SUMMARY
Cysteine chemoproteomics provides proteome-wide portraits of the ligandability or potential “druggability” for thousands of cysteine residues. Consequently, these studies are facilitating resources for closing the druggability gap, namely, achieving pharmacological manipulation of ~96% of the human proteome that remains untargeted by U.S. Food and Drug Administration (FDA) approved small molecules. Recent interactive datasets have enabled users to interface more readily with cysteine chemoproteomics datasets. However, these resources remain limited to single studies and therefore do not provide a mechanism to perform cross-study analyses. Here we report CysDB as a curated community-wide repository of human cysteine chemoproteomics data derived from nine high-coverage studies. CysDB is publicly available at https://backuslab.shinyapps.io/cysdb/ and features measures of identification for 62,888 cysteines (24% of the cysteinome), as well as annotations of functionality, druggability, disease relevance, genetic variation, and structural features. Most importantly, we have designed CysDB to incorporate new datasets to further support the continued growth of the druggable cysteinome.
In brief
Boatner et al. report the development of the CysDB resource, which provides annotations of function, human genetics, structure, and disease relevance for 24% of all cysteines the human proteome.
Graphical abstract
INTRODUCTION
Small-molecule chemical probes are useful tools for modulating protein function that can serve as leads for future medications. Therefore, ongoing efforts in the chemical biology community haveset ambitious goals inmatchingevery protein witha chemical probe.3 Complicating matters, <4% of the human proteome has been pharmacologically targeted by U.S. Food and Drug Administration (FDA)-approved small molecules. Cysteine chemoproteomics has emerged as an enabling technology that addresses this druggability gap by identifying thousands of functional and potentially druggable cysteines proteome-wide.1–25 Demonstrating this utility, prior cysteine chemoproteomic studies, including our own, have revealed a strikingly low overlap between proteins containing “ligandable” or potentially “druggable” cysteines and those that have been targeted by FDA-approved molecules.11
Cysteine proteomics experiments can be generally classified into four main categories: (1) identification, (2) measuring hyperreactivity, (3) measuring ligandability, and (4) measuring redox state. We consider identification studies as those aiming to increase coverage of cysteine-containing peptides.4–6 Hyperreactivity experiments measure the intrinsic reactivity of cysteines toward highly electrophilic probes,7–10 while ligandability experiments measure the intrinsic ligandability or potential “druggability” of cysteines using libraries of drug-like electrophilic molecules, natural products, and lipid-derived electrophiles.2,11,15–19 Finally, redox protocols are tailored to identify redox-sensitive cysteines.1,20–23
Although the overarching objectives of these studies are non-redundant, they do share general features, including conceptually similar workflows and, most important, shared targets. In a standard cysteine chemoproteomics experiment for example, the proteome is treated with a pan-cysteine-reactive probe, followed by enrichment on streptavidin resin, sequence-specific proteolysis, and tandem liquid chromatography-mass spectrometry analysis (LC-MS/MS).
Despite considerable recent advances in instrumentation, sample preparation, and data analysis, most cysteine chemoproteomics studies only sample a small fraction of all cysteines in the proteome, with the highest coverage studies sampling ~13% of all cysteines.1,7,9 Reasons for this gap include protein abundance and restricted expression profiles, location of cysteines in very long or very short tryptic peptides, which are not detected in standard trypsin digests, and unreactive cysteines, such as those buried within protein cores or located in structural disulfides. Despite these technical limitations, the cysteinome continues to grow, with the addition of multiple high-coverage studies in 2022 alone.6,10,14
The availability of easily searchable cysteine databases— including Oximouse,1 the Ligandable Cysteine Database, and previously reported Cysteinome24—has increased the general accessibility of these large proteomics datasets, allowing rapid queries for targets of interest.9,12,13 However, except for the Cysteinome database, which was launched in 2016 and is no longer publicly accessible, these databases are restricted to datasets derived from single publications.
To facilitate future studies aimed at global or target focused analyses of the cysteinome, we envisioned the establishment of a unified cysteine-focused database that would fulfill the following criteria. First, the database would incorporate datasets from many large-scale cysteinomic studies and therefore enable rapid and facile inter- and intra-dataset comparisons. Second, the database would include information about the reactivity and ligandability of cysteines together with the druggability of their corresponding proteins, as indicated by availability of FDA-approved drugs. Last, and most significant, the database would integrate functional and structural data from UniProtKB/Swiss-Prot, Cancer Gene Census (CGC), ClinVar, Human Protein Atlas (HPA), ChEMBL, DrugBank, and the Protein Data Bank (PDB),26–32 to enable prioritization of targets for future studies. Here we present CysDB, which is an interactive database that fulfills these criteria for 62,888 cysteines and 11,621 proteins. Importantly, to promote the continued growth of cysteine chemoproteomics, we also provide a straightforward route for addition of future datasets.
RESULTS
Data curation to establish a set of processed and aggregated chemoproteomics datasets to enable CysDB
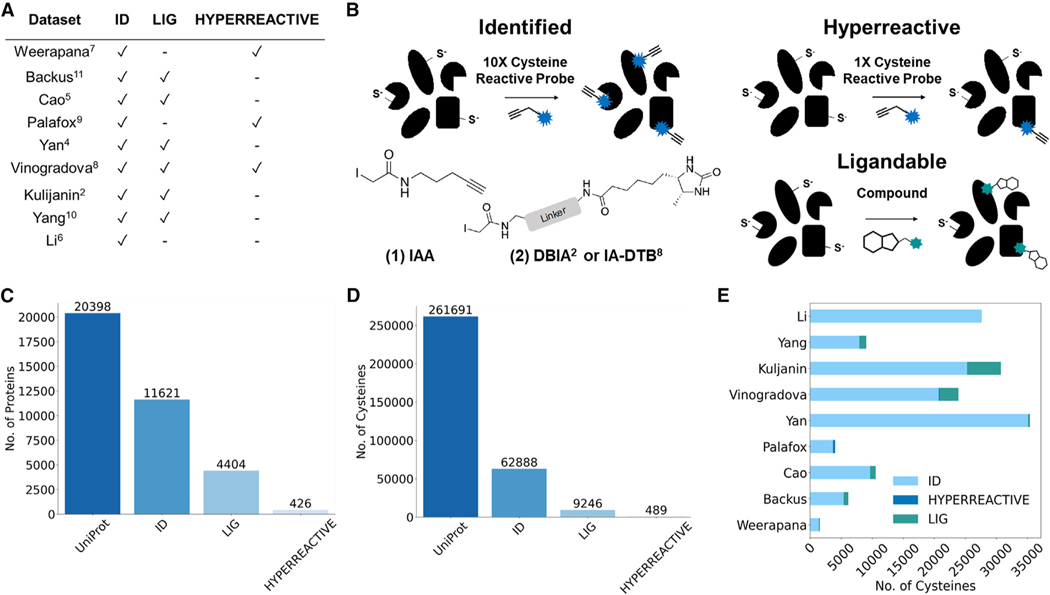
Our first step toward creating CysDB was to assemble a set of publicly available datasets. With the overarching goal of establishing a high coverage and highly curated database of human chemoproteomics studies to enable cross-dataset exploration, we opted to focus on a reduced set of available datasets. We prioritized studies that reported high-coverage datasets that measured one or more of the following parameters: (1) total number of cysteines identifiable by pan-cysteine-reactive probes, (2) measurement of cysteine intrinsic reactivity toward iodoacetamide alkyne (iodoacetamide alkyne [IAA, 1]; Figures 1A and S1), and (3) assaying cysteine ligandability (Figures 1A and S2). In total, we collected nine datasets that fulfilled our criteria (Figure 1B for all datasets used).2,4–11
Figure 1. Dataset selection and curation for the creation of CysDB.
(A) Table of datasets used as input for CysDB, including which datasets were used in each chemoproteomic category (identified, hyperreactive, and li-gandable).2,4–11
(B) General workflows for three categories of chemoproteomic methods included in CysDB that use iodoacetamide alkyne (IAA, 1) or an iodoacetamide desthiobiotin reagent (DBIA2 or IA-DTB8, 2) to capture cysteines for (1) high-coverage identification of cysteine-containing peptides, (2) quantitative profiling of intrinsic cysteine reactivity, and (3) assaying cysteine ligandability using an electrophile of interest.
(C and D) Quantification of the unique proteins (C) and cysteines (D) found in the Human UniProtKB/Swiss-Prot database, together with the identified, ligandable, and hyperreactive chemoproteomics subsets in CysDB.
(E) Study-specific breakdown of total number of unique cysteines, including those that are identified as hyperreactive and ligandable.
See also Figures S1–S5 and Data S1.
Notably, all these studies rely on the same general cysteine chemoproteomic workflow: cells or lysates are treated with a cysteine-reactive probe (Figure 1A; iodoacetamide alkyne) or an iodoacetamide desthiobiotin reagent (e.g., DBIA2 or IA-DTB8) to cap all accessible cysteines. Labeled proteins are subjected to enrichment on streptavidin or related resins together with sequence-specific proteolysis, followed by liquid chromatography-tandem mass spectrometry. Several of our included studies7–9 further classify cysteine intrinsic reactivity and pinpoint hyperreactive cysteines by comparing relative cysteine labeling by two concentrations (10× and 1×) of a cysteine enrichment handle (Figures 1A and S1). Signal intensity differences between 100 and 10 μM treated proteomes are reflected by a ratio (R[high]/[low]). Hyperreactive cysteines are defined as those with R10:1 values <2, indicating labeling events that are not concentration dependent. Most included studies provide a metric of cysteine ligandability or putative druggability,2,4,5,8,10,11 which is generated by comparing relative labeling by equimolar iodoacetamide in the presence and absence of electrophilic compound, with decreased labeling indicative of a high-occupancy labeling event (Figures 1A and S2).
To produce a rigorously curated database, we subjected our prioritized datasets to a series of data-processing steps. First, we aggregated all non-redundant cysteines published by all studies, using the unique identifier UniProtKBID_CYS#. For some studies2,4–9,11 residue positions and protein identifiers were provided in the supporting information. For a subset of studies, the supporting tables instead provided labeled peptide sequences and protein IDs.7,10 To merge these two data types, we mapped each peptide to the corresponding canonical protein sequence using the UniProtKB reference FASTA from January 2022; this approach recovered nearly all cysteines, with only 37 dropped because of mismapping (Data S1), likely caused by differences in UniProtKB releases used in dataset search, as observed in our prior study.9 In the event of proteomic analyses comparing cysteine labeling using different experimental conditions (e.g., unstimulated versus stimulated cells), we opted to incorporate only the datasets derived from control (no treatment) conditions, with the goal of limiting the potential impact of cell state-dependent differences of cysteine reactivity as a potential confounder to our downstream analyses. To address the many additional parameters, including data analysis pipeline differences, cysteines with incorrect residue numbers and peptides that match to multiple protein sequences (2,823 entries), we include the UniProtKB release and software used to process mass spectrometry data for each dataset in Data S1.7,18,33–38 Aggregation of all datasets, including results from using multiple cell lines,2,4–11 resulted in the chemoproteomic identification of 62,888 unique cysteines and 11,621 proteins (Figures 1C and 1D), which to our knowledge represents the most comprehensive cysteinome dataset reported to date.
