Abstract
Lysosomes have many roles, including degrading macromolecules and signaling to the nucleus1. Lysosomal dysfunction occurs in various human conditions, such as common neurodegenerative diseases and monogenic lysosomal storage disorders (LSDs)2–4. For most LSDs the causal genes have been identified, but in some the function of the implicated gene is unknown, in part because lysosomes occupy a small fraction of the cellular volume so that changes in lysosomal contents are difficult to detect. Here, we develop the LysoTag mouse for the tissue-specific isolation of intact lysosomes that are compatible with the multimodal profiling of their contents. We apply it to the study of CLN3, a lysosomal transmembrane protein of unclear function whose loss causes juvenile neuronal ceroid lipofuscinosis (Batten disease), a lethal neurodegenerative LSD. Untargeted metabolite profiling of lysosomes from the brains of mice lacking CLN3 revealed a massive accumulation of glycerophosphodiesters (GPDs), the end products of glycerophospholipid catabolism. GPDs also accumulate in the lysosomes of CLN3-deficient cultured cells and we show that CLN3 is required for their lysosomal egress. Loss of CLN3 also disrupts glycerophospholipid catabolism in the lysosome. Finally, we found elevated levels of glycerophosphoinositol in the cerebrospinal fluid of Batten disease patients, suggesting their potential use as a disease biomarker. Our results show that CLN3 is required for the lysosomal clearance of GPDs and reveal Batten disease as a neurodegenerative LSD with a defect in glycerophospholipid metabolism.
Lysosomes are membrane-bound organelles that degrade macromolecules and clear damaged organelles as well as regulate cellular signaling, such as the activation of the mTORC1 pathway by nutrients1,5,6. Mutations in genes encoding lysosomal proteins cause severe disorders collectively known as lysosomal storage diseases (LSDs)2,3,7–9. Many LSDs are associated with neurodegeneration, indicating a critical role for the lysosome in neuronal homeostasis3,4. Consistent with this, human genetic studies have implicated impaired lysosomal function in age-associated neurodegenerative diseases, including Alzheimer disease (AD), Parkinson disease (PD), and frontotemporal dementia (FTD)2,10,11. The function of several lysosomal proteins implicated in disease is incompletely understood, in part because lysosomes usually occupy only 1–3% of cellular volume so that changes in lysosomal contents can be difficult to ascertain in whole cells or tissues12.
The development of the LysoTag mouse
Given this and the increasing interest in the roles of lysosomes in physiology and disease, we adapted our LysoIP method for the rapid purification of lysosomes from cultured cells12 for use in mouse tissues. We generated a transgenic mouse line by integrating the cDNA for the lysosomally-localized TMEM192–3xHA fusion protein into the Rosa26 locus downstream of a lox-stop-lox (LSL) cassette13 (Fig. 1A). By crossing these mice with those expressing a Cre-recombinase driven by the CMV promoter, we generated “LysoTag” mice that constitutively express TMEM192–3xHA across tissues (Fig. 1A, Extended Data Fig. 1A). TMEM192–3xHA marks lysosomes with a triple HA epitope, enabling their immunoisolation with an anti-HA antibody. Immunofluorescence-based analyses of liver and brain sections, two tissues examined in this study, confirmed that TMEM192–3xHA colocalizes with the lysosomal marker LAMP1 (Fig. 1B, Extended Data Fig. 2B). Other than a 10–15% decrease in body weight, the constitutive LysoTag mice appear indistinguishable from their wild-type counterparts (Extended Data Fig.1B). Of importance, comprehensive characterizations confirm that LysoTag expression has no effect on lysosomal proteins, enzymatic activity, or lysosomal ultrastructure (Extended Data Fig. 1A, C–F).
Fig. 1: LysoTag mouse for proteomic and metabolite profiling of tissue lysosomes.
