Abstract
Amplification of human papillomavirus (HPV) DNA by L1 consensus primer systems (e.g., MY09/11 or GP5+/6+) can detect as few as 10 to 100 molecules of HPV targets from a genital sample. However, genotype determination by dot blot hybridization is laborious and requires at least 27 separate hybridizations for substantive HPV-type discrimination. A reverse blot method was developed which employs a biotin-labeled PCR product hybridized to an array of immobilized oligonucleotide probes. By the reverse blot strip analysis, genotype discrimination of multiple HPV types can be accomplished in a single hybridization and wash cycle. Twenty-seven HPV probe mixes, two control probe concentrations, and a single reference line were immobilized to 75- by 6-mm nylon strips. Each individual probe line contained a mixture of two bovine serum albumin-conjugated oligonucleotide probes specific to a unique HPV genotype. The genotype spectrum discriminated on this strip includes the high-risk, or cancer-associated, HPV genotypes 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 55, 56, 58, 59, 68 (ME180), MM4 (W13B), MM7 (P291), and MM9 (P238A) and the low-risk, or non-cancer-associated, genotypes 6, 11, 40, 42, 53, 54, 57, 66, and MM8 (P155). In addition, two concentrations of β-globin probes allowed for assessment of individual specimen adequacy following amplification. We have evaluated the performance of the strip method relative to that of a previously reported dot blot format (H. M. Bauer et al., p. 132–152, in C. S. Herrington and J. O. D. McGee (ed.), Diagnostic Molecular Pathology: a Practical Approach, (1992), by testing 328 cervical swab samples collected in Digene specimen transport medium (Digene Diagnostics, Silver Spring, Md.). We show excellent agreement between the two detection formats, with 92% concordance for HPV positivity (kappa = 0.78, P < 0.001). Nearly all of the discrepant HPV-positive samples resulted from weak signals and can be attributed to sampling error from specimens with low concentrations (<1 copy/μl) of HPV DNA. The primary advantage of the strip-based detection system is the ability to rapidly genotype HPVs present in genital samples with high sensitivity and specificity, minimizing the likelihood of misclassification.
Epidemiologic evidence identifying human papillomavirus (HPV) as the sexually transmitted, primary cause of cervical cancer is strong (12). It is clear from several large case-control and cohort studies that HPV infection is the main risk factor for the development of cervical intraepithelial neoplasia and that risk is significantly increased by persistent infection with high-risk, or cancer-associated, HPV genotypes (2, 3, 14, 15, 23). PCR technology, particularly with consensus, or general, primer systems such as MY09/11 (1) and GP5+/6+ (13), has been instrumental to these studies by elaborating the natural history of HPV infections. The recognition of HPV infection as a factor that is necessary, but not sufficient, for the development of cervical cancer has resulted in the initiation of several longitudinal studies and randomized clinical trials designed to examine the predictive value of HPV DNA testing (5, 6, 12, 17, 18, 24). Preliminary findings from these studies support the potential utility of HPV testing for the effective triage of Pap smears of atypical squamous cells of undetermined significance and atypical glandular cells of undetermined significance and suggest a potential role in primary screening of populations in which Pap smears have not been sufficiently effective. A rapid PCR-based test for HPV DNA is also important to accurately investigate the natural history of HPV infections. Furthermore, because of the high sensitivity and type specificity afforded only by amplified DNA detection methods, specific PCR-based HPV DNA typing may have a unique utility in the clinical management of cervical lesions.
We report a method that uses a sensitive and broad-spectrum amplification system (1), followed by a single hybridization with a reverse line blot detection method for complete HPV genotype discrimination (see Fig. 1). This method, like other PCR-based assays, avoids false negatives below the limit of detection of nonamplified methods and can readily detect a broad spectrum of HPV genotypes. Furthermore, HPV type-specific disease associations can be precisely defined, since genotypes are individually discriminated.
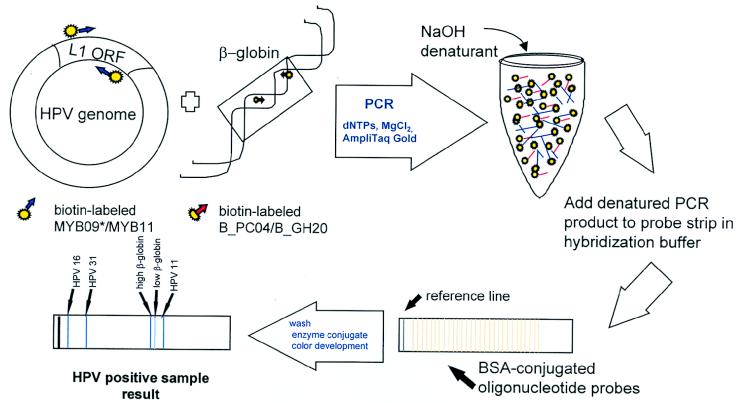
FIG. 1.
HPV genotyping of PCR product by reverse line blot method. Schematic of the reverse line blot genotyping assay from L1 consensus primer-generated PCR products. The drawing represents the detection of a hypothetical mixed infection of HPV 16, 31, and 11.
MATERIALS AND METHODS
Sample acquisition and preparation.
Cervical specimens were collected in 1.0 ml of specimen transport medium (Digene Diagnostics, Silver Spring, Md.) as part of an ongoing natural history study of HPV infection conducted at the University of New Mexico Health Sciences Center. The samples were processed by adding 30 μl of digestion solution to achieve a final concentration of 200 μg of proteinase K per ml and 0.1% Laureth-12. Digestion was conducted at 56°C for 1 h. A 300-μl aliquot of the digested material was added to 1.0 ml of absolute ethanol containing ammonium acetate and precipitated overnight at −20°C. The precipitated DNA was centrifuged for 30 min at 13,000 × g. The supernatant was immediately removed and discarded with a plugged Pasteur pipette. The crude DNA pellet was dried overnight at room temperature. The pellet was then resuspended in 150 μl of TE (10 mM Tris, 1 mM EDTA) and incubated for 15 min at 95°C to inactivate the proteinase K. The crude DNA extracts were then stored at −20°C until amplification.