Using the studies reporting measures of cysteine ligandability or labeling by electrophilic fragments or drug-like molecules, we further stratified our dataset to generate a master set of all ligandable cysteines. The datasets included in our database (Figure 1A) were all prepared using the same general workflow where samples (lysates or cells) were treated by either a vehicle (DMSO) or a cysteine-reactive electrophile functionalized compound and the compound-dependent changes in IAA, DBIA, or IA-DTB reactivity assayed using LC-MS/MS analysis. Prior analyses have revealed that comparable competition ratios can be calculated using either MS1 or MS2 level quantification.2,4,5,8,10,11 Therefore, we opted not to differentiate between samples analyzed using different quantification methods, including isotopic labeling strategy (TMT or isotopically enriched biotinylation reagents),2,6 label-free quantification, and data-independent acquisition (DIA) based MS2 level quantification (see Figure S2 for general workflow).4,8,10 The vast majority (97.2%) of all compounds screened were found to be functionalized with either a chloroacetamide or acrylamide moieties (Figure S3). A small data subset of compounds did, however, feature alternative electrophiles, including covalent reversible cyanoacrylamides,38 fumarates, and activated esters; although activated esters are primarily lysine reactive, our prior data indicate that they do also exhibit cysteine reactivity.40,41
All datasets included in our database relied on competition ratio cutoffs for what defines a cysteine as “ligandable.” Generally, cysteines were categorized as liganded if they had at least two ratios R ≥ 4 (hit fragments) and one ratio between 0.5 and 2 (control fragments). However, when processing the ligandability data for each dataset, we observed manuscript-specific differences in either the ratio cutoff value or number of minimum unique hit fragments (1 or 2) required to have the associated ratio cutoff value for designating a cysteine as ligandable. For example, Cao et al.5 implemented a slightly more permissive ratio cutoff of 3 to account for high-field asymmetric waveform ion mobility spectrometry (FAIMS)-induced ratio compression. By comparison, Vinogradova et al.8 implemented a more stringent ratio cutoff of 5. Another case we encountered was the inclusion of “ligandable” cysteines where the unique identifier contained multiple modified cysteine residues, such as UniProtKBID_CYS#1_CYS#2. These types of identifiers are derived from peptide sequences simultaneously labeled with capture reagents at multiple cysteine residues (C1*XXXC5*) within the same sequence. On the basis of our experience with such peptides yielding noisy ratios, we opted to remove them from CysDB; a total of 2,584 peptides were excluded because of this criterion. Otherwise, despite the differences in defining ligandability, we opted to retain all remaining liganded cysteines to accurately represent each study’s reported findings (the criteria for ligandability applied to each study are available in Data S1). In aggregate across all ligandability studies, a total of 43,475 unique cysteines (Data S2) had quantified ratios, and 9,246 unique cysteines were deemed ligandable. These cysteines were found in 4,404 proteins (Figures 1C and 1D).
Next, we parsed processed data from published datasets measuring cysteine hyperreactivity.7–9 The three hyperreactivity studies included in CysDB measured the relative IAA reactivity toward two concentrations of IAA (100 and 10 μM), where a quantitative isoTOP-ABPP ratio (R[high]/[low]) reflects the differences in signal intensities between the 100 and 10 μM treated proteomes. Highly reactive cysteines, termed “hyperreactive” residues, are identified as those that exhibit saturation or near saturation of labeling at the lower IAA concentration. All three publications used the same numerical ranges to delineate cysteines into “high,” “medium,” and “low” reactivity subsets, with high-reactivity, also termed “hyperreactive,” residues as those with R10:1 < 2, medium-reactivity cysteines between R100:10 ≥ 2 and R10:1 < 5, and low-reactivity cysteines R10:1 > 5. During dataset processing, we observed that Weerapana et al.7 and Palafox et al.9 reported median values of all the replicates for each individual measure of cysteine reactivity, as well as an overall mean of medians to quantify the average reactivity per cysteine. In contrast, Vinogradova et al.8 reported the average of medians across all measurements. To accommodate these dataset dependent differences, we opted to report the mean of median ratio values for each detected cysteine. In aggregate, 8,604 cysteines on 4,032 proteins were quantified by these three studies, which resulted in identification of 489 hyperreactive cysteines and 426 proteins containing hyperreactive cysteines (Figures 1C and 1D).
Collectively across all cysteines identified through our data aggregation efforts, 14.7% were deemed ligandable, and fewer than 1% were determined to be hyperreactive. Cross-dataset comparisons reveal the highest overall coverage dataset was reported by Yan et al.4 (Figures 1E and S4), where an optimized SP3-FAIMS strategy was applied to analyze the proteomes of seven cell lines, which in aggregate identified more than 34,000 cysteines on 9,714 proteins from 7 cell lines (Figures S4 and S5). A key outcome of the dataset aggregation required to build CysDB is an effective doubling of the size of the identified cysteinome. Collectively across all studies analyzed in CysDB, ~24% of all cysteines found on 57% of human proteins in UniProtKB have been assayed at least once by chemoproteomics (Figures 1C and 1D).
Establishing an SQL database with an RShiny user interface for CysDB
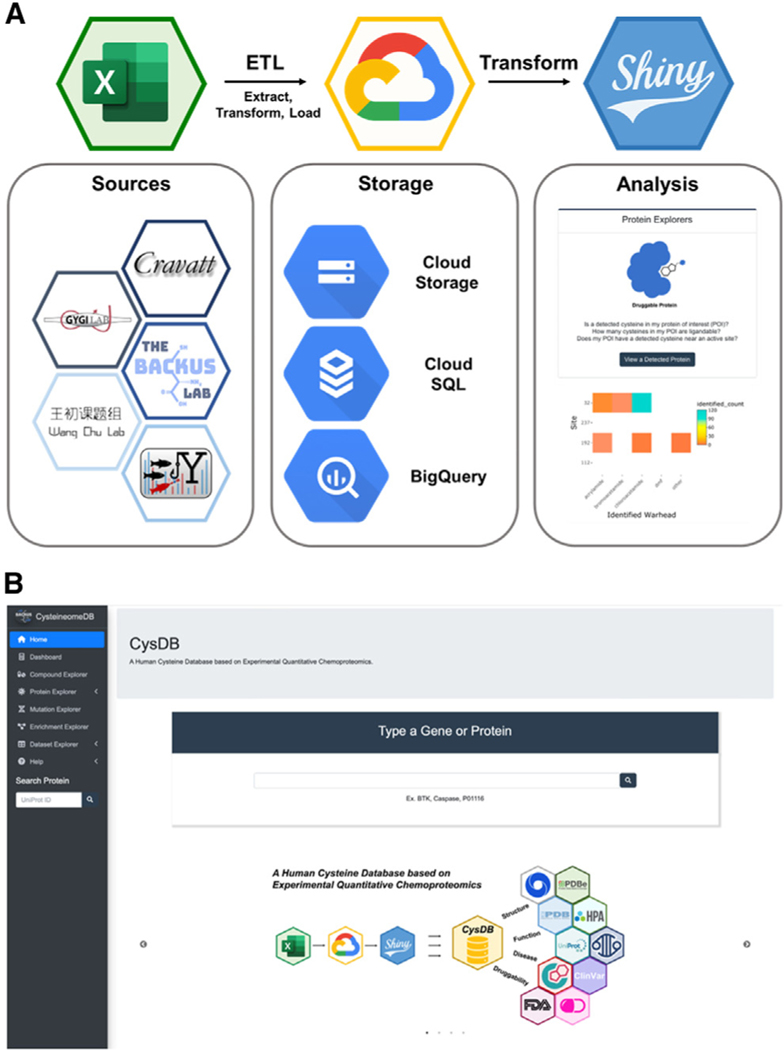
With a complete, curated dataset in hand, we constructed the CysDB database and web user interface outlined in Figure 2A. Processed data from prioritized studies (Data S1)2,4–11 were prepared into a standardized input format for SQL integration (see Data S1 for example data format and required information for future data integration to CysDB) and loaded into a database hosted in Google Cloud using MySQL version 8.0 (see STAR Methods for more details on data preparation and processing). CysDB is a relational database composed of six individual tables (Figure S6). For public accessibility of CysDB, we developed a front-end user interface powered by the Shiny framework (Figure 2B). Shiny converts queries from remote users into visualizations and results that are displayed on a web browser. Not only does our web application access the Cloud CysDB, but it additionally calls from both structural and functional external databases, including UniProtKB, COSMIC, ClinVar, and PDB.26–29,32
Figure 2. Workflow to generate CysDB SQL database.
(A) Data extracted from nine datasets (Data S1) was transformed and loaded into a MySQL relational database on the Google Cloud Platform. An accompanying front-end web interface was developed using RShiny to allow remote-user querying of the SQL database.
(B) Homepage of the CysDB app publicly available at https://backuslab.shinyapps.io/cysdb/.
One challenge we faced during our data processing, was one-to-one mapping of protein accessions to gene names for SQL querying. For gene-centric queries, not all HUGO Gene Nomenclature Committee (HGNC)42 or Entrez gene symbols are associated with a single protein. Gene sequences translated to the same protein sequence can lead to multi-mapping of various gene names to one UniProtKB accession.9 In CysDB, we found that 16 UniProtKB entries were associated with multiple gene names (Data S1; STAR Methods). To address this limitation, we included the capability to search entries using gene symbols or protein names. The user then selects one of the resulting UniProtKB accessions for CysDB search. The CysDB RShiny interface enables the user to interact with cysteine chemoproteomics datasets, generate personalized figures, and download their results. Anywhere in the app, a user can save graphs as an image by clicking on a camera button at the top right corner and export query results to a CSV (comma-separated value) file by clicking a download button at the bottom of a table. The CysDB app includes five sections: Protein, Mutation, Enrichment, Compound, Statistics, and Datasets.
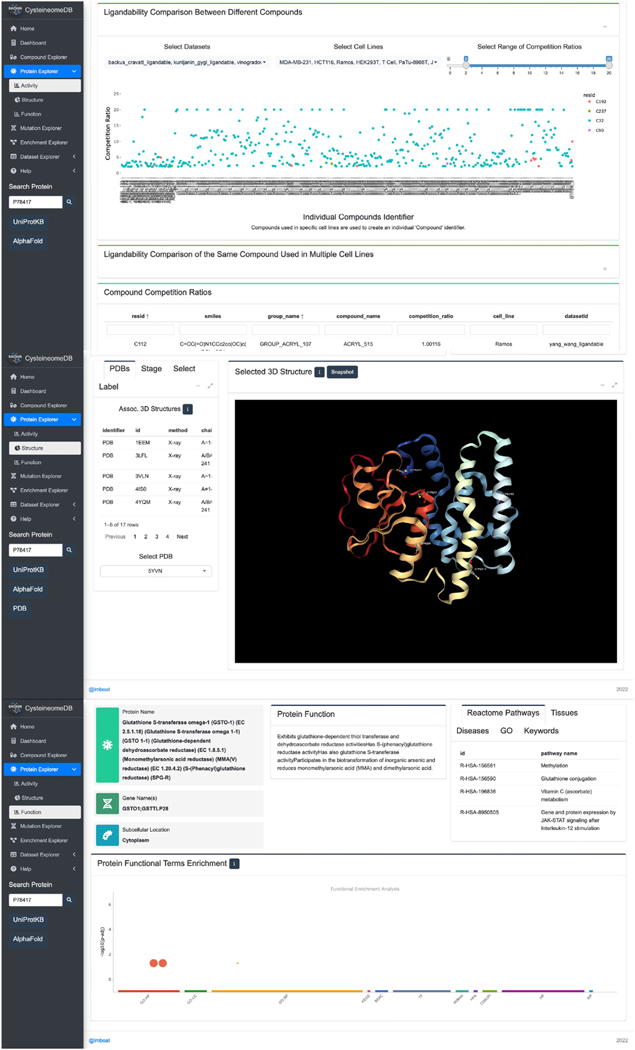
First, users can visualize the CysDB data in a protein-centric manner by selecting the protein explorer button, which is found on the homepage (Figure 3). Searching for a protein of interest (POI) by querying a UniProtKB ID returns the “Protein Section,” which is further broken up into three separate tabs detailing activity, structure, and function. The activity tab provides a “site map” indicating whether any cysteines in the POI are hyperreactive or ligandable together with the measured reactivity, measured competition ratios and the structures of all compounds that ligand the POI. The structure tab provides the user with annotations of proximal active site and binding site residues in both linear sequence and three-dimensional space and an easily accessible mechanism to visualize the three-dimensional protein microenvironment of chemoproteomic detected cysteines, including for structures reported in the PDB. Last, the function tab reports functional annotations for the POI generated from UniProtKB, Gene Ontology (GO), and Reactome.26,43,44
Figure 3. CysDB enables protein-centric queries.
Users can search for a protein of interest (POI) in the search bar on the protein page. Centered on the activity tab is a “site map,” indicating which cysteines have been identified, liganded, or hyperreactive by chemoproteomics. In addition, the activity tab allows users to assess the potential druggability of their POIs through scatterplots and heatmaps for quantitative chemoproteomic measurements. For a comprehensive view of the structural environment surrounding the chemoproteomic detected cysteines, publicly available 3D crystal structures are displayed in the structure tab. Users can choose which structure is shown and add customized labels. By clicking the function tab, one can view general information on the POI, including subcellular locations, functional pathways, and GO/Kyoto Encyclopedia of Genes and Genomes (KEGG) terms.