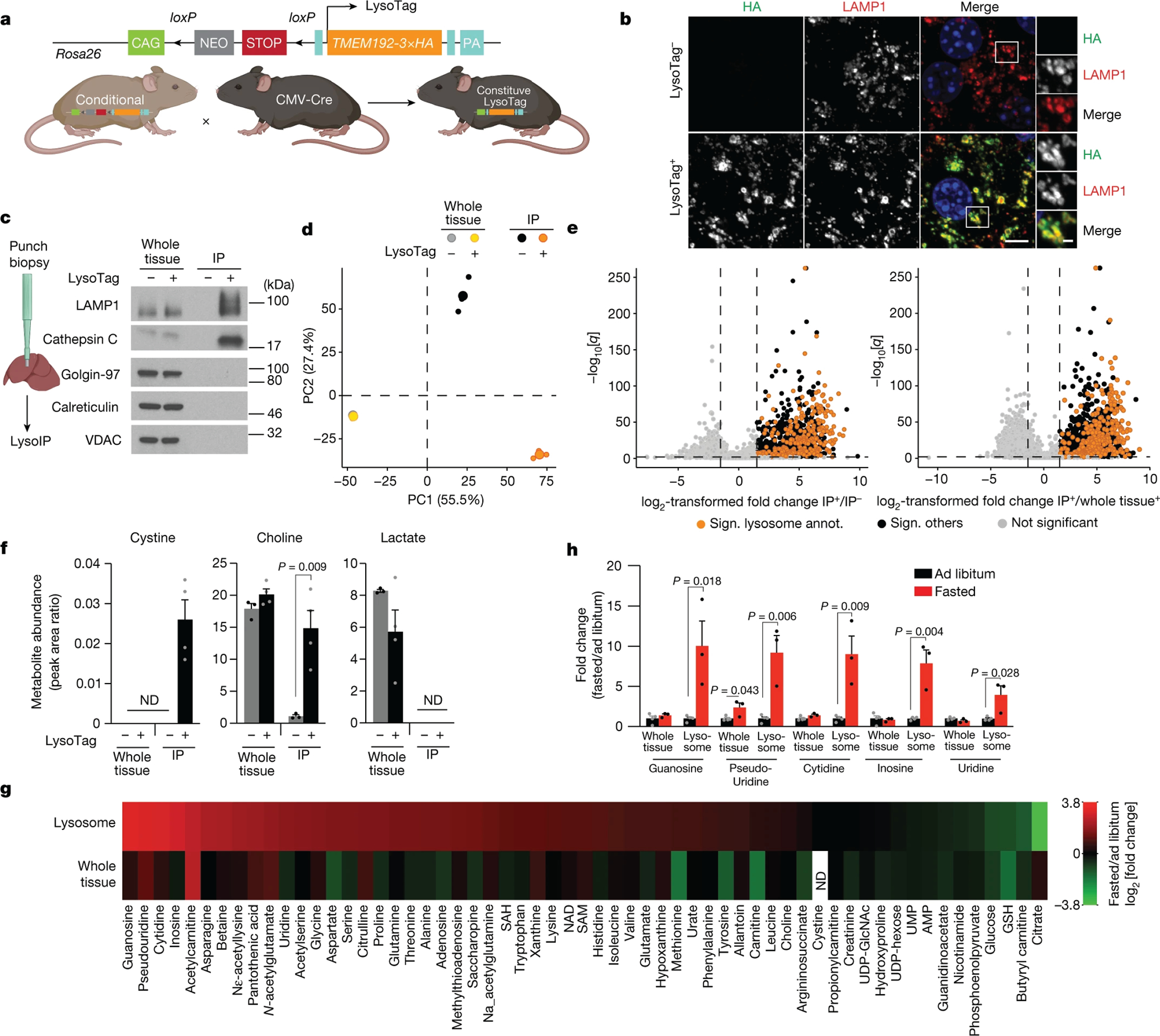
(A) Schematic of the TMEM192–3xHA fusion gene (LysoTag) in Rosa26 locus and the generation of the constitutive LysoTag line.
(B) Immunofluorescence analyses of TMEM192–3xHA (HA, green) and lysosomes (Lamp-1, red) in the liver. Blue marks nuclei. Scale bar = 5 μm and 1 μm in insets. Micrographs are representative of three independent experiments.
(C) Immunoblot analyses for protein markers of subcellular compartments in whole tissues, purified lysosomes (IP: LysoTag+), and control immunoprecipitates (IP: LysoTag−). Golgin-97, VDAC and Calreticulin were used for the Golgi, mitochondria and ER, respectively, and Lamp-1 and Cathepsin C for lysosomes. Immunoblots are representative of three independent experiments.
(D, E) Proteomic analyses of liver LysoIPs. Principal component analysis comparing levels of detected proteins (D). (E) Volcano plot showing that previously annotated lysosomal proteins (orange dots) are enriched in lysosomes. Enriched unannotated proteins are in black. Vertical dashed lines indicate log2 fold change > 1.5 in LysoTag+ IPs either over control IP (left) or whole tissue (right). Horizontal dashed lines indicate a q value < 0.01. n=3 male mice with average age of 6.5 weeks. Paired t-test performed at the peptide precursor level. P values were corrected for multiple testing as described by Storey42.
(F) Relative abundance of indicated metabolites in whole tissue and IPs. Not detected (N.D.). Values are mean ± SEM. LysoTag+ (n=4) and LysoTag− (n=3).
(G) Heat map of the mean of log2-transformed fold changes in metabolite levels after a 16-hour fast. See Supplementary Table 2.