PCR and dot blot-based HPV-testing methods.
Prior to amplification, the crude digests were allowed to reach room temperature and centrifuged briefly. Six microliters of each specimen was amplified with the MY09-MY11-HMB01 L1 consensus primer system (1) and AmpliTaq polymerase (Perkin-Elmer, Foster City, Calif.). To determine specimen adequacy, the GH20/PC04 human β-globin target was coamplified with the HPV consensus primers. The PCRs were amplified in a Perkin-Elmer GeneAmp PCR System 9600 for 40 cycles. The following ultrasensitive, or long, amplification profile was used: 95°C denaturation for 1 min, 55°C annealing for 1 min, and 72°C extension for 1 min for 40 cycles; followed by a 5-min terminal extension at 72°C. A subset of 56 specimens were amplified with an alternate amplification profile (rapid amplification) as follows: 95°C denaturation for 20 s, 55°C annealing for 20 s, and 72°C terminal extension for 30 s for 40 cycles; followed by a 5-min extension at 72°C.
HPV typing analyses were carried out by dot blot hybridization and biotinylated HPV type-specific oligonucleotide probes as previously described (1, 10). To each nylon membrane, 6 μl of each PCR product was denatured and applied to replicate membranes with dot blot apparatuses (Bio-Rad, Hercules, Calif.). Previously characterized PCR products were applied to 22 wells on each membrane as HPV type-specific controls (3.5 μl of PCR product per well). The membranes were hybridized at 53°C overnight with biotinylated HPV type-specific oligonucleotide probes (6/11, 16, 18, 31, 33, 35, 39, 45, 51 to 59, 66, 68, MM7, MM9, and MM4). Probes for HPV types 26 and MM8 and 40 and 42 were pooled as pairs during hybridization. A β-globin probe was used to assess specimen adequacy. Following hybridization, membranes were washed at 56 to 57°C to remove nonspecifically bound probe. The wash buffer was 56 to 57°C so that the wash stringency would be increased, given the salt and detergent concentrations and the selected oligonucleotide probes. This ensures efficient removal of the nonspecifically bound probe and optimal specific hybridization. The bound probes were detected with streptavidin-horseradish peroxidase (Vector, Burlingame, Calif.) and enhanced chemiluminescent substrate (ECL; Amersham, Arlington Heights, Ill.). Blots were exposed to Kodak X-OMAT AR 5 film initially for 10 min, followed by a second 2-h or overnight exposure. HPV positivity by dot blot was determined by establishment of a negative cutoff, and signals above the cutoff were scored based on four graded levels of intensity with visual standard references. In addition, these autoradiograms were read by two blinded independent observers. The discrepant results were resolved independently by a third observer.
PCR and line blot-based detection methods.
HPV DNA was amplified by the L1 consensus primer system previously described for dot blot detection, except each primer was labeled with a 5′ biotin molecule (denoted in the primer name by the inclusion of a capital B as follows: MYB09, MYB11, HMBB01, B GH20, B PC04). In brief, each amplification contained 10 mM Tris-HCl (pH 8.5), 50 mM KCl, 6 mM MgCl2, 200 μM (each) dCTP, dGTP, and dATP, 600 μM dUTP, 7 to 10 U of AmpliTaq Gold, 50 pmol of MYB09, 50 pmol of MYB11, 5 pmol of HMBB01, 5 pmol of B PC04, 5 pmol of B GH20, and 5 to 10 μl of sample (the MgCl2, dUTP, and AmpliTaq Gold were modifications from dot blot protocol). Modifications to the published L1 consensus amplification were made to obtain optimal sensitivity and to standardize the format to other RMS PCR assays. For eventual inclusion of uracil-N-glycosylase to prevent product carryover, dTTP was replaced with dUTP. It was empirically determined that the dUTP concentration must be increased threefold relative to the other dNTPs for efficient strand incorporation by a DNA polymerase. The MgCl2 was subsequently reoptimized to 6 mM to compensate for the increase in dNTP concentration. Reactions were amplified in a Perkin-Elmer TC9600 thermal cycler with the following ultrasensitive thermal profile: 9-min AmpliTaq Gold activation at 95°C, 40 cycles of 1-min denaturation at 95°C, 1-min annealing at 55°C, 1-min extension at 72°C; a 5-min final extension at 72°C; and a hold step at 15°C. A subset of 56 specimens were amplified with an alternate amplification profile (rapid amplification) as follows: a 9-min AmpliTaq Gold activation at 95°C followed by 40 cycles of denaturation for 20 s at 95°C, annealing for 20 s at 55°C, and terminal extension for 30 s at 72°C; followed by a 5-minute extension at 72°C and a hold step at 15°C. After removal from the thermal cycler, samples were stored at 4°C.