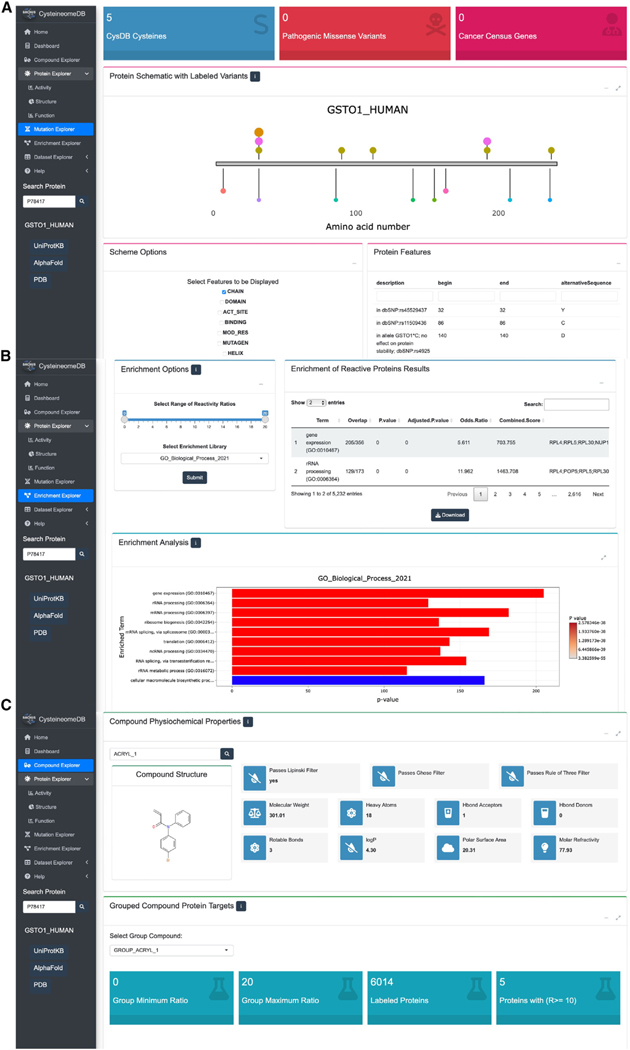
The “Mutation” section of CysDB, provides information complementary to that presented in the “Protein Explorer” section. Querying for a POI yields the aggregate number of CysDB cysteines, missense variants identified in ClinVar,28 the public repository of relationships between human genetic variation and phenotype, and CGC genes mapped to the POI. This page also generates a one-dimensional depiction of the corresponding protein sequence, decorated with the positions of ligandable and hyperreactive CysDB cysteines alongside individual missense variants, sequence elements, and known ligand binding sites (Figure 4A). To further enable pinpointing of cysteines relevant to human health, CysDB also provides CGC annotations of tumor types associated with POI, where relevant.
Figure 4. CysD enables disease, dataset, and cysteine-reactive compound wise queries.
(A) The disease relevance of a POI can be explored through the mutation page. Proximity of chemoproteomic detected cysteines, annotated small-molecule binders and variants of ranging clinical significance are visualized on a one-dimensional schematic of a protein sequence. Chemoproteomic cysteines are colored in gold for identified, pink for ligandable and orange for hyperreactive, while the remaining points are variant positions.
(B) Users can specify subsets of data available in CysDB, such as by compound chemotype or ranges of reactivity ratio, for pathway, ontology, and disease enrichment analyses. The results can then be downloaded as a CSV-formatted table or a bar graph as an image.
(C) Chemical structures and calculated “drug-likeness” properties of compounds used to ligand cysteines in CysDB can be accessed from the dropdown menu in the compound page.
Looking beyond individual POIs, the “Enrichment” section of CysDB was built for facile visualization and analysis of aggregated ligandable and hyperreactive CysDB subsets. Global analyses provided powered by the Enrichr package include functional pathways, ontologies, and disease enrichments of CysDB categories (Figure 4B).45,46 As with the dataset-wide meta-analysis provided by the Enrichment section, the “Compound” section of CysDB provides users with a global perspective of the electrophilic compounds employed in the CysDB cysteine ligandability studies. This portion of CysDB includes details of each molecule used in the ligandability experiments, including the publication name of each compound, corresponding CysDB names for each corresponding compound and dataset in an easily downloadable table.
For the “Compound” section, results can be searched on the basis of SMILES strings or newly created identifiers, defined by a unique combinations of SMILES strings, cell lines, and publication authors. Consistent with previous studies,47 we found that the molecular connectivity for a single two-dimensional (2D) chemical structure could be written in various forms (e.g., ethanol can be denoted as C(O)C, as well as CCO). Thus, we transformed the SMILES strings extracted from each publication into 2D chemical structures and converted these 2D chemical structures into new SMILES strings using RDKit. Selection of a compound identifier using the provided drop down menus, affords a two-dimensional rendering of the chemical structure and computed properties of “drug-likeness,” including the number of hydrogen bond donors and acceptors (Figure 4C).48–53 For this section, we created two separate CysDB compound identifiers to produce scatterplots showing the highest ratios collected for each compound.
The final “Statistics” section is accessible from the homepage both via the chemoproteomics explorer button and from the left menu. The Statistics section provides interested users with CysDB-wide metrics for hyperreactive and ligandable cysteine-containing proteins, proteins targeted by FDA-approved drugs, proteins associated with cancer, and proteins containing missense variants. In a user-centric manner, this section also allows interested users to compare individual datasets including by identification of unique and overlapping residues and proteins.
Understanding the scope of the CysDB ligandable or putative “druggable” proteome
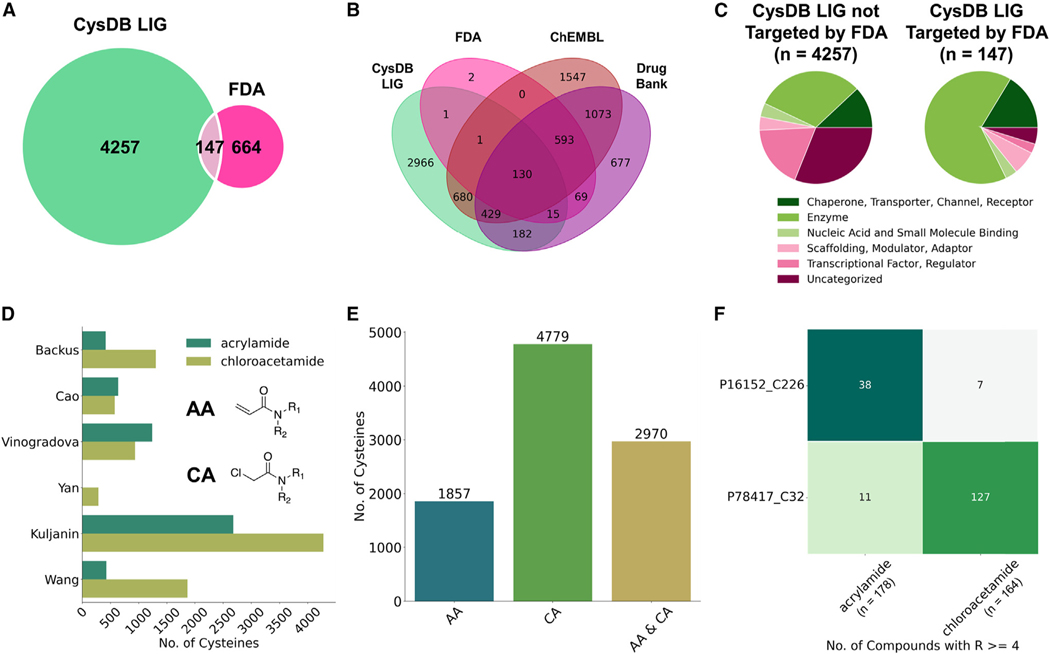
We further parsed the data available in CysDB to showcase features built into CysDB and to facilitate the identification of new potential targets for future chemical probe development campaigns. More broadly, we also seek to highlight future opportunities for the cysteine chemoproteomic community. Given the low overlap between FDA-approved drug targets and proteins labeled by cysteine-reactive compounds for prior smaller cysteine chemoproteomics studies,11 we next extended this analysis to CysDB. Fewer than 4% of all human proteins in UniProtKB have been targeted by FDA-approved small molecules (Figure S7). As only 14.7% of all cysteines in CysDB were reported as likely ligandable, we next performed the same analysis on the subset of proteins in CysDB that contain a ligandable cysteine. Again, consistent with the prior reports that have demonstrated a low overlap between targets of covalent compounds and FDA-approved drugs, we find that 3% of proteins that contain one or more ligandable cysteine have been targeted by FDA-approved drugs (Figure 5A). Broadening this analysis to a less restrictive set of compound-protein interactions, we find that 32.5% of proteins with ligandable cysteines have been targeted by small-molecules, as reported by ChEMBL, DrugBank, and the FDA (Figure 5B). These findings showcase the opportunities for targeting undrugged proteins using cysteine-reactive chemical probes.
Figure 5. Cysteines with available ligandability data.
(A) Overlap between CysDB ligandable (LIG) proteins and proteins targeted by FDA-approved drugs.
(B) Overlap between CysDB LIG proteins, proteins targeted by FDA-approved drugs, small molecules in DrugBank and ChEMBL.
(C) Distributions of protein functions for CysDB LIG proteins not targeted by FDA and CysDB LIG proteins targeted by FDA.
(D) Grouped bar graph showing the number of unique ligandable cysteines targeted by acrylamides or chloroacetamide for each dataset (R ≥ 4 for at least one compound).
(E) Bar graph of the overall number of unique cysteines targeted by acrylamides or chloroacetamide.
(F) Number of unique SMILES strings with an acrylamide and chloroacetamide moiety (on the basis of the “group compound identifier”), compounds with ratios ≥ 4 for protein carbonyl reductase (CBR1; UniProtKB: P16512) and protein glutathione s-transferase omega-1 (GSTO1; UniProtKB: P78417).
See also Figures S7–S10 and Data S2.
Prior studies have shown that drug and putative drug targets are highly enriched for protein classes featuring well-defined binding sites, including enzymes and receptors. Therefore, we characterized whether the CysDB members represent new druggable space by parsing UniProtKB keyword functional annotations of all ligandable proteins in CysDB. Stratification of the CysDB ligandable proteins into two categories, targeted and untargeted by FDA-approved compounds, acknowledged an enrichment for enzymes in the FDA-approved subset (Figure 5C). In contrast, the functions of the non-FDA subset of ligandable proteins in CysDB span important protein classes, including transcription factors (TFs), which are often categorized as a largely “undruggable” class of proteins, with the notable exception of TFs with well-defined small molecule ligand binding pockets, such as nuclear hormone receptors.
Next, we analyzed the compounds that target ligandable cysteine residues to further dissect the potential druggability of CysDB entries. Several different electrophilic moieties, often termed “warheads,” have been developed, which react with cysteine residues in both irreversible and covalent reversible modes of labeling.39,54–56 Examples of these electrophilic handles include compounds that react via a thiol-Michael addition (e.g., irreversible modifiers such as acrylamide, fumarate esters, vinyl sulfonamide together with reversible modifiers such as cyanoacrylamide), compounds that react via SN2 (e.g., alpha-halo compounds), as well as compounds that react via SNAr (e.g., halogen-substituted electron deficient heterocycles such as chlorotriazine). As prior studies have revealed varying proteome-wide reactivity and structure-activity relationships (SARs) for different cysteine-reactive electrophiles, we decided to quantify the number of cysteines detected as labeled by individual electrophile chemotypes. For this analysis, a cysteine was labeled by one of the five warheads if the cysteine had R ≥ 4 for at least one compound (Figures 5D, S8, and S9).2,27–62 Overall, we found that a large majority of the ligandability data were acquired for samples subjected to labeling by acrylamides (AAs) and chloroacetamide (CA)-substituted compounds across the panel of cell lines tested (Figures 5D and S10), with a small fraction derived from additional probes ranging from cyanoacry-lamides to dimethylfumarate listed in Data S2. Interestingly, we noticed that some cysteines react promiscuously with both AA and CA electrophiles, whereas others show an electrophile preference (Figure 5E). The proteins glutathione S-transferase omega-1 (GSTO1) and carbonyl reductase (CBR1) exemplify the striking electrophile preference observed for some proteins (Figure 5F). For GSTO1, the highly ligandable cysteine (Cys 32) exhibits strong preference for reacting with chloroacetamide-substituted compounds (1 to 11.5 in favor of CA electrophiles, with respect to unique SMILES strings with the CA moiety). In contrast, cysteine 226 of CBR1 shows marked acrylamide bias (5 to 1 in preference of AA warheads, with respect to unique SMILES strings with the AA moiety).
