK, Simon MI, et al. Gz, a guanine nucleotide-binding protein with unique biochemical properties. J Biol Chem 1990; 265: 2383–2390. [PubMed] [Google Scholar]
- 20. Stewart A, Fisher RA. Introduction: G protein-coupled receptors and RGS proteins. Prog Mol Biol Transl Sci 2015; 133: 1–11. [DOI] [PubMed] [Google Scholar]
- 21. Arshavsky VY, Wensel TG. Timing is everything: GTPase regulation in phototransduction. Investig Ophthalmol Vis Sci 2013; 54: 7725–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zerangue N, Jan LY. G-protein signaling: fine-tuning signaling kinetics. Curr Biol 1998; 8: R313–R316. [DOI] [PubMed] [Google Scholar]
- 23. Dohlman HG, Apaniesk D, Chen Y, et al. Inhibition of G-protein signaling by dominant gain-of-function mutations in sst2p, a pheromone desensitization factor in Saccharomyces cerevisiae. Mol Cell Biol 1995; 15: 3635–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dohlman HG, Song J, Ma D, et al. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with gpa1 (the G-protein alpha subunit). Mol Cell Biol 1996; 16: 5194–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koelle MR, Horvitz HR. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell 1996; 84: 115–125. [DOI] [PubMed] [Google Scholar]
- 26. Lymperopoulos A, Suster MS, Borges JI. Cardiovascular GPCR regulation by regulator of G protein signaling proteins. Prog Mol Biol Transl Sci 2022; 193: 145–166. [DOI] [PubMed] [Google Scholar]
- 27. Riddle EL, Schwartzman RA, Bond M, et al. Multi-tasking RGS proteins in the heart: the next therapeutic target? Circ Res 2005; 96: 401–411. [DOI] [PubMed] [Google Scholar]
- 28. Ingi T, Krumins AM, Chidiac P, et al. Dynamic regulation of RGS2 suggests a novel mechanism in G-protein signaling and neuronal plasticity. J Neurosci 1998; 18: 7178–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koelle MR. A new family of G-protein regulators – the RGS proteins. Curr Opin Cell Biol 1997; 9: 143–147. [DOI] [PubMed] [Google Scholar]
- 30. Tesmer JJ, Berman DM, Gilman AG, et al. Structure of RGS4 bound to AlF4-activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell 1997; 89: 251–261. [DOI] [PubMed] [Google Scholar]
- 31. Zhang P, Mende U. Regulators of G-protein signaling in the heart and their potential as therapeutic targets. Circ Res 2011; 109: 320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perschbacher KJ, Deng G, Fisher RA, et al. Regulators of G protein signaling in cardiovascular function during pregnancy. Physiol Genomics 2018; 50: 590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Squires KE, Montañez-Miranda C, Pandya RR, et al. Genetic analysis of rare human variants of regulators of G protein signaling proteins and their role in human physiology and disease. Pharmacol Rev 2018; 70: 446–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bansal G, Druey KM, Xie Z. R4 RGS proteins: regulation of G-protein signaling and beyond. Pharmacol Ther 2007; 116: 473–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem 2000; 69: 795–827. [DOI] [PubMed] [Google Scholar]
- 36. Masuho I, Balaji S, Muntean BS, et al. A global map of G protein signaling regulation by RGS proteins. Cell 2020; 183: 503–521.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cho H, Harrison K, Schwartz O, et al. The aorta and heart differentially express RGS (regulators of G-protein signalling) proteins that selectively regulate sphingosine 1-phosphate, angiotensin II and endothelin-1 signalling. Biochem J 2003; 371: 973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neill JD, Duck LW, Sellers JC, et al. Potential role for a regulator of G protein signaling (RGS3) in gonadotropin-releasing hormone (GnRH) stimulated desensitization. Endocrinology 1997; 138: 843–846. [DOI] [PubMed] [Google Scholar]
- 39. Osei-Owusu P, Sabharwal R, Kaltenbronn KM, et al. Regulator of G protein signaling 2 deficiency causes endothelial dysfunction and impaired endothelium-derived hyperpolarizing factor-mediated relaxation by dysregulating Gi/o signaling. J Biol Chem 2012; 287: 12541–12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O’Brien JB, Wilkinson JC, Roman DL. Regulator of G-protein signaling (RGS) proteins as drug targets: progress and future potentials. J Biol Chem 2019; 294: 18571–18585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Erdely HA, Lahti RA, Lopez MB, et al. Regional expression of RGS4 mRNA in human brain. Eur J Neurosci 2004; 19: 3125–3128. [DOI] [PubMed] [Google Scholar]