The general principle of immobilized probe hybridization has been described elsewhere (4, 21), and a schematic of the procedure is presented in Fig. 1. Generally, the probes were diluted into a coating buffer (50 mM 3-[cyclohexylamino]-1-propanesulfonic acid [CAPS] and 0.1 g of orange dye II per liter) and applied to a plastic-backed nylon membrane strip with a pump mechanism which delivers controlled amounts of probe to the membrane. The HPV genotyping strip contains 29 probe lines plus one reference ink line, detecting 27 individual HPV genotypes and two concentrations of the β-globin control probe. Two bovine serum albumin (BSA)-conjugated probes per HPV type, corresponding to each of two hypervariable regions within the MY09/MY11 amplicon, are deposited in a single line for each of the following HPV types: 16, 18, 26, 31, 33, 35, 39, 42, 45, 51 to 59, 66, 68, MM4, MM7, MM8, and MM9. HPV types 6, 11, 40, and the β-globin controls have a single probe deposited per line. Subsequent to this study, HPV 51A has been removed from the HPV 51 pool, due to apparent cross-reactivity with nonspecific amplicon. The configuration of the genotyping strip is diagrammed in Fig. 2, and the probe sequences are listed in Table 1. The high- and low-risk HPV types are visually separated by β-globin control lines such that all types between the reference and β-globin control lines are associated with high cancer risk and all types beyond the control lines are associated with low or no cancer risk. Disease association was defined according to the International Biological Study on Cervical Cancer (3). In the International Biological Study on Cervical Cancer, HPV types were considered high risk if detected as a single HPV infection within an invasive cancer. One exception is an HPV 6 which was found alone in a single invasive tumor; we still considered HPV 6 to be an HPV with low oncogenic potential, and it remains in the low-risk category within our present study.
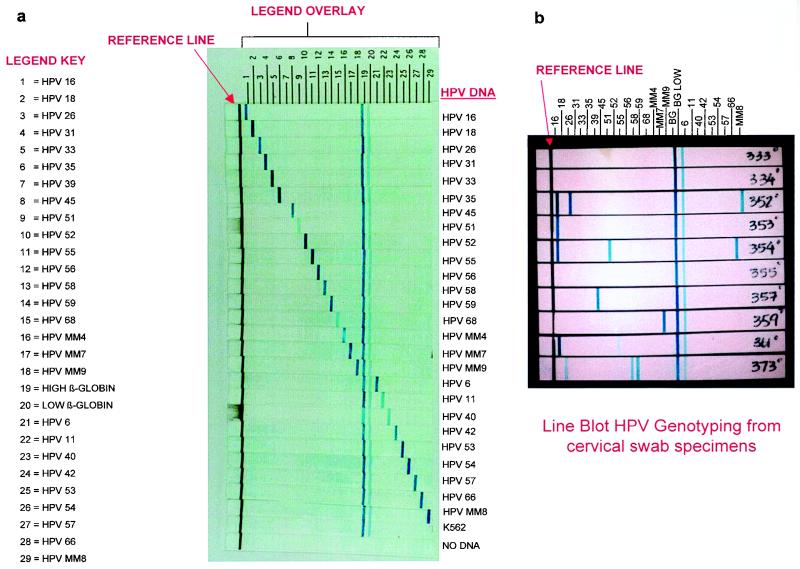
FIG. 2.
Probe layout of the HPV genotyping strip. (a) HPV genotyping strips (n = 28) hybridized with the HPV L1 consensus PCR product generated from the HPV targets indicated to the right. Fifty microliters of PCR product generated from amplification of 106 HPV plasmid targets (with the exception of HPV 51 and 68, which were amplified with 103 plasmid targets) in a background of human cellular DNA (12.5 ng/PCR) was hybridized to the HPV genotyping strips and detected by the previously described reverse line blot method. (b) Line blot genotyping hybridization results for 10 clinical specimens in the previously described study. Fifty microliters of denatured PCR product was hybridized to each strip. The genotyping results for the specimens are as follows: no. 333, HPV negative; no. 334, HPV negative; no. 352, HPV 16, 26, and MM8; no. 353, HPV 16; no. 354, HPV 16, 51, and 66; no. 355, HPV negative; no. 357, HPV 39; no. 359, HPV MM7; no. 361, HPV 16 and 52; and no. 373, HPV 18, 56, and 58.
TABLE 1.
BSA-conjugated HPV line blot probe sequences
| HPV type | BSA probe | Biotinylated probe | Sequencea |
|---|---|---|---|
| 6 | HPV611A | MYB12 | CATCCGTAACTACATCTTCCA (1) |
| 11 | HPV611B | MYB13 | TCTGTGTCTAAATCTGCTACA (1) |
| 16 | HPV16A | MYB95 | GATATGGCAGCACATAATGAC (1) |
| HPV16B | MYB133 | GTAACATCCCAGGCAATTG (1) | |
| 18 | HPV18C | CTTAAATTTGGTAGCATCATATTGb | |
| HPV18D | TCAGCCGGTGCAGCATCC | ||
| 26 | HPV26A | MYB186 | GCTGACAGGTAGTAGCAGAGTT (10) |
| HPV26B | MYB187 | GCCATAACATCTGTTGTAAGTG (10) | |
| 31 | HPV31C | GATCTTCCTTGGGCTTTTGG | |
| HPV31D | TGTCTGTTTGTGCTGCAATT | ||
| 33 | HPV33C | CTGTCACTAGTTACTTGTGTGCAT | |
| HPV33E | TTTGGAGGTACTGTTTTTTGA | ||
| 35 | HPV35A | MYB115 | CTGCTGTGTCTTCTAGTGACAG (1) |
| HPV35B | MYB117 | ATCATCTTTAGGTTTTGGTGC (1) | |
| 39 | HPV39A | MYB89 | TAGAGTCTTCCATACCTTCTAC (1) |
| HPV39B | MYB90 | CTGTAGCTCCTCCACCATCT (1) | |
| 40 | HPV40A | MYB176 | CCCAAGGTACGGGAGGATCC (10) |
| 42 | HPV42C | GCGTTGTTACCTTAGCCTGA | |
| HPV42D | ATCACCAGATGTTGCAGTG | ||
| 45 | HPV45A | MYB69 | ATACTACACCTCCAGAAAAGC (1) |
| HPV45B | MYB129 | GCACAGGATTTTGTGTAGAG (10) | |
| 51 | HPV51Ab | MYB87 | TATTAGCACTGCCACTGCTG (1) |
| HPV51D | CATCCTCCAAACTAGCAGAC | ||
| 52 | HPV52A | MYB81 | CACTTCTACTGCTATAACTTGT (1) |
| HPV52B | MYB82 | ACACACCACCTAAAGGAAAGG (1) | |
| 53 | HPV53A | MYB102 | TTCTACCTTACTGGAAGACTGG (10) |
| HPV53B | MYB182 | GCAACCACACAGTCTATGTC (10) | |
| 54 | HPV54C | TTATTAAAGCTATCCTGCGTGG | |
| HPV54D | TCCTCCAAACTACTTGTAGCTG | ||
| 55 | HPV55A | MYB151 | GTGCTGCTACAACTCAGTCT (10) |
| HPV55D | CGCATGTATTGTTTATATTCTGTA | ||
| 56 | HPV56A | MYB197 | GCACAGCTATAACATGTCAACG (10) |
| HPV56C | CGTGCATCATATTTACTTAACTG | ||