Characterizing CysDB proteins on the basis of structural, activity, and functional annotations
Given the sheer scope of available chemoproteomics datasets, one of the foremost ongoing challenges with cysteine chemoproteomic studies is delineating the functional impact of covalent cysteine modification in a high throughput manner. Although for some cysteines, such as catalytic nucleophiles, covalent modification will almost invariably afford a defined functional outcome, the impact of modifying other less well annotated cysteines, such as those in proteins or protein domains of unknown function, remains less clear. To encourage discovery of likely functional and disease-relevant cysteines, CysDB includes metrics of functionality from UniProtKB, known CGC, and genetic variants in ClinVar. These databases were chosen to provide measures of relevance to functional biology and human disease.
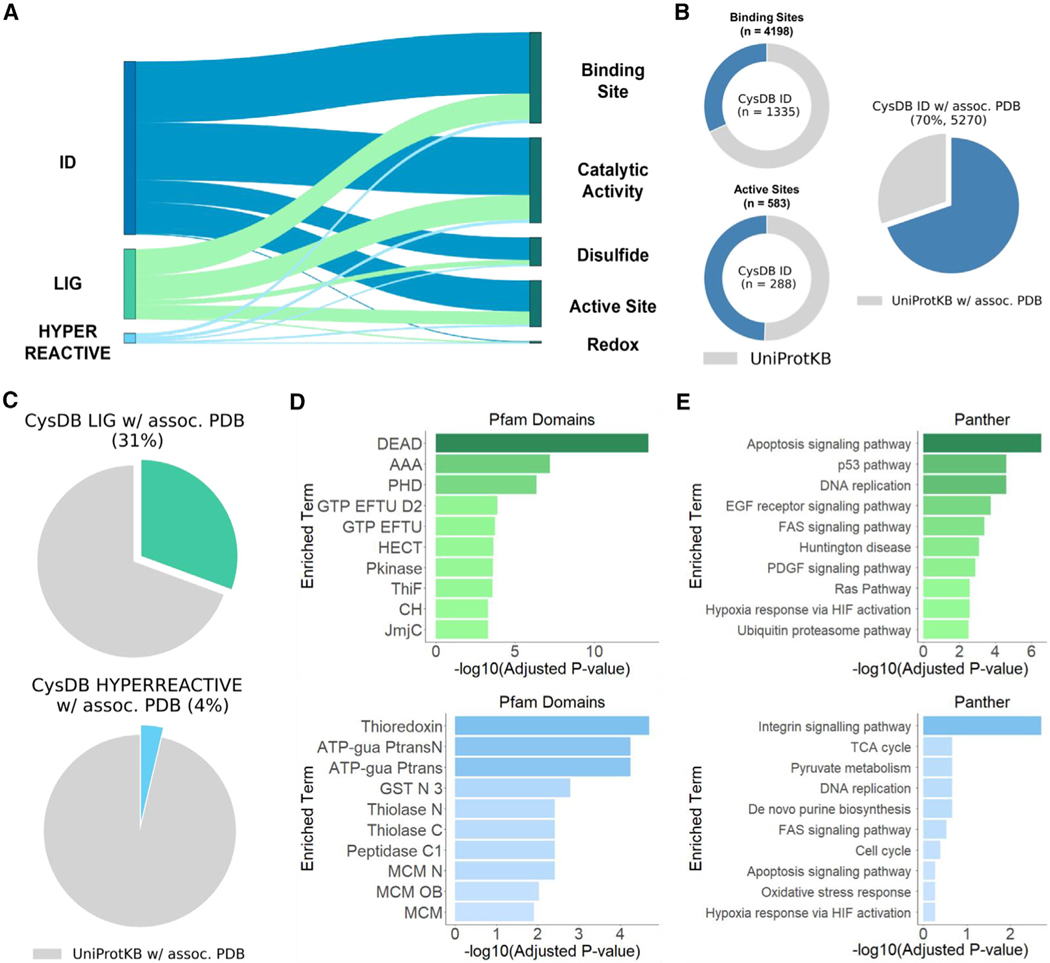
We first harnessed UniProtKB annotations to determine which CysDB proteins had functional annotations of the following active sites, binding sites, catalytic activity, disulfide bonds, and redox potentials. Analysis concluded 1,505 CysDB proteins possess an active site, 2,961 possess a binding site, 2,784 have experimental evidence for catalytic activity, 1,077 have annotated disulfide bonds and 52 have experimental evidence for redox potentials (Figure 6A). Comparable distribution of functional annotations was observed when stratifying the CysDB dataset to consider hyperreactive and ligandable proteins.
Figure 6. Cysteines with available functional and structural annotations.
(A) CysDB-identified, ligandable, and hyperreactive proteins with annotated active sites, binding sites, catalytic activity, disulfide bonds, and redox potentials.
(B) Distribution of identified cysteines in CysDB ID annotated as cysteine-specific binding sites or active sites (left). The total number of cysteines in UniProtKB annotated as binding or active sites are shown in gray. Percentage of proteins associated with a PDB structure and containing an identified cysteine.
(C) Percentage of proteins associated with a PDB structure and containing a ligandable (CysDB LIG) or hyperreactive (CysDB HYPERREACTIVE) cysteine.
(D) Top 10 enriched protein domains from Pfam term enrichment analysis of liganded (green) and hyperreactive (light blue) proteins.
(E) Top 10 enriched pathways from Panther term enrichment analysis of liganded (green) and hyperreactive (light blue) proteins.
See also Figures S11–S20 and Data S3.
To assess whether any CysDB cysteines were annotated as known active or binding sites, we parsed the UniProtKB site annotations for residue positions. This analysis uncovered that, while cysteine is a relatively rare amino acid (2.3% of all proteinacious amino acids are cysteines1), cysteine is the second most abundant binding site amino acid and the third most abundant active site amino acid (Figures S11 and S12). Overall, CysDB reports identification of 1,335 (31.8%) of all known cysteine matching UniProtKB annotated binding sites and 288 (49%) of all known cysteine-active sites (Figure 6B). Of the 4,198 cysteine specific binding sites, 178 of them have been liganded by a compound in CysDB. In addition, 98 out of the 583 cysteine-active sites have been liganded by a compound in CysDB and 41 out of the 583 cysteine-active sites were deemed hyperreactive (Figure S13).
Extending this analysis to look for cysteines “in or near” annotated active or binding sites using protein sequences, we searched 10 amino acids upstream and downstream of a CysDB-identified cysteine. Using this method, we were able to increase the number of cysteines proximal to these functional sites. In total, 2,602 CysDB cysteines are near binding sites, including 396 ligandable and 41 hyperreactive CysDB cysteines (Figure S14), and 496 CysDB cysteines are near active sites, including 56 ligandable and 12 hyperreactive cysteines (Figure S15).
As the UniProtKB dataset is limited to 1D analysis, we asked whether CysDB could also provide insight into the three-dimensional (3D) microenvironment of identified cysteines, using structures reported in the PDB. In total, 5,270 CysDB ID proteins are associated with an available PDB structure, which represents 70% of all human genes with available crystallographic structures (Figure S16). Of these, 2,314 (31%) contain one or more ligandable cysteines and 279 feature at least one hyperreactive cysteine (Figure 6C). To confirm whether a CysDB cysteine was resolved in a PDB structure, we parsed the residue numbers and coordinates from PDB files. To account for discrepancies between UniProtKB and PDB residue numbers, residue to protein sequence numbering was mapped using SIFT annotations63 (Figure S16). This systematic analysis of residue-level mapping established that out of all the proteins with annotated binding or active sites, 2,684 and 1,315 proteins, respectively, are associated with PDB structures (Figures S17 and S18; STAR Methods). Of these, 1,007 proteins have cysteine-binding sites resolved in a corresponding structure, while 338 proteins have cysteine-active sites resolved in a corresponding structure. In aggregate, 18,959 (30.1%) of CysDB-identified cysteines are resolved in a corresponding crystal structure. Further inspection of this dataset revealed that 1,212 CysDB cysteines are proximal (within 10 Å) to binding site residues and 704 CysDB cysteines are proximal to active site residues in 3D space (Figures S19 and S20; STAR Methods). To assist structure-guided analysis of cysteine datasets, CysDB provides users with 3D interactive renderings of cysteine-containing structures that include known functional annotations.
Notably, 8,214 proteins (71%) identified by chemoproteomics do not have highly supported evidence in UniProtKB for binding or active sites. Therefore, we next asked whether the CysDB platform could provide additional information about these proteins and corresponding identified cysteines to further aid in delineation of functionally significant cysteines. To guide our platform development efforts, we tested whether the ligandable and hyperreactive cysteine-containing protein subsets are enriched for particular structural domains and functional pathways. Enrichment analysis ofprotein family(Pfam)64 domains elucidateda 13-fold enrichment of liganded proteins in the DEAD/DEAH box helicase family, which is consistent with our prior observation of enrichment for RNA binding proteins in chemoproteomics datasets (Figure 6D).65 Responsible for unwinding the duplex of double-stranded RNA, mutations in DEAD/DEAH proteins have been linked to autoimmune disease and some cancers, such as DEAD-box helicase 3 X-linked (DDX3X) in medulloblastoma.66–69 Pfam domain enrichment analysis for the hyperreactive cysteine subset, revealed an enrichment of thioredoxin and arginine kinase families. These findings are consistent with prior reports of redox enzymes featuring highly reactive cysteines.7 Notably, creatine kinase enzymes are members of the arginine kinase family of enzymes, which are known to have highly reactive active site cysteines.7
We then extended these studies to Panther70 pathway analysis to assess if pathways are enriched for reactive or ligandable cysteines. We observe an enrichment of ligandable cysteine-containing proteins implicated in apoptosis (Figure 6E). Examples of ligandable cysteine-containing proteins include TP53, caspase-8, and APBB2. Given the central relevance in modulating cell death to treat numerous disorders, including cancers and neurodegenerative disorders, we expect that this observed notable enrichment indicates untapped opportunities for the development of probes targeting cell death.71,72 The hyperreactive cysteine-containing protein set, by contrast, was distinctly enriched for proteins involved in integrin signaling. These findings are consistent with the enrichment for hyperreactive cysteines in thioredoxin proteins and related antioxidant systems that are critical for regulation of integrin abundance, secretion, and disulfide formation.73,74
Stratifying CysDB proteins on the basis of disease-relevant annotations, including cancer association and measures of genetic variation
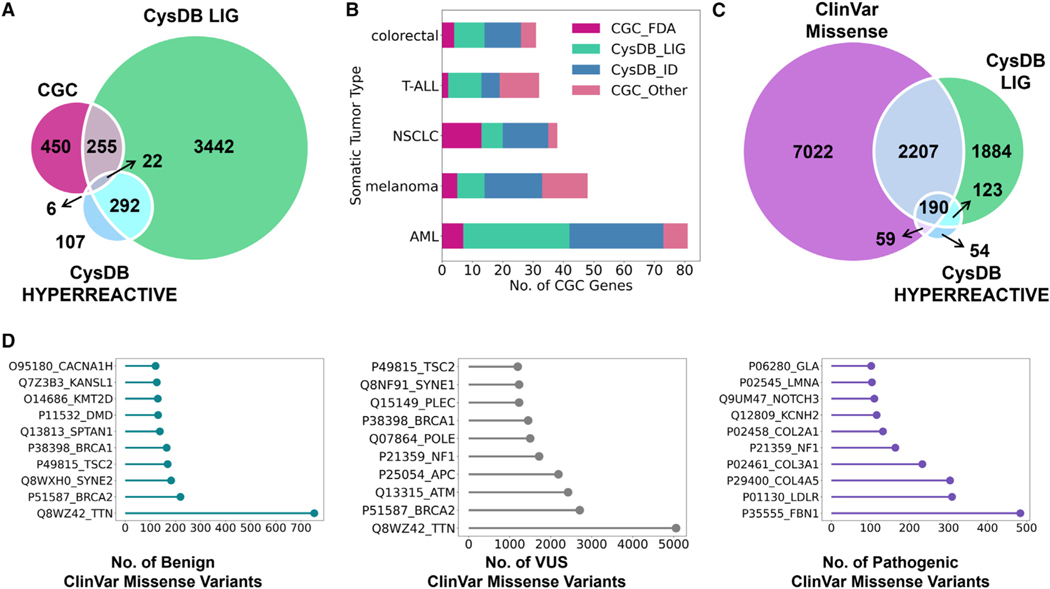
Building upon our analyses of protein function, we assessed the human disease relevance of the CysDB proteins. Restricting our analysis to the ligandable and hyperreactive subsets, we analyzed which phenotypes were associated with CysDB proteins. Using disease annotations from the Online Mendelian Inheritance in Man (OMIM)75 knowledge base, ligandable cysteine-containing proteins showed terms related to a broad range of cancers, including colorectal, breast, and leukemia. The hyperreactive cysteine-containing protein subset was enriched for terms associated with immune-relevant diseases, specifically those affecting the lymphatic system (Figure S25). Next, we determined how many CysDB proteins are annotated as cancer-driving genes, as dictated by the CGC.27 Seventy-six percent of CGC genes have been identified by CysDB (559/733) (Figure S28). Of all the CGC genes, 38% are annotated as ligandable in CysDB, indicating untapped opportunities for the development of tailored therapies targeting driver mutations (Figure 7A; Data S4). These results compare favorably with the 11% of cancer-driving genes that have been targeted by FDA-approved small molecules (Figure S29; Data S2). We observed a considerable difference in the number of available therapies for different cancers during our enrichment analysis for CysDB proteins associated with different tumor types. Although acute myeloid leukemia (AML) genes are the most represented somatic tumor type in CGC, only 5% of these genes are targets of FDA-approved small molecules. By contrast, 13 out of 38 (34%) of non-small cell lung cancer (NSLC) genes have been targeted by FDA-approved drugs. Toward addressing this therapy gap, CysDB detects most CGC genes associated with AML, 71 out of 81 (88%) (Figure 7B). In fact, 36 of these AML genes have been liganded by a compound in CysDB, such as the class 2 AML genes nucleophosmin 1 (NPM1) and core-binding factor subunit beta (CBFB).