- 42. Li X, Chen L, Ji C, et al. Isolation and expression pattern of RGS21 gene, a novel RGS member. Acta Biochim Pol 2005; 52: 943–946. [PubMed] [Google Scholar]
- 43. Nagata Y, Oda M, Nakata H, et al. A novel regulator of G-protein signaling bearing GAP activity for Galphai and Galphaq in megakaryocytes. Blood 2001; 97: 3051–3060. [DOI] [PubMed] [Google Scholar]
- 44. Park IK, Klug CA, Li K, et al. Molecular cloning and characterization of a novel regulator of G-protein signaling from mouse hematopoietic stem cells. J Biol Chem 2001; 276: 915–923. [DOI] [PubMed] [Google Scholar]
- 45. Liu Y, Huang H, Zhang Y, et al. Regulator of G protein signaling 3 protects against cardiac hypertrophy in mice. J Cell Biochem 2014; 115: 977–986. [DOI] [PubMed] [Google Scholar]
- 46. Zhang S, Watson N, Zahner J, et al. RGS3 and RGS4 are GTPase activating proteins in the heart. J Mol Cell Cardiol 1998; 30: 269–276. [DOI] [PubMed] [Google Scholar]
- 47. Owen VJ, Burton PB, Mullen AJ, et al. Expression of RGS3, RGS4 and gi alpha 2 in acutely failing donor hearts and end-stage heart failure. Eur Heart J 2001; 22: 1015–1020. [DOI] [PubMed] [Google Scholar]
- 48. Cifelli C, Rose RA, Zhang H, et al. RGS4 regulates parasympathetic signaling and heart rate control in the sinoatrial node. Circ Res 2008; 103: 527–535. [DOI] [PubMed] [Google Scholar]
- 49. Stewart A, Huang J, Fisher RA. RGS proteins in heart: brakes on the vagus. Front Physiol 2012; 3: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takimoto E, Koitabashi N, Hsu S, et al. Regulator of G protein signaling 2 mediates cardiac compensation to pressure overload and antihypertrophic effects of PDE5 inhibition in mice. J Clin Investig 2009; 119: 408–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Klaiber M, Kruse M, Völker K, et al. Novel insights into the mechanisms mediating the local antihypertrophic effects of cardiac atrial natriuretic peptide: role of cGMP-dependent protein kinase and RGS2. Basic Res Cardiol 2010; 105: 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Salim S, Sinnarajah S, Kehrl JH, et al. Identification of RGS2 and type V adenylyl cyclase interaction sites. J Biol Chem 2003; 278: 15842–15849. [DOI] [PubMed] [Google Scholar]
- 53. Sinnarajah S, Dessauer CW, Srikumar D, et al. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III. Nature 2001; 409: 1051–1055. [DOI] [PubMed] [Google Scholar]
- 54. Salim S, Dessauer CW. Analysis of the interaction between RGS2 and adenylyl cyclase. Meth Enzymol 2004; 390: 83–99. [DOI] [PubMed] [Google Scholar]
- 55. Li H, He C, Feng J, et al. Regulator of G protein signaling 5 protects against cardiac hypertrophy and fibrosis during biomechanical stress of pressure overload. Proc Natl Acad Sci U S A 2010; 107: 13818–13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Porter KE, Turner NA. Cardiac fibroblasts: at the heart of myocardial remodeling. Pharmacol Ther 2009; 123: 255–278. [DOI] [PubMed] [Google Scholar]
- 57. Kawano H, Do YS, Kawano Y, et al. Angiotensin II has multiple profibrotic effects in human cardiac fibroblasts. Circulation 2000; 101: 1130–1137. [DOI] [PubMed] [Google Scholar]
- 58. Hafizi S, Wharton J, Chester AH, et al. Profibrotic effects of endothelin-1 via the ETA receptor in cultured human cardiac fibroblasts. Cell Physiol Biochem 2004; 14: 285–292. [DOI] [PubMed] [Google Scholar]
- 59. Zhang P, Su J, King ME, et al. Regulator of G protein signaling 2 is a functionally important negative regulator of angiotensin II-induced cardiac fibroblast responses. Am J Physiol Heart Circ Physiol 2011; 301: H147–H156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Johnson EN, Druey KM. Functional characterization of the G protein regulator RGS13. J Biol Chem 2002; 277: 16768–16774. [DOI] [PubMed] [Google Scholar]
- 61. Xie Z, Geiger TR, Johnson EN, et al. RGS13 acts as a nuclear repressor of CREB. Mol Cell 2008; 31: 660–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kardestuncer T, Wu H, Lim AL, et al. Cardiac myocytes express mRNA for ten RGS proteins: changes in RGS mRNA expression in ventricular myocytes and cultured atria. FEBS Lett 1998; 438: 285–288. [DOI] [PubMed] [Google Scholar]