| 57 | HPV57A | MYB154 | AATGTCTCTTTGTGTGCCAC (10) |
| HPV57B | MYB156 | GGATCAGTAGGGGTCTTAGG (10) | |
| 58 | HPV58A | MYB94 | AGCACCCCCTAAAGAAAAGGA (10) |
| HPV58B | MYB179 | GACATTATGCACTGAAGTAACTAAG (10) | |
| 59 | HPV59A | MYB123 | GCCAGTTAAACAGGACCC (10) |
| HPV59B | MYB162 | CCTAATGWATACACACCTACCAG (10) | |
| 66 | HPV66A | MYB83 | ATTAATGCAGCTAAAAGCACATT (10) |
| HPV66B | MYB178 | CATGTCAGAGGGAACAGCC (10) | |
| 68 | HPV68A | MYB191 | CATACCGCTATCTGCAATCAG (10) |
| HPV68B | MYB194 | CTACTACTGAATCAGCTGTACC (10) | |
| MM4 | HPVMM4A | MYB164 | CTCAATCTGTTGCACAAACA (10) |
| HPVMM4B | MYB165 | TAACCTTGCCCCCCTCAG (10) | |
| MM7 | HPVMM7A | MYB166 | GGCTAATGAATACACAGCCTC (10) |
| HPVMM7C | TCCTTCCACCAGCCTTGAT | ||
| MM8 | HPVMM8A | MYB85 | CCAACACCGAATCAGAATATAAA (10) |
| HPVMM8B | MYB163 | GTTGTGCCCCCTCCCTCCA (10) | |
| MM9 | HPVMM9A | MYB104 | GTAGGTACACAGGCTAGTAGCTC (10) |
| HPVMM9B | MYB106 | AGTTGCCAACGTCCTCAAC (10) | |
| β-Globin | B PCO3 | PC03 | ACACAACTGTGTTCACTAGC (1) |
Bold-faced sequences have not been previously published.
HPV 51A was subsequently removed from the assay; see Materials and Methods.
All liquid detection reagents used for the line blot assay were from Amplicor strip detection reagent kits (Dynal, Oslo, Norway). PCR products were denatured with 0.13 N NaOH (1:2 dilution of 0.4 N NaOH in PCR product). HPV genotyping strips were placed into individual wells of the typing trays (Perkin-Elmer) and covered with 3 ml of hybridization buffer (4× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA, pH 7.7], 0.1% sodium dodecyl sulfate) prewarmed to 53°C. Seventy microliters of denatured, biotinylated product was added to each well and incubated in a shallow, shaking (60 rpm) water bath at 53°C for 30 min. Following hybridization, trays were removed from the water bath, and hybridization solution was removed with a vacuum aspirator. Strips were briefly rinsed in the trays with ambient wash buffer (1× SSPE, 0.1% sodium dodecyl sulfate). After removal of the rinse by aspiration, 3 ml of prewarmed (53°C) wash buffer was added to each well and the trays were incubated in a shaking water bath at 53°C for 15 min. After the stringent wash, buffer was removed, 3 ml of streptavidin-horseradish peroxidase conjugate was added to each well, and the tray was placed on a rotating platform at room temperature for 30 min, with shaking at 70 rpm. Unbound conjugate was removed by a quick rinse with ambient wash buffer followed by two 10-min washes in ambient wash buffer. After the final wash, buffer was removed by vacuum aspiration, and strips were rinsed in 0.1 M sodium citrate. Color development was activated by incubation in a 4:1 mixture of substrates A (hydrogen peroxide in sodium citrate buffer) and B (3,3′,5,5′-tetramethylbenzidine in dimethylformamide) for 5 min on a rotating platform (70 rpm). Strips were rinsed in deionized water and stored in citrate buffer until interpretation. Developed strips were interpreted or photographed within 2 h of color development for accurate analysis of the results. Strips can be stored in citrate buffer in a sealed plastic bag in the dark. Any prolonged exposure to light results in fading of the signal and darkening of the membrane. Alternatively, the strips can be dried immediately following the final citrate buffer wash and taped directly into a research notebook. Strip interpretation was performed with a labeled acetate overlay, with lines indicating the position of each probe relative to the reference mark.
RESULTS
Analytic sensitivity of the HPV-type spectrum detected for both dot blot and line blot assays was determined by serial dilution of HPV plasmid or M13 phage clones amplified in a background of 12.5 ng of human cellular DNA from the K562 cell line (ATCC CCL243). HPV types 58, 59, 61, 62, 64, and 67 were provided by T. Matsukura; HPVs 33, 39, 42, 54, 55, 66, 68, and 70 were from G. Orth; HPVs 6, 11, 16, 18, 53, and 57 were from E. M. de Villiers; HPV 52 was from W. Lancaster; HPV 26 was from R. Ostrow; HPV 45 was from K. Shah; and HPV 51 was from S. Silverstein. Clinical HPV types, including MM4 (W13B), MM7 (P291), MM8 (P155), and MM9 (P238A) had been previously cloned as PCR fragments of approximately 450 bp (16). Additional sensitivities were determined in precharacterized cervical specimens. Sensitivities were virtually identical by both dot and line blot assays, ranging from 10 to 100 genomes per PCR for HPV types 6, 11, 16, 18, 31, 33, 39, 45, 51, 52, 58, 59, 66, and 68 and from ∼500 to 1,000 genomes per PCR for HPV types 26, 35, 40, 42, and 53 to 57. Variation in sensitivity among the genotypes reflects the number and position of mismatched bases in the primer-binding region at nondegenerate sites.