Figure 7. Assessment of the scope of disease-relevant proteins contained in CysDB of biologically relevant proteins using cysteine chemoproteomics.
(A) Overlap between genes associated with cancer by the Cancer Gene Census (CGC), genes associated with CysDB ligandable proteins, and genes associated with CysDB hyperreactive proteins.
(B) For the five most abundant tumor types in CGC, the number of CGC genes targeted by FDA-approved drugs (CGC_FDA), non-FDA targeted CGC genes identified in CysDB (CysDB_ID), non-FDA targeted CGC genes liganded in CysDB (CysDB_LIG), and non-FDA targeted CGC genes not identified in CysDB (CGC_Other).
(C) Overlap between unique proteins associated with ClinVar genes containing missense variants (9,951 genes mapped to 9,478 proteins), CysDB ligandable proteins, and CysDB hyperreactive proteins.
(D) Top ten CysDB identified proteins with the highest number of benign missense variants (teal), missense variants of unknown significance (VUS) (gray), and pathogenic missense variants (purple).
See also Figures S21–S31 and Data S4.
Genetic variants, along with wild-type genes, can contribute toward harmful disease phenotypes. The ClinVar28 database provides a curated set of clinical significance for more than 1 million genetic variants, which are classified as benign, pathogenic, or variants of unknown significance (VUS). Of 12,858 unique UniProtKB proteins associated with ClinVar variants (mapped to 31,685 unique genes), 9,478 proteins (73.7%) have a missense variant (Figure S30). Overall, more than half of the proteins identified in CysDB have an associated ClinVar missense variant, of which 3,075 contain liganded cysteines and 330 contain hyperreactive cysteines (Figure 7C). Previously we reported a trend between chemoproteomic identified cysteines and missense pathogenicity, where chemoproteomic detected cysteine codons were predicted to be more deleterious than undetected cysteine codons.9 Consistent with the ubiquity of missense variants in ClinVar, the most common mutation associated with CysDB ID CGC genes are missense mutations.27 Of the CysDB ID proteins that have a ClinVar missense variant, 4,418 proteins have a benign variant, 2,524 proteins have a pathogenic variant, and 3,333 proteins have a variant of unknown significance (Figure S31). The proteins with the highest number of pathogenic variants are fibrillin-1 (FBN1; UniProtKB: P35555) and low-density lipoprotein receptor (LDLRl UniProtKB: P01130) (Figure 7D). Mutations in FBN1 are known to frequently cause Marfan syndrome by destabilizing disulfide bonds of conserved cysteine residues in epidermal growth factor (EGF)-like domains.76–78 Additionally, LDLR contains cysteine-rich repeats that bind lipoproteins. Loss-of-function mutations in these regions result in the disruption of cholesterol transport, leading to an increased risk for heart disease.79,80 In addition to enabling human genotype-guided target prioritization, targeting variantcontaining chemoproteomic detected proteins may also prove useful precision therapy development in a manner akin to the recent Gly12Cys-directed KRAS compounds, including FDA-approved Sotorasib.81–83
DISCUSSION
Leading groups in cysteine chemoproteomics have discovered thousands of functional and potentially druggable cysteines proteome-wide.1–9 These studies have yielded global measures of the SAR of compounds that target specific cysteines together with the intrinsic reactivity toward promiscuous electrophilic probes. Given the functional and clinical significance of identification of reactive and ligandable cysteines, the development of strategies that enable rapid cross-dataset comparisons between these studies represents an important opportunity for the cysteine chemoproteomics community that will enable a more comprehensive understanding of the cysteinome. Here we present CysDB as such a tool that unites high-coverage chemoproteomic measures of identification, ligandability, and hyperreactivity across multiple studies, together with integration with relevant resources to provide metrics of functionality and disease relevance. CysDB achieves identification of an impressive 62,888 unique cysteines and 11,621 proteins, which represents a ~100% increase in total number of identified cysteine residues compared with individual prior studies, with added potential for further growth as new datasets become available.
For our first step toward constructing CysDB, we accumulated and curated a selected set of cysteine chemoproteomics studies, which were prioritized because of the high coverage of identified cysteines. During our stringent data curation, we observed study-dependent differences in conventions for designating a cysteine as hyperreactive and/or ligandable. To account for the potential uncertainty caused by a general absence of field-wide data analysis conventions, we retained all hyperreactive and/or liganded cysteines to accurately represent each study’s reported findings. The development of statistically rigorous conventions for the field will aid in normalizing future cross-dataset comparison efforts. Recently, in our studies we have required comparable ratios with low SDs identified across multiple biological replicates together with inclusion of inactive control datasets to further simplify removal of potentially spurious elevated ratios. For studies that rely on MS1-based quantification, so-called singleton values, should be treated with an additional level of stringency, as these can prove more prone to yielding spurious ratios. These ratios are derived from peptides with precursor ions that have only been identified with either a heavy or light isotopic modification. Therefore, we followed general conventions for filtering singletons, by setting a maximum ratio value of log2(ratio) equivalent to 20 requiring identification of additional lower ratio ions. Future studies, including our own, will benefit significantly from harnessing advances in data acquisition and analysis to improve reproducibility, including imputation and data-independent acquisition, as showcased by recent efforts by the Wang group.84
Illustrating the utility of CysDB, we find that by combining datasets generated across multiple cell lines and using different labeling reagents, we substantially increased aggregate coverage of the cysteinome. Alongside cysteine coverage, CysDB reveals that cell line selection can impact not only which cysteines are identified in proteomes derived from different cell lines (Figure S5), but also the hyperreactivity and ligandability of individual cysteines. We ascribe these differences in part to both cell state specific expression as well as the stochastic nature of datadependent acquisition (DDA), which is the acquisition method used to generate nearly all datasets analyzed.
In its current iteration, CysDB provides a low-throughput mechanism to assess reproducible ligandability of cysteines across studies, including those that analyze identical compounds. To enable such comparisons, we grouped identical compounds shared across multiple publication datasets under a shared identifier, termed “group compound ID.” The group compound ID allows users to easily visualize the reproducibility of cysteine ligandability across studies. The relative rarity of shared compounds used across multiple studies (25 in total in CysDB) remains a limitation for reproducibility analysis at the level of specific compounds. One notable exception to this paradigm is the recent work by Yang et al.10 that validates many compounds assayed by DDA using a DIA approach. We hope that future studies will consider inclusion of several benchmark scout fragments to stimulate efforts in assessing the reproducibility of ligandable ratios across studies. In addition, these cross-dataset comparisons revealed a marked bias toward chemoproteomic analysis of chloroacetamide and acrylamides, which points to largely untapped opportunities in expanding the scope of the ligandable cysteinome through assaying additional classes of electrophiles.
A key feature of CysDB is the inclusion of functional and disease annotations from UniProtKB, CGC, and ClinVar. We expect that the centralization of the annotations should allow rapid prioritization of ligandable cysteines for future studies. Showcasing the utility of cysteine chemoproteomics to access tough-to-drug classes of proteins, we find a considerable enrichment in transcription factors containing ligandable cysteines (Figure 5C). We also observe that many Cancer Gene Census driver genes contain a cysteine identified in a chemoproteomics study. These findings together with our observation that a smaller but still substantial 38% of all Cancer Gene Census genes contain a ligandable cysteine suggests opportunities for future studies to more comprehensively assess the ligandability of these genes.
During our efforts to map annotations generated from genomics data (e.g., ClinVar/Cancer Gene Census data), we encountered issues with mismapping for a subset of identifiers. While processing all datasets included in CysDB, we observed that a handful (16) of gene names did not map to UniProtKB protein accession numbers in a one-to-one manner during SQL querying; multiple HGNC or Gene Entrez symbols can be associated with a single protein identifier if the translated gene products are identical protein sequences.26 Given the utility of a gene-centric search, we have incorporated such identifiers in this release of CysDB to aid future proteogenomic analysis.
An ongoing goal of CysDB is to facilitate expanding the scope of the ligandable and potentially druggable cysteinome, particularly for functional and disease-relevant proteins. Given our observed bias in CysDB ligandability datasets toward chloroacetamide and acrylamide moieties, we expect that future expansions of the ligandable cysteinome may stem in part from chemoproteomic studies using additional classes of electrophiles. In a similar manner, we expect that inclusion of datasets generated using alternatives to iodoacetamide as promiscuous cysteine-reactive capping agents, including, for example, hypervalent iodine-based probes,19 should further increase coverage of labeled cysteines. In this first iteration of CysDB, we have opted to restrict our datasets to those generated through lysate-based proteomic studies, which eliminates challenges associated with deconvolving changes in protein abundance from direct cysteine labeling. Given the importance of cell-based studies for target discovery and hit-to-lead optimization, we look forward to including such datasets in future releases, particularly when combined with bulk measures of protein abundance. In a similar manner, we look forward to incorporating redox proteomics datasets in subsequent iterations of CysDB, alongside generalized strategies to merge the diverse data formats generated by these studies. Looking ahead, we are enthusiastic about the continued growth of CysDB and encourage all interested users to consider submission of relevant chemoproteomics datasets thatcomply with oursubmission format (Data S1)and thatinclude spectral files deposited in a public data repository, such as Pride.85
Limitations of this study
Data integration and identifier mismapping were central challenges that we encountered during CysDB construction. To faithfully reflect the datasets reported by each publication, we opted to maintain each UniProtKB identifier and residue number as reported in the original study. Because of UniProtKB update cycles, some identifiers may have been deleted or merged with new protein codes. Additionally, a small subset of cysteine amino acid numbers may not match the cysteine residue numbers in the most current version of UniProtKB. These dataset-dependent differences will likely complicate cross-dataset comparisons for a handful of proteins identified. To address these limitations going forwards, we would encourage users to include the specific UniProtKB release data and reference FASTA to facilitate data integration. The latter is particularly relevant for datasets searched against legacy reference files no longer publicly available via UniProtKB. As only the annual UniProtKB January release is archived, to facilitate future data integration, we would recommend search against a reference database built from a January release. Continued growth of the CysDB ecosystem will improve the rigor and reproducibility of chemoproteomics and will facilitate the continued growth of the cysteineome. These benefits will require community-wide adoption of data deposition as a component of the chemoproteomics publishing process.