- 63. Patten M, Stübe S, Thoma B, et al. Interleukin-1beta mediates endotoxin- and tumor necrosis factor alpha-induced RGS16 protein expression in cultured cardiac myocytes. Naunyn Schmiedebergs Arch Pharmacol 2003; 368: 360–365. [DOI] [PubMed] [Google Scholar]
- 64. Johnson EN, Seasholtz TM, Waheed AA, et al. RGS16 inhibits signalling through the Gα13–Rho axis. Nat Cell Biol 2003; 5: 1095–1103. [DOI] [PubMed] [Google Scholar]
- 65. Drosatos K, Lymperopoulos A, Kennel PJ, et al. Pathophysiology of sepsis-related cardiac dysfunction: driven by inflammation, energy mismanagement, or both? Curr Heart Fail Rep 2015; 12: 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stuebe S, Wieland T, Kraemer E, et al. Sphingosine-1-phosphate and endothelin-1 induce the expression of rgs16 protein in cardiac myocytes by transcriptional activation of the rgs16 gene. Naunyn Schmiedebergs Arch Pharmacol 2008; 376: 363–373. [DOI] [PubMed] [Google Scholar]
- 67. Tamirisa P, Blumer KJ, Muslin AJ. RGS4 inhibits G-protein signaling in cardiomyocytes. Circulation 1999; 99: 441–447. [DOI] [PubMed] [Google Scholar]
- 68. Rogers JH, Tamirisa P, Kovacs A, et al. RGS4 causes increased mortality and reduced cardiac hypertrophy in response to pressure overload. J Clin Investig 1999; 104: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rogers JH, Tsirka A, Kovacs A, et al. RGS4 reduces contractile dysfunction and hypertrophic gene induction in Galpha q overexpressing mice. J Mol Cell Cardiol 2001; 33: 209–218. [DOI] [PubMed] [Google Scholar]
- 70. Communal C, Singh K, Sawyer DB, et al. Opposing effects of beta(1)- and beta(2)-adrenergic receptors on cardiac myocyte apoptosis: role of a pertussis toxin-sensitive G protein. Circulation 1999; 100: 2210–2212. [DOI] [PubMed] [Google Scholar]
- 71. Chesley A, Lundberg MS, Asai T, et al. The β2-adrenergic receptor delivers an antiapoptotic signal to cardiac myocytes through Gi-dependent coupling to phosphatidylinositol 3′-kinase. Circ Res 2000; 87: 1172–1179. [DOI] [PubMed] [Google Scholar]
- 72. Mittmann C, Chung CH, Höppner G, et al. Expression of ten RGS proteins in human myocardium: functional characterization of an upregulation of RGS4 in heart failure. Cardiovasc Res 2002; 55: 778–786. [DOI] [PubMed] [Google Scholar]
- 73. Carbone AM, Borges JI, Suster MS, et al. Regulator of G-protein signaling-4 attenuates cardiac adverse remodeling and neuronal norepinephrine release-promoting free fatty acid receptor FFAR3 signaling. Int J Mol Sci 2022; 23: 5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kimura I, Ichimura A, Ohue-Kitano R, et al. Free fatty acid receptors in health and disease. Physiol Rev 2020; 100: 171–210. [DOI] [PubMed] [Google Scholar]
- 75. Lymperopoulos A, Suster MS, Borges JI. Short-chain fatty acid receptors and cardiovascular function. Int J Mol Sci 2022; 23: 3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kimura I, Inoue D, Maeda T, et al. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc Natl Acad Sci U S A 2011; 108: 8030–8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lymperopoulos A, Borges JI, Cora N, et al. Sympatholytic mechanisms for the beneficial cardiovascular effects of SGLT2 inhibitors: a research hypothesis for Dapagliflozin’s effects in the adrenal gland. Int J Mol Sci 2021; 22: 7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rutting S, Xenaki D, Malouf M, et al. Short-chain fatty acids increase TNFα-induced inflammation in primary human lung mesenchymal cells through the activation of p38 MAPK. Am J Physiol Lung Cell Mol Physiol 2019; 316: L157–L174. [DOI] [PubMed] [Google Scholar]