The specificity of HPV genotype discrimination was tested by hybridization of 500 ng (determined by gel quantitation) of amplified product to the HPV genotyping strips. Specificity of typing was excellent, with negligible background or cross-reactivity.
To test the utility of the line blot HPV detection method in clinical samples, we analyzed 359 specimens collected in Digene specimen transport medium. Type-specific oligonucleotide probe results obtained by the standard MY09-MY11-HMB01 dot blot hybridization method were compared to those obtained using the MYB09-MYB11-BHMB01 reverse line blot method. Two separate aliquots of each digested STM sample were taken and processed independently at the two participating laboratory sites, where all aliquots were amplified by using the ultrasensitive amplification profile (see Materials and Methods). Thirty-two HPV-positive and 24 HPV-negative samples determined by the ultrasensitive cycle system were randomly chosen for analysis by the short cycle profile, in which the time at each temperature step in the thermal profile was shortened. Samples were amplified separately for dot and line blot detection because of the requirement of unlabeled versus labeled primers in the dot and line blot detection methods, respectively. Investigators performing the two assays were blinded to results until all interpretations were final.
Of the 359 samples evaluated, 30 were excluded because of false signal generation from the ECL substrate (1), presumably caused by pseudoperoxidases in the sample, thus precluding interpretation of the dot blot results. The results from the remaining 329 samples are presented.
The HPV prevalence in this population was 24.0 and 25.5% by the dot blot and line blot detection methods, respectively. Table 2 represents the overall HPV concordance between the two detection formats. Agreement for HPV-positive results was good, with a kappa statistic of 0.78. Type-specific agreement between the two methods was good, with total concordance ranging from 97 to 100%. Within the HPV-positive samples, multiple HPV types were detected in 10.7 and 8.5% of specimens by the dot blot and line blot detection methods, respectively. A comparison of the results from 56 samples that were amplified and detected with a long and short cycle profile is presented in Table 3. Only 49 of the 56 samples were included in the final analysis due to false ECL signals on the dot blot. As expected, the agreement between the two amplification profiles reflected the increase in detection of low levels of HPV with the longer, ultrasensitive profile. Only the line blot results for rapid versus ultrasensitive amplification profiles are shown; however, the dot blot results were virtually identical (87.8% agreement, kappa = 0.75 for both line and dot blot). Further analysis of HPV data for both line and dot HPV assay was conducted based on recorded intensities. Signal intensity scores were as follows: 1, strong; 2, medium; 3, weak; 4, very weak; and 0, negative. Stratified analyses by signal intensity revealed that a short versus a long profile resulted in discordance within HPV-positive specimens designated 3, 4, and 0 (i.e., weak or negative) for both the line and dot blot assays.
TABLE 2.
Correlation between results for dot and line blot assays (n = 329)
| HPV type | Dot blot positive n (%) | Line blot positive n (%) | Dot or line positive n (%) | Dot and strip positive n (%) | % overall agreement | % agreement among positives | Kappa |
|---|---|---|---|---|---|---|---|
| Any | 79 (24.0) | 84 (25.5) | 95 (28.9) | 68 (20.7) | 91.8 | 71.6 | 0.780 |
| 6 or 11 | 5 (1.5) | 6 (1.8) | 6 (1.8) | 5 (1.5) | 99.7 | 83.3 | 0.908 |
| 16 | 16 (4.9) | 22 (6.7) | 22 (6.7) | 16 (4.9) | 98.2 | 72.7 | 0.883 |
| 18 | 5 (1.5) | 6 (1.8) | 7 (2.1) | 4 (1.2) | 99.1 | 57.1 | 0.723 |
| 26 or MM8 | 4 (1.2) | 5 (1.5) | 5 (1.5) | 4 (1.2) | 99.7 | 80.0 | 0.887 |
| 31 | 10 (3.0) | 9 (2.7) | 12 (3.6) | 7 (2.1) | 98.5 | 58.3 | 0.729 |
| 33 | 2 (0.6) | 2 (0.6) | 2 (0.6) | 2 (0.6) | 100 | 100 | 1.000 |
| 35 | 1 (0.3) | 1 (0.3) | 1 (0.3) | 1 (0.3) | 100 | 100 | 1.000 |
| 39 | 5 (1.5) | 9 (2.7) | 10 (3.0) | 4 (1.2) | 98.2 | 40.0 | 0.563 |
| 40 or 42 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||
| 45 | 3 (0.9) | 3 (0.9) | 4 (1.2) | 2 (0.6) | 99.4 | 50.0 | 0.664 |
| 51 | 6 (1.8) | 13 (4.0) | 13 (4.0) | 6 (1.8) | 97.9 | 46.2 | 0.622 |
| 52 | 14 (4.3) | 5 (1.5) | 15 (4.6) | 4 (1.2) | 96.7 | 26.7 | 0.408 |
| 53 | 10 (3.0) | 12 (3.6) | 13 (4.0) | 9 (2.7) | 98.8 | 69.2 | 0.812 |
| 54 | 3 (0.9) | 7 (2.1) | 7 (2.1) | 3 (0.9) | 98.9 | 42.9 | 0.595 |
| 55 | 2 (0.6) | 0 (0.0) | 2 (0.6) | 0 (0.0) | 99.4 | 0.0 | |
| 56 | 8 (2.4) | 6 (1.8) | 9 (2.7) | 5 (1.5) | 98.8 | 55.6 | 0.708 |
| 57 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||
| 58 | 4 (1.2) | 4 (1.2) | 5 (1.5) | 3 (0.9) | 99.4 | 60.0 | 0.747 |
| 59 | 7 (2.1) | 4 (1.2) | 7 (2.1) | 4 (1.2) | 99.1 | 57.1 | 0.723 |
| 66 | 5 (1.5) | 6 (1.8) | 7 (2.1) | 4 (1.2) | 99.1 | 57.1 | 0.723 |
| 68 | 3 (0.9) | 3 (0.9) | 3 (0.9) | 3 (0.9) | 100 | 100 | 1.000 |
| MM4 | 1 (0.3) | 1 (0.3) | 1 (0.3) | 1 (0.3) | 100 | 100 | 1.000 |
| MM7 | 4 (1.2) | 6 (1.8) | 6 (1.8) | 4 (1.2) | 99.4 | 66.7 | 0.797 |
| MM9 | 1 (0.3) | 2 (0.6) | 3 (0.9) | 0 (0.0) | 99.1 | 0.0 | −0.004 |
TABLE 3.