SIGNIFICANCE
Chemoproteomics has emerged as highly enabling technology capable of pinpointing functional and potentially druggable cysteine residues proteome-wide. Although many cysteine chemoproteomic datasets are now available, the overlap and reproducibility between studies remains unknown because of a lack of mechanisms for data integration. Here we report CysDB, a comprehensive database of cysteine chemoproteomics data that facilitates rapid discovery of the reactivity, ligandability, potential therapeutic relevance for 62,888 cysteines and 11,621 proteins. By including available data from multiple published studies together with annotations of function, disease relevance, and structural information, CysDB represents an unparalleled resource for understanding the scope and functional significance of the cysteinome. Given the emerging value of cysteine-reactive molecules as clinical candidates and chemical probes, CysDB also provides a resource for ongoing and future electrophilic probe development campaigns. Showcasing the utility of CysDB, we report the enrichment of ligandable cysteines in undruggable classes of proteins, observation of a subset of cysteines showing marked preferences for specific classes of electrophiles, and that ligandable cysteines are present in numerous undrugged disease-relevant proteins.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and data should be directed to and will be fulfilled by the lead contact, Keriann Backus (kbackus@mednet.ucla.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This paper analyzes existing, publicly available data These accession numbers for the datasets are listed in the key resources table.
Original code has been deposited at https://github.com/lmboat/cysdb_app and is publicly available as of the date of publication.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Software and algorithms | ||
|
| ||
| CysDB | This paper | https://backuslab.shinyapps.io/cysdb/,https://github.com/lmboat/cysdb_app |
| Human proteome | UniProtKB | UP000005640 |
| UniProtKB/Swiss-Prot Fasta | 2201-release | https://www.uniprot.org/ |
| UniProtKB/Swiss-Prot | 2209-release | https://www.uniprot.org/ |
| COSMIC | 2209-release | https://cancer.sanger.ac.uk/census |
| ClinVar | 2209-release | https://cancer.sanger.ac.uk/census |
| Human Protein Atlas (HPA) | Version 21.1 | https://www.proteinatlas.org |
| Enrichr Panther | 2016 | http://www.pantherdb.org/pathway/ |
| Enrichr Pfam Domains | 2019 | https://pfam.xfam.org/ |
| Enrichr OMIM Disease | https://www.omim.org/downloads | |
| R version 4.2.1 | R project | https://www.r-project.org |
| RStudio Version 2022.07.1 | https://rstudio.com | |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Enrichr | Accessed Sept. 2022 | https://maayanlab.cloud/Enrichr/ |
|
| ||
| Other | ||
|
| ||
| Quantitative reactivity profiling predicts functional cysteines in proteomes | 41586_2010_BFnature09472_ MOESM204_ESM.xlsx |
PMID: 21085121 |
| Proteome-wide covalent ligand discovery in native biological systems | 41586_2016_BFnature18002_ MOESM54_ESM.xlsx |
PMID: 27309814 |
| An activity-guided map of electrophile-cysteine interactions in primary human T cells | NIHMS1616434-supplement-mmc4.xlsx | PMID: 32730809 |
| Reimagining high-throughput profiling of reactive cysteines for cell-based screening of large electrophile libraries | 41587_2020_778_S4_ESM.xlsx, 41587_2020_778_S6_ESM.xlsx, 41587_2020_778_S7_ESM.xlsx, 41587_2020_778_S8_ESM.xlsx |
PMID: 33398154 |
| SP3-FAIMS Chemoproteomics for High-Coverage Profiling of the Human Cysteinome |
cbic202000870-sup-0001-table_s4.xlsx, cbic202000870-sup-0001-table_s6.xlsx | PMID: 33442901 |
| Multiplexed CuAAC Suzuki–Miyaura Labeling for Tandem Activity-Based Chemoproteomic Profiling |
ac0c04726_si_002.xlsx, ac0c04726_si_003.xlsx | PMID: 33470097 |
| From chemoproteomic-detected amino acids to genomic coordinates: insights into precise multi-omic data integration | msb20209840-sup-0020-datasetev18.xlsx | PMID: 33599394 |
| Quantitative Chemoproteomic Profiling with Data-Independent Acquisition-Based Mass Spectrometry |
ja1c11053_si_002.xlsx, ja1c11053_si_003.xlsx | PMID: 34986311 |
| Benchmarking Cleavable Biotin Tags for Peptide-Centric Chemoproteomics |
pr2c00174_si_002.xlsx, pr2c00174_si_003.xlsx, pr2c00174_si_004.xlsx, pr2c00174_si_005.xlsx, pr2c00174_si_006.xlsx | PMID: 35467356 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All data are generated from the datasets provided in the key resources table.
METHOD DETAILS
Proteomics data analysis
Chemoproteomics data was collected from publicly accessible supplementary tables of previous literature.2,4–11 Columns were parsed for UniProtKB protein identifiers and locations of the corresponding modified cysteine amino acid numbers to create a new identifier for CysDB: UniProtKBID_CYS#. Any cysteine classified as ‘ligandable’ or ‘hyperreactive’ is listed in CysDB as ligandable or hyperreactive. Individual ligandability and reactivity ratios found from each publication are listed in Data S1 and Data S2. In some cases, for the ligandability and reactivity datasets, publications listed ratios for peptides simultaneously modified at multiple cysteines such as UniProtKBID_CYS#1_CYS#2, where the ratios provided for UniProtKBID_CYS#1_CYS#2 differed from UniProtKBID_CYS#1. Thus, ratios for peptides modified at multiple cysteines were not included in further analyses.
Compounds found in ligandability studies were stratified according to their cell line and chemotype. Unique identifiers for each compound were constructed based on their chemotype within the five categories: acrylamide, bromoacetamide, chloroacetamide, dimethyl fumarate (dmf) and others, such as ACRYL_#. Unique group identification numbers were constructed for compounds based on their chemotype and SMILES string, such as GROUP_ACRYL_# Publication names for each compound and CysDB names are provided in Data S2.
SMILES strings listed in the Supplementary Tables for each publication were copied and pasted into a new document. To obtain a uniform SMILES format for all the compounds in CysDB, published SMILES strings were converted into molecules and converted back into SMILES strings using RDKit.53
In the event amino acid numbers were not provided by the author, python scripts (available on GitHub) were utilized to map the listed peptide sequences to the canonical protein sequences of the 2201-release UniProtKB human fasta reference file, as this release is the only version saved in the UniProtKB archive for future mapping. Cysteines from unmatched peptides were removed prior to subsequent analyses. To inspect the extent of mismapped identifiers in CysDB, we collected peptides mapped to multiple proteins or peptides labeled at multiple cysteine sites from each publication (Data S1). Peptides labeled at multiple cysteines were dropped from our ligandability and hyperreactivity data aggregation.
Cancer Gene Census (CGC) website reports were downloaded Sept. 2022 and mapped to CysDB data using UniProtKB accessions. Due to frequent UniProtKB updates, Gene symbols reported in the Cancer Gene Census were mapped to gene names in UniProtKB to identify the updated UniProtKB codes (2209-release).
Functional, structural, and druggability annotations data analysis
Custom Python scripts classified protein functions based on annotations in the UniProtKB/Swiss-Prot26 database (2209-release). UniProtKB accessions were collected from proteins with available ChEMBL and DrugBank UniProtKB annotations. Data from the Human Protein Atlas29 (HPA) version 21.1 was downloaded and parsed to obtain genes targeted by FDA approved drugs. HGNC gene symbols were mapped to UniProtKB accessions to collect proteins targeted by FDA approved drugs.
Custom Python scripts classified protein functions based on annotations in the UniProtKB/Swiss-Prot database (2209-release), HPA version 21.1 and the ScaPD database.86 UniProtKB keywords were parsed to classify proteins into five broad functional categories: chaperones/transporter/channel/receptor, enzyme, nucleic acid and small molecule binding, scaffolding/modulator/adaptor, transcription factor/regulator and uncategorized. Transcription factors, channels and transporters were also found using protein class descriptions from the HPA. In addition, examples of experimentally validated scaffolding proteins were collected from the ScaPD database. For proteins in more than one category, annotations were prioritized based on the following: enzyme > chaperones/transporter/channel/receptor > scaffolding/modulator/adaptor > transcription factor/regulator > nucleic acid and small molecule binding.
Counts of how many CysDB proteins had UniProtKB annotations for active sites, binding sites, catalytic activity, disulfide bonds and redox potentials were calculated based on matches between the position of the identified residue and UniProtKB functional annotation. Further parsing of UniProtKB active and binding site annotations were extracted to obtain specific residues and amino acid numbers. Positions of binding and active sites that were not cysteine residues were discarded. Exact amino acid positions of UniProtKB cysteine active and binding sites were cross-referenced with CysDB cysteine identifiers.
CysDB cysteines ‘in or near’ UniProtKB annotated active or binding sites were assessed using primary protein sequences. Positions of identified cysteines were found via their amino acid numbering. Annotated active or binding sites within +/−10 amino acids from the identified cysteine were considered as a cysteine ‘in or near’ an active or binding site.
Protein Data Bank32 identifiers were found from UniProtKB annotations. Proteins without PDB structures were filtered out. PDB structures for proteins with PDB annotations were downloaded and parsed for amino acid numbering and residue names. A list of cysteines resolved in each PDB was stored for further processing. SIFTS63 files, providing residue level mapping between PDB sequences and protein sequences, were downloaded for each PDB. Cysteines resolved in each PDB were mapped to their appropriate UniProtKB protein sequence and identifiers for PDB to UniProtKB pairs were created: PDB_C#_UniProtKBID_C#. From these paired identifiers, the number of unique UniProtKBID_C# records were counted to determine the number of UniProtKB cysteines resolved in PDBs.
CysDB cysteines ‘in or near’ UniProtKB annotated active or binding sites were assessed using 3D PDB structures. From the workflow described below (determining cysteines in PDB structures), PDB structures were parsed to find all neighboring residues within a 10 Angstrom distance of a cysteine residue. PDB_UniProtKB identifiers were created for each cysteine and corresponding list of neighboring residues. If the UniProtKB annotated active or binding sites were resolved in an associated crystal structure and found within the 10 Angstroms net, it was classified as a cysteine proximal to a known active or binding site.
CysDB database
CysDB was created as a relational database using MySQL v.8.0. Overall, the database contains six tables and is hosted on Google Cloud. The major parent tables, ‘Datasets’ and ‘Identifiers’, were further broken down into child tables, such as ‘Ligandable’, ‘Reactive’, ‘Compound’ and ‘Warheads’ (Figure S6). The Datasets table contains information specific to each of the nine publications, while the Identifiers table contains information specific to each modified cysteine or protein identifier. Columns within Datasets and Identifiers include binary results for the following three categories: identified, hyperreactive and ligandable. However, individual competition ratios are listed in the Ligandable table and individual reactivity ratios are listed in the Reactive table. Calculated molecular properties for ‘drug-likeness’ were acquired using RDKit53 and are stored in the ‘Compounds’ table. This table also contains the CysDB compound identifier mapped to their associated publication abbreviation or designated name. Group compound identifiers (“GROUP_WARHEAD_#”) were defined by unique standardized SMILES strings and individual compound identifiers (“WARHEAD_#”) were defined by unique standardized SMILES string, cell line and publication author combinations. Finally, the warhead table holds chemotype classifications for each compound. The five chemotype classifications were as follows: acrylamide, bromoacetamide, chloroacetamide, dimethyl fumarate and other.