(H) Fold changes in the levels of nucleosides upon fasting (mean ± SEM, ad libitum (n=4) or fasted (n=3) for G and H. Two-tailed unpaired t-test (F, H). Male mice with an average age of 5.5 weeks were used (F-H).
We modified our protocol12 for preparing anti-HA immunoprecipitates (IPs) from cultured cells for use with tissues and applied it to livers from control or LysoTag mice. IPs from LysoTag mice (which we call LysoIPs), but not from control mice, were heavily enriched for the lysosomal membrane and lumenal proteins Lamp1 and Cathepsin C, respectively, but free of markers of other organelles, including the Golgi, endoplasmic reticulum (ER), and mitochondria (Fig. 1C). Furthermore, data independent acquisition (DIA)-based proteomics confirmed that the LysoIPs (Fig. 1D, E, Supplementary Table 1) were highly enriched for proteins previously annotated as lysosomal regardless of whether we determined lysosomal enrichment relative to the abundance of these proteins in control IPs or whole-liver lysates (Fig. 1E). The LysoIPs were mostly depleted for proteins from other compartments, except for a select set of proteins mostly associated with the Golgi, suggesting a possible interaction between organelles (Supplementary Table 1). Lysosomal membrane and lumenal proteins were enriched to similar degrees, indicating that lysosomes purified by the LysoIP protocol are intact (Supplementary Table 1). Finally, we observed enrichment of some endosomal markers, although to a lower extent than those for lysosomes (Supplementary Table 1, Extended Data Fig. 1G).
To test the feasibility of our method for studying lysosomal metabolites, we measured cystine, which is known to be enriched in lysosomes12,14. Indeed, the liver LysoIPs had much higher cystine levels than the control IPs or whole-liver samples, in which it was below the limit of detection (Fig. 1F). The LysoIPs also contained metabolites that we previously detected in lysosomes from cultured cells12, including choline (Fig. 1F, Supplementary Table 2). Importantly, abundant cytosolic metabolites, such as lactate, were either not detected in the LysoIPs or equally abundant in the control IPs that serve to reveal metabolites that bind non-specifically to the magnetic beads (Fig. 1F, Supplementary Table 2).
To test how lysosomal contents change under different physiological states, we generated targeted metabolite profiles of lysosomes from the livers of mice fed ad libitum or fasted. While some of the fasting-induced changes in lysosomal metabolites were also detected at the whole-tissue level (e.g., acetyl-carnitine), others were observed only in lysosomes (Fig. 1G, Supplementary Table 2). For example, fasting increased the levels in lysosomes of all nucleosides, without impacting, except for pseudouridine, those in whole-liver samples (Fig. 1G, H, Supplementary Table 2). Collectively, these data show that the LysoIP can be used to rapidly isolate intact and highly pure lysosomes from mouse organs and to study metabolite changes not detectable with traditional tissue-based metabolite profiling.
CLN3 loss alters lysosomal metabolome
To determine the utility of the LysoIP approach for deorphaning gene function, we turned to CLN3, the gene mutated in the lethal neurodegenerative disorder Batten disease15,16, and whose function remains obscure17,18. The CLN3 gene product is a multi-pass membrane protein that localizes to the lysosome19–23 and its loss is associated with defects in multiple cellular functions including autophagy, membrane fusion, vesicular trafficking, and arginine transport17,18. While its localization might suggest it transports molecules across the lysosomal membrane, only recently has CLN3 been suggested to be an atypical solute carrier based on structural predictions24.
To identify changes in the lysosomal metabolome caused by CLN3 loss we crossed the LysoTag mice with Cln3−/− mice25,26 (Extended Data Fig. 2A). As in the liver, TMEM192–3xHA colocalized with Lamp1-positive lysosomes in the brains of all genotypes (Extended Data Fig. 2B). Consistent with previous reports, Lamp1-positive compartments in neurons lacking CLN3 were noticeably more abundant than in control cells (Extended Data Fig. 2B)27. In addition, our protocol successfully yielded pure and intact lysosomes from the brain, regardless of CLN3 status (Fig. 2A).
Fig. 2: Loss of CLN3 leads to significant alterations in metabolite levels in brain lysosomes.