Correlation between ultrasensitive and rapid PCR resultsa
| HPV type | Ultrasensitive positive n (%) | Rapid positive n (%) | Ultrasensitive or rapid positive n (%) | Ultrasensitive and rapid positive n (%) | % overall agreement | % agreement among positives | Kappa |
|---|---|---|---|---|---|---|---|
| Any | 24 (49.0) | 22 (44.9) | 26 (53.1) | 20 (40.8) | 87.8 | 76.9 | 0.755 |
| 6 or 11 | 1 (2.0) | 1 (2.0) | 1 (2.0) | 1 (2.0) | 100 | 100 | 1.000 |
| 16 | 12 (24.5) | 10 (20.4) | 12 (24.5) | 10 (20.4) | 95.9 | 83.3 | 0.883 |
| 18 | 2 (4.1) | 2 (4.1) | 3 (6.1) | 1 (2.0) | 95.9 | 33.3 | 0.479 |
| 26 or MM8 | 1 (2.0) | 3 (6.1) | 3 (6.1) | 1 (2.0) | 95.9 | 33.3 | 0.484 |
| 31 | 2 (4.1) | 2 (4.1) | 2 (4.1) | 2 (4.1) | 100 | 100 | 1.000 |
| 33 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 100 | ||
| 35 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 100 | ||
| 39 | 1 (2.0) | 1 (2.0) | 1 (2.0) | 1 (2.0) | 100 | 100 | 1.000 |
| 40 or 42 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 100 | ||
| 45 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 100 | ||
| 51 | 5 (10.2) | 2 (4.1) | 5 (10.2) | 2 (4.1) | 93.9 | 40.0 | 0.545 |
| 52 | 3 (6.1) | 0 (0.0) | 3 (6.1) | 0 (0.0) | 93.9 | 0.0 | |
| 53 | 3 (6.1) | 1 (2.0) | 3 (6.1) | 1 (2.0) | 95.9 | 33.3 | 0.484 |
| 54 | 2 (4.1) | 2 (4.1) | 3 (6.1) | 1 (2.0) | 95.9 | 33.3 | 0.479 |
| 55 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 100 | ||
| 56 | 2 (4.1) | 2 (4.1) | 2 (4.1) | 2 (4.1) | 100 | 100 | 1.000 |
| 57 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 100 | ||
| 58 | 2 (4.1) | 2 (4.1) | 2 (4.1) | 2 (4.1) | 100 | 100 | 1.000 |
| 59 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 100 | ||
| 66 | 3 (6.1) | 2 (4.1) | 3 (6.1) | 2 (4.1) | 98.0 | 66.7 | 0.790 |
| 68 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 100 | ||
| MM4 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 100 | ||
| MM7 | 2 (4.1) | 2 (4.1) | 2 (4.1) | 2 (4.1) | 100 | 100 | 1.000 |
| MM9 | 1 (2.0) | 0 (0.0) | 1 (2.0) | 0 (0.0) | 98.0 | 0.0 |
n = 49 for line blot results.
Assays of samples with discrepant results were repeated by line blot. Results by line blot were consistent after repeat analysis, with the exception of weak-positive signals, which were inconsistently amplified. To ascertain the possibility of irreproducibility due to sampling error from low concentrations of viral DNA, we added HPV 16 plasmid DNA to a PCR premix for a final concentration of 1.27 × 10−4 fg/μl, the equivalent of a single target per 100-μl reaction mixture. This mixture was aliquotted to 80 PCR tubes and amplified under sensitive amplification profiles. Analysis of the products by strip analysis indicated that only 42 of 80, or 52.5%, were positive for HPV DNA. Human DNA was included at a concentration of 2.5 ng per PCR and was amplified in all 80 reactions.
DISCUSSION
We compared our reformatted line blot system to the established dot blot assay to evaluate its performance. In general, the results from this comparison are highly concordant, both for overall HPV DNA detection and for genotype-specific discrimination. Most of the signals from the few discrepant samples were weak, suggesting low concentration of viral DNA, with disagreement likely attributable to sampling error and variable amplification of low levels of HPV DNA. This explanation is substantiated by the following observations. First, the design of the study required each laboratory to prepare, amplify, and detect each sample separately. This procedure creates at least three separate circumstances wherein subaliquots of each sample were transferred to a subsequent step in the protocol. The likelihood of each transfer containing equivalent concentrations of HPV DNA is low. Second, the discrepant results were evenly distributed between the two methods, indicating that neither method had a propensity toward false-negative or false-positive results. Third, we demonstrated in a controlled experiment that a homogeneous mixture of low-copy DNA yielded a positive result in only 52.5% (42 of 80) of the reactions tested. Based on these results, we attribute most discrepancies to random sampling error, except those for HPV types 51, 52, 54, and MM9. In these cases, the more-discordant detection rates were attributed to differences in type-specific amplification efficiencies among degenerate primer lots (data not shown).