CysDB web application
The CysDB web application was developed using the Shiny R package (https://shiny.rstudio.com/). Schematics of protein sequence chains, domains and motifs on the CysDB web server are constructed using the drawProteins R package (https://github.com/brennanpincardiff/drawProteins). Interactive viewing of PDB crystal structures is performed using NGLViewR (https://github.com/nglviewer/nglview). Protein protein interaction networks are accessed via the STRING database (https://string-db.org/). Gene set library enrichment analyses are provided with the Enrichr R package (https://maayanlab.cloud/Enrichr/) and ontology enrichment plots are produced with the gprofiler2 R package (https://biit.cs.ut.ee/gprofiler/gost). All plots are generated with the ggplot2 and plotly (https://plotly.com/r/) R libraries.
QUANTIFICATION AND STATISTICAL ANALYSIS
Enrichment of Panther 2016, Pfam Domains 2019 and OMIM Disease gene set library terms were performed using the GSEApy package.87 Proteins identified by chemoproteomics studies in CysDB were utilized as the background protein set. UniProtKB protein identifiers were mapped to Entrez gene symbols as input for Enrichr. P-values were computed from Fisher’s exact test to determine the significance of each enriched term. The negative log of these p-values was calculated using R.
ADDITIONAL RESOURCES
The CysDB dataset is provided as an interactive web resource at https://backuslab.shinyapps.io/cysdb/.
Dataset addition to CysDB guidelines
Email submission materials to cysteineomedb@gmail.com with the following information: copy of publication, supplementalinformation, additional details for data filtering and note the version of UniProt used to obtain protein accessions. Proteins must be identified through UniProtKB accessions. Please use the format, UniProtKBID_CYS#, to indicate which residues have been labeled. For ligandability experiments using a variety of electrophiles, inclusion of SMILES strings and criteriafor ‘ligandability’ classification is required (ex. R>=4 for at least n number of compounds). Table templates and additional information for submission requests can be found in Data S1.
Supplementary Material
Highlights.
Comprehensive repository for human cysteine chemoproteomics data
Enrichment of ligandable cysteines in undruggable classes of proteins
Visualization of lead compounds for 9,246 cysteines
Web app includes annotations of functionality, druggability, and structural motifs
ACKNOWLEDGMENTS
This study was supported by a Beckman Young Investigator Award (K.M.B.), DOD-Advanced Research Projects Agency (DARPA) grant D19AP00041 (K.M.B.), and NIGMS System and Integrative Biology grant 5T32GM 008185-33 (L.M.B.). We thank all members of the Backus lab for helpful suggestions. We thank S. Forli and J. Eberhardt for helpful suggestions.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.chembiol.2023.04.004.
DECLARATION OF INTERESTS
K.M.B. is a paid consultant for Oncovalent Therapeutics and Matchpoint Therapeutics.
REFERENCES
- 1.Xiao H, Jedrychowski MP, Schweppe DK, Huttlin EL, Yu Q, Heppner DE, Li J, Long J, Mills EL, Szpyt J, et al. (2020). A quantitative tissue-specific landscape of protein redox regulation during aging. Cell 180, 968–983.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuljanin M, Mitchell DC, Schweppe DK, Gikandi AS, Nusinow DP, Bulloch NJ, Vinogradova EV, Wilson DL, Kool ET, Mancias JD, et al. (2021). Reimagining high-throughput profiling of reactive cysteines for cell-based screening of large electrophile libraries. Nat. Biotechnol. 39, 630–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Müller S, Ackloo S, Al Chawaf A, Al-Lazikani B, Antolin A, Baell JB, Beck H, Beedie S, Betz UAK, Bezerra GA, et al. (2022). Target 2035–update on the quest for a probe for every protein. RSC Med. Chem 13, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan T, Desai HS, Boatner LM, Yen SL, Cao J, Palafox MF, Jami-Alahmadi Y, and Backus KM (2021). SP3-FAIMS chemoproteomics for high coverage profiling of the human cysteinome. Chembiochem 22, 1841–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao J, Boatner LM, Desai HS, Burton NR, Armenta E, Chan NJ,Castelló n JO, and Backus KM (2021). Multiplexed CuAAC Suzuki-Miyaura labeling for tandem activity-based chemoproteomic profiling. Anal. Chem 93, 2610–2618. 10.1021/acs.analchem.0c04726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, Liu K, Xu P, and Yang J. (2022). Benchmarking cleavable biotintags for peptide-centric chemoproteomics. J. Proteome Res 21, 1349–1358. 10.1021/acs.jproteome.2c00174. [DOI] [PubMed] [Google Scholar]
- 7.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MBD, Bachovchin DA, Mowen K, Baker D, and Cravatt BF (2010). Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795. 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vinogradova EV, Zhang X, Remillard D, Lazar DC, Suciu RM, Wang Y, Bianco G, Yamashita Y, Crowley VM, Schafroth MA, et al. (2020). An activity-guided map of electrophile-cysteine interactions in primary human T cells. Cell 182, 1009–1026.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palafox MF, Desai HS, Arboleda VA, and Backus KM (2021). From chemoproteomic-detected amino acids to genomic coordinates: insights into precise multi-omic data integration. Mol. Syst. Biol 17, e9840. 10.15252/msb.20209840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang F, Jia G, Guo J, Liu Y, and Wang C. (2022). Quantitative chemoproteomic profiling with data-independent acquisition-based mass spectrometry. J. Am. Chem. Soc 144, 901–911. 10.1021/jacs.1c11053. [DOI] [PubMed] [Google Scholar]
- 11.Backus KM, Correia BE, Lum KM, Forli S, Horning BD, González-Páez GE, Chatterjee S, Lanning BR, Teijaro JR, Olson AJ, et al. (2016). Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Peled L, Kemper EK, Suciu RM, Vinogradova EV, Backus KM, Horning BD, Paul TA, Ichu TA, Svensson RU, Olucha J, et al. (2017). Chemical proteomics identifies druggable vulnerabilities in a genetically defined cancer. Cell 171, 696–709.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Backus KM (2018). Applications of Reactive Cysteine Profiling (Activity-Based Protein Profiling). [DOI] [PubMed] [Google Scholar]
- 14.Abegg D, Frei R, Cerato L, Prasad Hari D, Wang C, Waser J, and Adibekian A. (2015). Proteome-wide profiling of targets of cysteine reactive small molecules by using ethynyl benziodoxolone reagents. Angew. Chem 54, 10852–10857. [DOI] [PubMed] [Google Scholar]
- 15.Kulkarni RA, Bak DW, Wei D, Bergholtz SE, Briney CA, Shrimp JH, Alpsoy A, Thorpe AL, Bavari AE, Crooks DR, et al. (2019). A chemoproteomic portrait of the oncometabolite fumarate. Nat. Chem. Biol 15, 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grossman EA, Ward CC, Spradlin JN, Bateman LA, Huffman TR, Miyamoto DK, Kleinman JI, and Nomura DK (2017). Covalent ligand discovery against druggable hotspots targeted by anti-cancer natural products. Cell Chem. Biol 24, 1368–1376.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian C, Sun R, Liu K, Fu L, Liu X, Zhou W, Yang Y, and Yang J. (2017). Multiplexed thiol reactivity profiling for target discovery of electrophilic natural products. Cell Chem. Biol 24, 1416–1427.e5. [DOI] [PubMed] [Google Scholar]
- 18.Wang C, Weerapana E, Blewett MM, and Cravatt BF (2014). A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 11, 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abegg D, Tomanik M, Qiu N, Pechalrieu D, Shuster A, Commare B, Togni A, Herzon SB, and Adibekian A. (2021). Chemoproteomic profiling by cysteine fluoroalkylation reveals Myrocin G as an inhibitor of the nonhomologous end joining DNA repair pathway. J. Am. Chem. Soc 143, 20332–20342. [DOI] [PubMed] [Google Scholar]
- 20.Fu L, Li Z, Liu K, Tian C, He J, He J, He F, Xu P, and Yang J(2020). A quantitative thiol reactivity profiling platform to analyze redox and electrophile reactive cysteine proteomes. Nat. Protoc 15, 2891–2919. 10.1038/s41596-020-0352-2. [DOI] [PubMed] [Google Scholar]
- 21.Desai HS, Yan T, Yu F, Sun AW, Villanueva M, Nesvizhskii AI, and Backus KM (2022). SP3-Enabled rapid and high coverage chemoproteomic identification of cell-state–dependent redox-sensitive cysteines. Mol. Cell. Proteomics 21, 100218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Y, Fu L, Yang J, and Carroll KS (2021). Wittig reagents for chemoselective sulfenic acid ligation enables global site stoichiometry analysis and redox-controlled mitochondrial targeting. Nat. Chem 13, 1140–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mnatsakanyan R, Markoutsa S, Walbrunn K, Roos A, Verhelst SHL, and Zahedi RP (2019). Proteome-wide detection of S-nitrosylation targets and motifs using bioorthogonal cleavable-linker-based enrichment and switch technique. Nat. Commun 10, 2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu S, Luo Howard H, Wang H, Zhao W, Hu Q, and Yang Y. (2016). Cysteinome: the first comprehensive database for proteins with targetable cysteine and their covalent inhibitors. Biochem. Biophys. Res. Commun 478, 1268–1273. [DOI] [PubMed] [Google Scholar]
- 25.Yan T, Palmer AB, Geiszler DJ, Polasky DA, Boatner LM, Burton NR, Armenta E, Nesvizhskii AI, and Backus KM (2022). Enhancing cysteine chemoproteomic coverage through systematic assessment of click chemistry product Fragmentation. Anal. Chem 94, 3800–3810. 10.1021/acs.analchem.1c04402. [DOI] [PubMed] [Google Scholar]
- 26.Consortium UniProt (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, 506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, and Forbes SA (2018). The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 18, 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, et al. (2018). ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, 1062–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. (2010). Towards a knowledge-based human protein atlas. Nat. Biotechnol 28, 1248–1250. [DOI] [PubMed] [Google Scholar]
- 30.Mendez D, Gaulton A, Bento AP, Chambers J, De Veij M, Félix E, Magariños MP, Mosquera JF, Mutowo P, Nowotka M, et al. (2019). ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 47, 930–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, et al. (2018). DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, 1074–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose PW, Prlić A, Altunkaya A, Bi C, Bradley AR, Christie CH, and Burley SK (2016). The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eng JK, McCormack AL, and Yates JR (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom 5, 976–989. 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 34.Yu F, Teo GC, Kong AT, Haynes SE, Avtonomov DM, Geiszler DJ, and Nesvizhskii AI (2020). Identification of modified peptides using localization-aware open search. Nat. Commun 11, 4065. 10.1038/s41467-020-17921-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Integrated Proteomics Pipeline (IP2). http://www.integratedproteomics.com/