- 79. Martin-Gallausiaux C, Béguet-Crespel F, Marinelli L, et al. Butyrate produced by gut commensal bacteria activates TGF-beta1 expression through the transcription factor SP1 in human intestinal epithelial cells. Sci Rep 2018; 8: 9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhang Y, Lei Y, Honarpisheh M, et al. Butyrate and class I histone deacetylase inhibitors promote differentiation of neonatal porcine islet cells into beta cells. Cells 2021; 10: 3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mighiu AS, Heximer SP. Controlling parasympathetic regulation of heart rate: a gatekeeper role for RGS proteins in the sinoatrial node. Front Physiol 2012; 3: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bastin G, Luu L, Batchuluun B, et al. RGS4-deficiency alters intracellular calcium and PKA-mediated control of insulin secretion in glucose-stimulated beta islets. Biomedicines 2021; 9: 1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Owen VJ, Burton PB, Michel MC, et al. Myocardial dysfunction in donor hearts. A possible etiology. Circulation 1999; 99: 2565–2570. [DOI] [PubMed] [Google Scholar]
- 84. Eschenhagen T, Mende U, Diederich M, et al. Long term beta-adrenoceptor-mediated up-regulation of Gi alpha and G(o) alpha mRNA levels and pertussis toxin-sensitive guanine nucleotide-binding proteins in rat heart. Mol Pharmacol 1992; 42: 773–783. [PubMed] [Google Scholar]
- 85. Reithmann C, Gierschik P, Sidiropoulos D, et al. Mechanism of noradrenaline-induced heterologous desensitization of adenylate cyclase stimulation in rat heart muscle cells: increase in the level of inhibitory G-protein alpha-subunits. Eur J Pharmacol 1989; 172: 211–221. [DOI] [PubMed] [Google Scholar]
- 86. Feldman AM, Cates AE, Veazey WB, et al. Increase of the 40,000-mol wt pertussis toxin substrate (G protein) in the failing human heart. J Clin Investig 1988; 82: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Böhm M, Eschenhagen T, Gierschik P, et al. Radioimmunochemical quantification of Giα in right and left ventricles from patients with ischemic and dilated cardiomyopathy and predominant left ventricular failure. J Mol Cell Cardiol 1994; 26: 133–149. [DOI] [PubMed] [Google Scholar]
- 88. Böhm M, Gierschik P, Jakobs KH, et al. Increase of Gi alpha in human hearts with dilated but not ischemic cardiomyopathy. Circulation 1990; 82: 1249–1265. [DOI] [PubMed] [Google Scholar]
- 89. Brodde OE, Michel MC. Adrenergic and muscarinic receptors in the human heart. Pharmacol Rev 1999; 51: 651–690. [PubMed] [Google Scholar]
- 90. Feldman MD, Copelas L, Gwathmey JK, et al. Deficient production of cyclic AMP: pharmacologic evidence of an important cause of contractile dysfunction in patients with end-stage heart failure. Circulation 1987; 75: 331–339. [DOI] [PubMed] [Google Scholar]
- 91. Mehel H, Emons J, Vettel C, et al. Phosphodiesterase-2 is up-regulated in human failing hearts and blunts β-adrenergic responses in cardiomyocytes. J Am Coll Cardiol 2013; 62: 1596–1606. [DOI] [PubMed] [Google Scholar]
- 92. Guellich A, Mehel H, Fischmeister R. Cyclic AMP synthesis and hydrolysis in the normal and failing heart. Pflugers Arch 2014; 466: 1163–1175. [DOI] [PubMed] [Google Scholar]
- 93. El-Armouche A, Eschenhagen T. Beta-adrenergic stimulation and myocardial function in the failing heart. Heart Fail Rev 2009; 14: 225–241. [DOI] [PubMed] [Google Scholar]
- 94. Bers DM. Calcium cycling and signaling in cardiac myocytes. Annu Rev Physiol 2008; 70: 23–49. [DOI] [PubMed] [Google Scholar]
- 95. Han F, Bossuyt J, Martin JL, et al. Role of phospholemman phosphorylation sites in mediating kinase-dependent regulation of the Na+-K+-ATPase. Am J Physiol Cell Physiol 2010; 299: C1363–C1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Moss RL, Fitzsimons DP, Ralphe JC. Cardiac MyBP-C regulates the rate and force of contraction in mammalian myocardium. Circ Res 2015; 116: 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Pohlmann L, Kröger I, Vignier N, et al. Cardiac myosin-binding protein C is required for complete relaxation in intact myocytes. Circ Res 2007; 101: 928–938. [DOI] [PubMed] [Google Scholar]
- 98. Packer M. Diastolic function as a target of therapeutic interventions in chronic heart failure. Eur Heart J 1990; 11: 35–40. [DOI] [PubMed] [Google Scholar]
- 99. Garcia MJ. Left ventricular filling. Heart Fail Clin 2008; 4: 47–56. [DOI] [PubMed] [Google Scholar]
- 100. Rengo G, Lymperopoulos A, Zincarelli C, et al. Myocardial adeno-associated virus serotype 6-betaARKct gene therapy improves cardiac function and normalizes the neurohormonal axis in chronic heart failure. Circulation 2009; 119: 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lymperopoulos A, Rengo G, Funakoshi H, et al. Adrenal GRK2 upregulation mediates sympathetic overdrive in heart failure. Nat Med 2007; 13: 315–323. [DOI] [PubMed] [Google Scholar]