We also evaluated the effect of amplification conditions on the low-end sensitivity of the assay by decreasing the time spent at each thermal cycling step in the amplification profile. The results confirm that the discrepancies predominate among the low-copy, or weak, positives, while all other results are consistent, independent of the profile used. These results reflect the inherent variability in sensitivity that results from seemingly minor changes in protocol. Thus, it is recommended that changes to standardized protocols be accompanied by revalidated assays and appropriately redefined performance criteria.
It has been clearly demonstrated that accurate measurement of even minute levels of HPV DNA is critical for a comprehensible evaluation of the natural history of HPV infection (7). Use of a nonamplified method can dramatically skew the strength and even the existence of important epidemiologic associations. Thus, amplification methods, including consensus, or general, primer PCR have been adopted for the majority of epidemiological studies (11, 20, 22). While the use of PCR methods can increase the molecular sensitivity of HPV DNA detection, the issue of misclassification remains (8, 9, 19). It is important not only to increase the sensitivity relative to detectable levels of virus but also to increase sensitivity by increasing the spectrum of HPV genotypes detected, a goal met by consensus primer PCR assays. However, previously described consensus PCR methods that utilized dot blot formats are too laborious to allow for the rapid evaluation of genotype-specific infections in large population studies. As a result, some investigators have combined genotyping probes into mixtures of presumably related HPV types in an attempt to reduce the number of hybridizations required. For example, HPV 18 and 45 are often combined, the HR HPV 50s are combined, the HR HPV 30s are combined, etc. Assumptions have to be made regarding the validity of these groupings. Groupings have historically been made according to anecdotal observations of disease relatedness and more recently according to phylogenetic relatedness. While these assumptions are reasonable for grouping related viruses, the information obtained from such groupings is inherently biased. Given that slight misclassification of HPV-type status may have dramatic effects on the interpretation of far-more-subtle associations, some of which are also prone to measurement error, it is clearly important to develop accurate discriminatory HPV-typing systems.
The PCR-based line blot HPV detection method described here allows sensitive amplification of a broad spectrum of HPV genotypes and accurate discrimination between 27 of those types. Subsequent to this report, our collaborative efforts have extended the HPV type-specific discrimination of this novel line blot to an additional 12 HPV types (data not shown). Thus, disease associations can be more accurately defined, since discrete and comprehensive genotyping information is available, without confounding due to potential misclassification. Also, since the central role of HPVs in the etiology of cervical cancer has been defined, natural history studies of HPV infection will now address more difficult issues, such as persistence, transmissibility, and immunologic responses. All these parameters are adequately studied only in an HPV type-specific or perhaps even in an HPV variant-specific manner.
In addition to natural history studies of HPV infection, the success of HPV DNA testing in patient management and cancer screening strategies is dependent upon the methodology used. The key to improving the current standard of Pap screening may be to significantly increase the specificity of cytology-based screening, without a concomitant decrease in sensitivity. It is believed that an HPV DNA testing method with both high-positive and negative predictive value will serve to increase both the sensitivity and specificity of cervical cancer screening. Furthermore, the amplification and detection protocols used with the line blot detection method are compatible with automation, facilitating the use of this method in large-scale studies or screening. The ability to visually categorize high- versus low-risk HPV infection rapidly by the line blot supports the use of a detailed and informative research assay for routine clinical screening and patient management purposes.
ACKNOWLEDGMENTS
This work was funded in part by a grant to C.M.W. from the National Institutes of Health (AI32917).
We thank William C. Hunt for performing the statistical analysis, Susan Eaton for excellent technical support, and the Roche Molecular Systems DNA synthesis group for oligonucleotide support.
REFERENCES
- 1.Bauer H M, Greer C E, Manos M M. Determination of genital human papillomavirus infection using consensus PCR. In: Herrington C S, McGee J O D, editors. Diagnostic molecular pathology: a practical approach. Oxford, United Kingdom: Oxford University Press; 1992. pp. 132–152. [Google Scholar]
- 2.Bosch F X, Munoz N, de Sanjose S, Navarro C, Moreo P, Ascunce N, Gonzalez L C, Tafur L, Gili M, Larranaga I, et al. Human papillomavirus and cervical intraepithelial neoplasia grade III/carcinoma in situ: a case-control study in Spain and Colombia. Cancer Epidemiol Biomarkers Prev. 1993;2:415–422. [PubMed] [Google Scholar]
- 3.Bosch F X, Manos M M, Munoz N, Sherman M, Jansen A M, Peto J, Schiffman M H, Moreno V, Kurman R, Shah K V the IBSCC Study Group. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 4.Bugawan T L, Apple R, Erlich H. A method for typing polymorphism at the HLA-A locus using PCR amplification and immobilized oligonucleotide probes. Tissue Antigens. 1994;44:137–147. doi: 10.1111/j.1399-0039.1994.tb02371.x. [DOI] [PubMed] [Google Scholar]