- 36.Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK,Cociorva D, Lu B, Liao L, Hewel J, Han X, et al. (2015). ProLuCID: an improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteomics 129, 16–24. 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, andNesvizhskii AI (2017). MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry–based proteomics. Nat. Methods 14, 513–520. 10.1038/nmeth.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eng JK, Jahan TA, and Hoopmann MR (2013). Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24. 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 39.Serafimova IM, Pufall MA, Krishnan S, Duda K, Cohen MS, Maglathlin RL, McFarland JM, Miller RM, Frö din M, and Taunton J. (2012). Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles. Nat. Chem. Biol 8, 471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hacker SM, Backus KM, Lazear MR, Forli S, Correia BE, and Cravatt BF (2017). Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem 9, 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbasov ME, Kavanagh ME, Ichu TA, Lazear MR, Tao Y, Crowley VM, Am Ende CW, Hacker SM, Ho J, Dix MM, et al. (2021). A proteome-wide atlas of lysine-reactive chemistry. Nat. Chem 13, 1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braschi B, Denny P, Gray K, Jones T, Seal R, Tweedie S, Yates B, and Bruford E. (2019). Genenames. org: the HGNC and VGNC resources in 2019. Nucleic Acids Res. 47, 786–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The Gene Ontology Consortium (2019). The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47, 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, D’Eustachio P, Jassal B, Korninger F, May B, et al. (2018). The reactome pathway knowledgebase. Nucleic Acids Res. 46, 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoenmaker L, Béquignon OJ, Jespers W, and Westen GJ (2023). UnCorrupt SMILES: a novel approach to de novo design. Journal of Cheminformatics 15, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bickerton GR, Paolini GV, Besnard J, Muresan S, and Hopkins AL (2012). Quantifying the chemical beauty of drugs. Nat. Chem 4, 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benet LZ, Hosey CM, Ursu O, and Oprea TI (2016). BDDCS, the Rule of 5 and druggability. Adv. Drug Deliv. Rev 101, 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lipinski CA, Lombardo F, Dominy BW, and Feeney PJ (2012). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev 64, 4–17. [DOI] [PubMed] [Google Scholar]
- 51.Ghose AK, Viswanadhan VN, and Wendoloski JJ (1999). A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem 1, 55–68. [DOI] [PubMed] [Google Scholar]
- 52.Congreve M, Carr R, Murray C, and Jhoti H. (2003). A’rule of three’for fragment-based lead discovery? Drug Discov. Today 8, 876–877. [DOI] [PubMed] [Google Scholar]
- 53.Landrum G. (2013). Rdkit documentation. Release 1, 1–79. [Google Scholar]
- 54.Senkane K, Vinogradova EV, Suciu RM, Crowley VM, Zaro BW, Bradshaw JM, Brameld KA, and Cravatt BF (2019). The proteome--wide potential for reversible covalency at cysteine. Angew. Chem 58, 11385–11389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krishnan S, Miller RM, Tian B, Mullins RD, Jacobson MP, and Taunton J. (2014). Design of reversible, cysteine-targeted Michael acceptors guided by kinetic and computational analysis. J. Am. Chem. Soc 136, 12624–12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zambaldo C, Vinogradova EV, Qi X, Iaconelli J, Suciu RM, Koh M, Senkane K, Chadwick SR, Sanchez BB, Chen JS, et al. (2020). 2-Sulfonylpyridines as tunable, cysteine-reactive electrophiles. J. Am. Chem. Soc 142, 8972–8979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du X, Guo C, Hansell E, Doyle PS, Caffrey CR, Holler TP, McKerrow JH, and Cohen FE (2002). Synthesis and structure activity relationship study of potent trypanocidal thio semicarbazone inhibitors of the trypanosomal cysteine protease cruzain. J. Med. Chem 45, 2695–2707. [DOI] [PubMed] [Google Scholar]
- 58.Greenbaum DC, Mackey Z, Hansell E, Doyle P, Gut J, Caffrey CR, Lehrman J, Rosenthal PJ, McKerrow JH, and Chibale K. (2004). Synthesis and structure activity relationships of parasiticidal thiosemicarbazone cysteine protease inhibitors against Plasmodium falciparum, Trypanosoma brucei, and Trypanosoma cruzi. J. Med. Chem 47, 3212–3219. [DOI] [PubMed] [Google Scholar]
- 59.Shenai BR, Lee BJ, Alvarez-Hernandez A, Chong PY, Emal CD, Neitz RJ, Roush WR, and Rosenthal PJ (2003). Structure-activity relationships for inhibition of cysteine protease activity and development of Plasmodium falciparum by peptidyl vinyl sulfones. Antimicrob. Agents Chemother. 47, 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klüver E, Schulz-Maronde S, Scheid S, Meyer B, Forssmann WG, and Adermann K. (2005). Structure-activity relation of human β-defensin 3: influence of disulfide bonds and cysteine substitution on antimicrobial activity and cytotoxicity. Biochemistry 44, 9804–9816. [DOI] [PubMed] [Google Scholar]
- 61.Grzonka Z, Jankowska E, Kasprzykowski F, Kasprzykowska R, Lankiewicz L, Wiczk W, Wieczerzak E, Ciarkowski J, Drabik P, Janowski R, et al. (2001). Structural studies of cysteine proteases and their inhibitors. Acta Biochim. Pol 48, 1–20. [PubMed] [Google Scholar]
- 62.Zanon PR, Yu F, Musacchio P, Lewald L, Zollo M, Krauskopf K, andHacker SM (2021). Profiling the Proteome-wide Selectivity of Diverse Electrophiles. ChemRxiv. 10.26434/chemrxiv.14186561.v1. [DOI] [Google Scholar]
- 63.Dana JM, Gutmanas A, Tyagi N, Qi G, O’Donovan C, Martin M, and Velankar S. (2019). SIFTS: updated Structure Integration with Function, Taxonomy and Sequences resource allows 40-fold increase in coverage of structure-based annotations for proteins. Nucleic Acids Res. 47, 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Julio AR, and Backus KM (2021). New approaches to target RNA binding proteins. Curr. Opin. Chem. Biol 62, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de la Cruz J, Kressler D, and Linder P. (1999). Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci 24, 192–198. [DOI] [PubMed] [Google Scholar]
- 67.Aubourg S, Kreis M, and Lecharny A. (1999). The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res. 27, 628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patmore DM, Jassim A, Nathan E, Tong Y, Tahan D, Hoffmann N, Gilbertson RJ, Smith KS, Kanneganti TD, Suzuki H, et al. (2020). DDX3X suppresses the susceptibility of hindbrain lineages to medulloblastoma. Dev. Cell 54, 455–470.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andrisani O, Liu Q, Kehn P, Leitner WW, Moon K, Vazquez-Maldonado N, and Gale M. (2022). Biological Functions of DEAD/ DEAH-box RNA Helicases in Health and Disease. Nature Immunology 23, 354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mi H, Muruganujan A, Ebert D, Huang X, and Thomas PD (2019). PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fesik SW (2005). Promoting apoptosis as a strategy for cancer drug discovery. Nat. Rev. Cancer 5, 876–885. [DOI] [PubMed] [Google Scholar]
- 72.Aguilar A, Lu J, Liu L, Du D, Bernard D, McEachern D, Przybranowski S, Li X, Luo R, Wen B, et al. (2017). Discovery of 4-((3′ R, 4′ S, 5′ R)-6 ′′-Chloro-4′-(3-chloro-2-fluorophenyl)-1′-ethyl-2 ′′--oxodispiro [cyclohexane-1, 2′-pyrrolidine-3′, 3 ′′-indoline]-5′-carboxamido) bicyclo [2.2. 2] octane-1-carboxylic acid (AA-115/APG-115): a potent and orally active murine double minute 2 (MDM2) inhibitor in clinical development. J. Med. Chem 60, 2819–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giancotti FG, and Ruoslahti E. (1999). Integrin signaling. Science 285, 1028–1032. [DOI] [PubMed] [Google Scholar]
- 74.Cooper J, and Giancotti FG (2019). Integrin signaling in cancer: mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell 35, 347–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hamosh A, Scott AF, Amberger JS, Bocchini CA, and McKusick VA (2005). Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33, D514–D517. 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schrijver I, Liu W, Brenn T, Furthmayr H, and Francke U. (1999). Cysteine substitutions in epidermal growth factor–like domains of fibrillin-1: distinct effects on biochemical and clinical phenotypes. Am. J. Hum. Genet 65, 1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Russell DW, Brown MS, and Goldstein JL (1989). Different combinations of cysteine-rich repeats mediate binding of low density lipoprotein receptor to two different proteins. J. Biol. Chem 264, 21682–21688. [PubMed] [Google Scholar]
- 78.Daly NL, Scanlon MJ, Djordjevic JT, Kroon PA, and Smith R. (1995). Three-dimensional structure of a cysteine-rich repeat from the low-density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 92, 6334–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Esser V, Limbird LE, Brown MS, Goldstein JL, and Russell DW (1988). Mutational analysis of the ligand binding domain of the low density lipoprotein receptor. J. Biol. Chem 263, 13282–13290. [PubMed] [Google Scholar]
- 80.Lanman BA, Allen JR, Allen JG, Amegadzie AK, Ashton KS, Booker SK, and Cee VJ (2020). Discovery of a Covalent Inhibitor of KRASG12C (AMG 510) for the Treatment of Solid Tumors. J. Med. Chem 63, 52–65. [DOI] [PubMed] [Google Scholar]
- 81.Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, Chen Y, Babbar A, Firdaus SJ, Darjania L, et al. (2018). Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589.e17. [DOI] [PubMed] [Google Scholar]
- 82.Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, Chen Y, Kucharski JM, Feng J, Ely T, et al. (2016). Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive StateTargeting inactive KRASG12C suppresses oncogenic signaling. Cancer Discov. 6, 316–329. [DOI] [PubMed] [Google Scholar]
- 83.Ostrem JM, Peters U, Sos ML, Wells JA, and Shokat KM (2013). K-Ras (G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, and Cox J. (2016). The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- 85.Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Vizcaíno JA, Prakash A, Frericks-Zipper A, Eisenacher M, et al. (2022). The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han X, Wang J, Wang J, Liu S, Hu J, Zhu H, and Qian J. (2017). ScaPD: a database for human scaffold proteins. BMC Bioinf. 18, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, et al. (2021). Gene set knowledge discovery with enrichr. Curr. Protoc 1, e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available data These accession numbers for the datasets are listed in the key resources table.
Original code has been deposited at https://github.com/lmboat/cysdb_app and is publicly available as of the date of publication.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Software and algorithms | ||
|
| ||
| CysDB | This paper | https://backuslab.shinyapps.io/cysdb/,https://github.com/lmboat/cysdb_app |
| Human proteome | UniProtKB | UP000005640 |
| UniProtKB/Swiss-Prot Fasta | 2201-release | https://www.uniprot.org/ |
| UniProtKB/Swiss-Prot | 2209-release | https://www.uniprot.org/ |
| COSMIC | 2209-release | https://cancer.sanger.ac.uk/census |
| ClinVar | 2209-release | https://cancer.sanger.ac.uk/census |
| Human Protein Atlas (HPA) | Version 21.1 | https://www.proteinatlas.org |
| Enrichr Panther | 2016 | http://www.pantherdb.org/pathway/ |
| Enrichr Pfam Domains | 2019 | https://pfam.xfam.org/ |
| Enrichr OMIM Disease | https://www.omim.org/downloads | |
| R version 4.2.1 | R project | https://www.r-project.org |
| RStudio Version 2022.07.1 | https://rstudio.com | |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Enrichr | Accessed Sept. 2022 | https://maayanlab.cloud/Enrichr/ |
|
| ||
| Other | ||
|
| ||
| Quantitative reactivity profiling predicts functional cysteines in proteomes | 41586_2010_BFnature09472_ MOESM204_ESM.xlsx |
PMID: 21085121 |
| Proteome-wide covalent ligand discovery in native biological systems | 41586_2016_BFnature18002_ MOESM54_ESM.xlsx |
PMID: 27309814 |
| An activity-guided map of electrophile-cysteine interactions in primary human T cells | NIHMS1616434-supplement-mmc4.xlsx | PMID: 32730809 |
| Reimagining high-throughput profiling of reactive cysteines for cell-based screening of large electrophile libraries | 41587_2020_778_S4_ESM.xlsx, 41587_2020_778_S6_ESM.xlsx, 41587_2020_778_S7_ESM.xlsx, 41587_2020_778_S8_ESM.xlsx |
PMID: 33398154 |
| SP3-FAIMS Chemoproteomics for High-Coverage Profiling of the Human Cysteinome |
cbic202000870-sup-0001-table_s4.xlsx, cbic202000870-sup-0001-table_s6.xlsx | PMID: 33442901 |
| Multiplexed CuAAC Suzuki–Miyaura Labeling for Tandem Activity-Based Chemoproteomic Profiling |
ac0c04726_si_002.xlsx, ac0c04726_si_003.xlsx | PMID: 33470097 |
| From chemoproteomic-detected amino acids to genomic coordinates: insights into precise multi-omic data integration | msb20209840-sup-0020-datasetev18.xlsx | PMID: 33599394 |
| Quantitative Chemoproteomic Profiling with Data-Independent Acquisition-Based Mass Spectrometry |
ja1c11053_si_002.xlsx, ja1c11053_si_003.xlsx | PMID: 34986311 |
| Benchmarking Cleavable Biotin Tags for Peptide-Centric Chemoproteomics |
pr2c00174_si_002.xlsx, pr2c00174_si_003.xlsx, pr2c00174_si_004.xlsx, pr2c00174_si_005.xlsx, pr2c00174_si_006.xlsx | PMID: 35467356 |