- 102. Lymperopoulos A, Cora N, Maning J, et al. Signaling and function of cardiac autonomic nervous system receptors: insights from the GPCR signalling universe. FEBS J 2021; 288: 2645–2659. [DOI] [PubMed] [Google Scholar]
- 103. Fenske S, Hennis K, Rötzer RD, et al. CAMP-dependent regulation of HCN4 controls the tonic entrainment process in sinoatrial node pacemaker cells. Nat Commun 2020; 11: 5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Mika D, Fischmeister R. Cyclic nucleotide signaling and pacemaker activity. Prog Biophys Mol Biol 2021; 166: 29–38. [DOI] [PubMed] [Google Scholar]
- 105. Capote LA, Mendez Perez R, Lymperopoulos A. GPCR signaling and cardiac function. Eur J Pharmacol 2015; 763: 143–148. [DOI] [PubMed] [Google Scholar]
- 106. Posokhova E, Wydeven N, Allen KL, et al. RGS6/Gβ5 complex accelerates IKACh gating kinetics in atrial myocytes and modulates parasympathetic regulation of heart rate. Circ Res 2010; 107: 1350–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Yang J, Huang J, Maity B, et al. RGS6, a modulator of parasympathetic activation in heart. Circ Res 2010; 107: 1345–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Neubig RR. And the winner is . . . Rgs4! Circ Res 2008; 103: 444–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wydeven N, Posokhova E, Xia Z, et al. RGS6, but not RGS4, is the dominant regulator of G protein signaling (RGS) modulator of the parasympathetic regulation of mouse heart rate. J Biol Chem 2014; 289: 2440–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Guasch E, Benito B, Qi X, et al. Atrial fibrillation promotion by endurance exercise: demonstration and mechanistic exploration in an animal model. J Am Coll Cardiol 2013; 62: 68–77. [DOI] [PubMed] [Google Scholar]
- 111. Bender K, Nasrollahzadeh P, Timpert M, et al. A role for RGS10 in beta-adrenergic modulation of G-protein-activated K+ (GIRK) channel current in rat atrial myocytes. J Physiol 2008; 586: 2049–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Opel A, Nobles M, Montaigne D, et al. Absence of the regulator of G-protein signaling, RGS4, predisposes to atrial fibrillation and is associated with abnormal calcium handling. J Biol Chem 2015; 290: 19233–19244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Sebastian S, Weinstein LS, Ludwig A, et al. Slowing heart rate protects against pathological cardiac hypertrophy. Function (Oxf) 2022; 4: zqac055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Siedlecki A, Anderson JR, Jin X, et al. RGS4 controls renal blood flow and inhibits cyclosporine-mediated nephrotoxicity. Am J Transplant 2010; 10: 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Xie Y, Wolff DW, Wei T, et al. Breast cancer migration and invasion depend on proteasome degradation of regulator of G-protein signaling 4. Cancer Res 2009; 69: 5743–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Mu XM, Shi W, Sun LX, et al. Pristimerin inhibits breast cancer cell migration by up- regulating regulator of G protein signaling 4 expression. Asian Pac J Cancer Prev 2012; 13: 1097–1104. [DOI] [PubMed] [Google Scholar]
- 117. Madigan LA, Wong GS, Gordon EM, et al. RGS4 overexpression in lung attenuates airway hyperresponsiveness in mice. Am J Respir Cell Mol Biol 2018; 58: 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Xu FL, Yao J, Wang BJ. Association between RGS4 gene polymorphisms and schizophrenia: a protocol for systematic review and meta-analysis. Medicine (Baltimore) 2021; 100: e27607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lee PC, Sowa ME, Gygi SP, et al. Alternative ubiquitin activation/conjugation cascades interact with N-end rule ubiquitin ligases to control degradation of RGS proteins. Mol Cell 2011; 43: 392–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Jiang Y, Choi WH, Lee JH, et al. A neurostimulant para-chloroamphetamine inhibits the arginylation branch of the N-end rule pathway. Sci Rep 2014; 4: 6344. [DOI] [PMC free article] [PubMed] [Google Scholar]