- 5.Cox J T, Lorincz A T, Schiffman M H, Sherman M E, Cullen A, Kurman R J. Human papillomavirus testing by hybrid capture appears to be useful in triaging women with a cytologic diagnosis of atypical squamous cells of undetermined significance. Am J Obstet Gynecol. 1995;172:946. doi: 10.1016/0002-9378(95)90026-8. [DOI] [PubMed] [Google Scholar]
- 6.Cuzick J, Szarewski A, Terry G, Ho L, Hanby A, Maddox P, Anderson M, Kocjan G, Steele S T, Guillebaud J. Human papillomavirus testing in primary cervical screening. Lancet. 1995;345:1533. doi: 10.1016/s0140-6736(95)91086-7. [DOI] [PubMed] [Google Scholar]
- 7.Franco E L. The sexually transmitted disease model for cervical cancer: incoherent epidemiologic findings and the role of misclassification of human papillomavirus infection. Epidemiology. 1991;2:98–106. [PubMed] [Google Scholar]
- 8.Franco E L. Measurement errors in epidemiological studies of human papillomavirus and cervical cancer. In: Munoz N, Bosch F X, Shah K J, Meheus A, editors. The epidemiology of human papillomavirus and cervical cancer. Oxford, United Kingdom: Oxford University Press; 1992. pp. 181–197. [PubMed] [Google Scholar]
- 9.Franco E L. Statistical issues in studies of human papillomavirus infection and cervical cancer. In: Franco E, Monsonego J, editors. New developments in cervical cancer screening and prevention. Oxford, United Kingdom: Blackwell Science Ltd.; 1997. pp. 39–50. [Google Scholar]
- 10.Hildesheim A, Schiffman M H, Gravitt P E, Glass A G, Greer C E, Zhang T, Scott D R, Rush B B, Lawler P, Sherman M, Kuman R J, Manos M M. Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis. 1994;169:235–240. doi: 10.1093/infdis/169.2.235. [DOI] [PubMed] [Google Scholar]
- 11.Ho G Y F, Burk R D, Klein S, Kadish A S, Chang C J, Palan P, Bassu J, Tachezy R, Lewis R, Romney S. Persistent genital human papillomavirus infection as a risk factor for persistent cervical dysplasia. J Natl Cancer Inst. 1995;87:1365–1371. doi: 10.1093/jnci/87.18.1365. [DOI] [PubMed] [Google Scholar]
- 12.International Agency for Research on Cancer. Monographs on the evaluation of the carcinogenic risk of chemicals to humans. 64. Human papillomaviruses. Lyon, France: International Agency for Research on Cancer; 1995. [Google Scholar]
- 13.Jacobs M V, Snijders P J F, van den Bruie A J C, Helmerhorst T J M, Meijer C J L M, Walboomers J M M. A general primer GP5+/6+-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J Clin Microbiol. 1997;35:791–795. doi: 10.1128/jcm.35.3.791-795.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koutsky L A, Holmes K K, Critchlow C W, Stevens C E, Paavonen J, Beckmann A M, DeRouen T A, Galloway D A, Vernon D, Kiviat N B. A cohort study of the risk of cervical intraepithelial neoplasia grade 2 or 3 in relation to papillomavirus infection. N Engl J Med. 1992;327:272–278. doi: 10.1056/NEJM199210293271804. [DOI] [PubMed] [Google Scholar]
- 15.Liaw K-L, Hsing A W, Chen C J, Schiffman M H, Zhang T Y, Hsieh C Y, Greer C E, You S L, Huang T W, Wu T C, et al. Human papillomavirus and cervical neoplasia: a case-control study in Taiwan. Int J Cancer. 1995;62:565–571. doi: 10.1002/ijc.2910620513. [DOI] [PubMed] [Google Scholar]
- 16.Manos M M, Waldman J, Zhang T Y, Greer C E, Eichinger G, Schiffman M H, Wheeler C M. Epidemiology and partial nucleotide sequence of four novel genital human papillomaviruses. J Infect Dis. 1994;170:1096–1099. doi: 10.1093/infdis/170.5.1096. [DOI] [PubMed] [Google Scholar]
- 17.Meijer C J L M, Rozendaal L, van der Linden J C, et al. Human papillomavirus testing for primary cervical screening. In: Franco E, Monsonego J, editors. New developments in cervical cancer screening and prevention. Oxford, United Kingdom: Blackwell Science Ltd.; 1997. pp. 338–347. [Google Scholar]
- 18.Mould T A, Singer A. Human papillomavirus testing for diagnostic triage of minor-grade cytological abnormalities: the European perspective. In: Franco E, Monsonego J, editors. New developments in cervical cancer screening and prevention. Oxford, United Kingdom: Blackwell Science Ltd.; 1997. pp. 354–359. [Google Scholar]
- 19.Qu W, Jiang G, Cruz Y, Chang C J, Ho G Y F, Klein R S, Burk R D. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J Clin Microbiol. 1997;35:1304–1310. doi: 10.1128/jcm.35.6.1304-1310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Remmink A J, Walboomers J M M, Helmerhorst T J M, Voorhorst F J, Rozendaal L, Risse E K J, Meijer C J L M, Kenemans P. The presence of persistent high-risk HPV genotypes in dysplastic cervical lesions is associated with progressive disease: natural history up to 36 months. Int J Cancer. 1995;61:306–311. doi: 10.1002/ijc.2910610305. [DOI] [PubMed] [Google Scholar]
- 21.Saiki R K, Walsh P S, Levenson C H, Erlich H A. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc Natl Acad Sci USA. 1989;86:6230–6234. doi: 10.1073/pnas.86.16.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schiffman M H. New epidemiology of human papillomavirus infection and cervical neoplasia. J Natl Cancer Inst. 1995;87:1345–1347. doi: 10.1093/jnci/87.18.1345. [DOI] [PubMed] [Google Scholar]
- 23.Schiffman M, Bauer H, Hoover R, Glass A G, Cadell D M, Rush B B, Scott D R, Sherman M E, Kurman R J, Wacholder S, Stanton C K, Manos M M. Epidemiologic evidence showing that HPV infection causes most cervical intraepithelial neoplasia. J Natl Cancer Inst. 1993;85:958–964. doi: 10.1093/jnci/85.12.958. [DOI] [PubMed] [Google Scholar]
- 24.Sherman M E, Schiffman M H, Lorincz A T, Manos M M, Scott D R, Kurman R J, Kiviat N B, Stoler M, Glass A G, Rush B B. Toward objective quality assurance in cervical cytopathology: correlation of cytopathologic diagnoses with detection of high-risk human papillomavirus types. Am J Clin Pathol. 1994;2:182–187. doi: 10.1093/ajcp/102.2.182. [DOI] [PubMed] [Google Scholar]