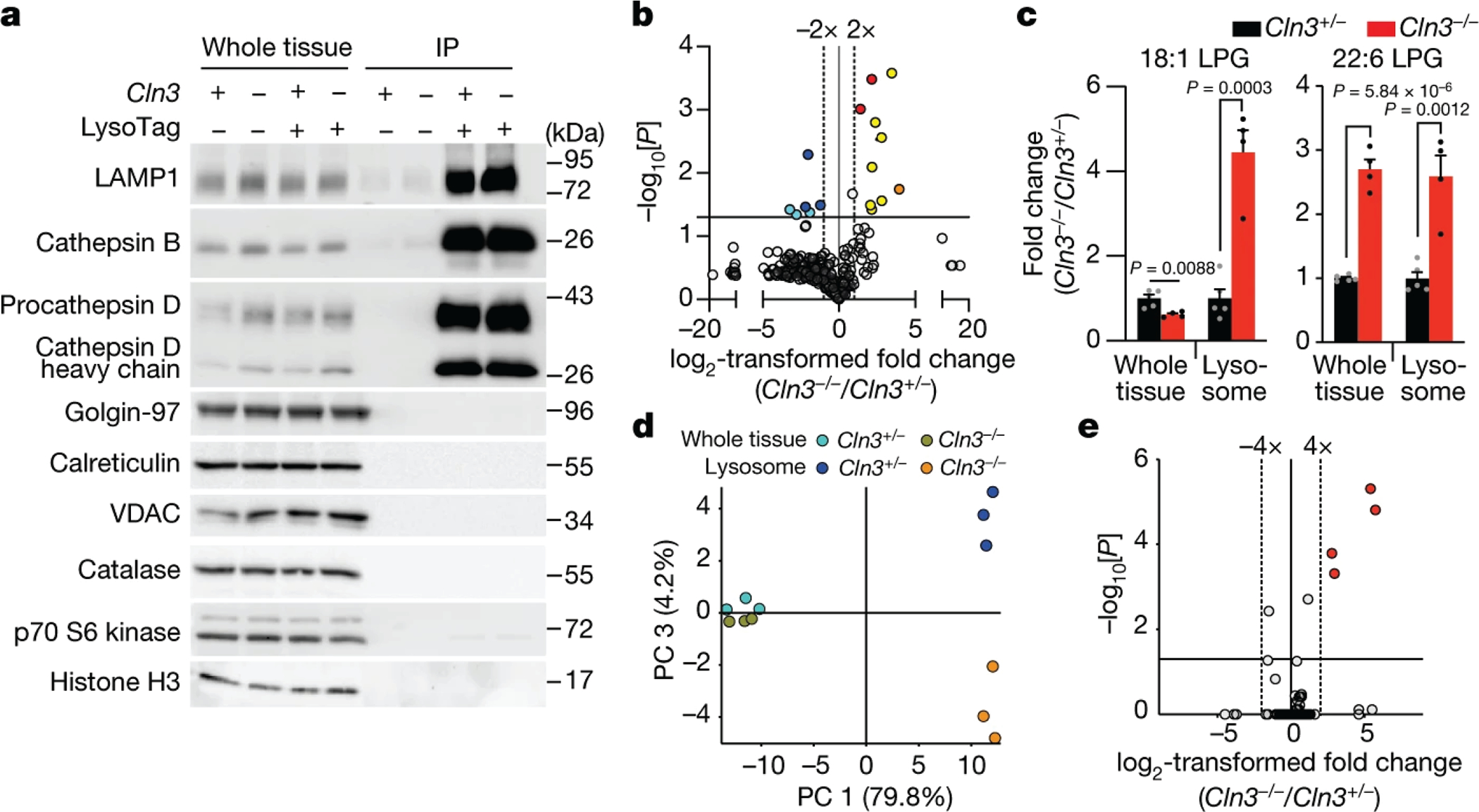
(A) Immunoblot analyses of organellar protein markers in whole-tissue fractions, purified lysosomes, and control IP. Lamp-1, Cathepsin B and D were used for lysosomes. Golgin-97, Calreticulin, VDAC, Catalase, S6 Kinase and Histone H3 were used for the Golgi, ER, mitochondria, peroxisomes, the cytosol and the nucleus, respectively. Immunoblots are representative of three independent experiments.
(B) Volcano plot comparing untargeted LysoIP lipidomic data from lysosomes derived from brains of Cln3+/− and Cln3−/− mice. Significantly changing lipids belong to phospholipids (yellow), lysophosphatidylglycerol (LPG) (red), lysophosphatidylcholine (orange), bis(monoacylglycero)phosphate (BMP) (blue). BMP/PG (cyan) annotation was used when MS/MS fragmentation was not acquired. Data in Supplementary Table 3. Horizontal line indicates a p-value of 0.05, and vertical dotted lines a fold change of 2. Two-tailed unpaired t-test was used.
(C) Targeted analyses of LPGs. Fold changes in LPGs in lysosomes from Cln3+/− and Cln3−/− mice were calculated after subtracting background from control IP. Values represent mean ± SEM (Two-tailed unpaired t-test). For B and C, n=5 and 4 female mice (7 months of age) for Cln3+/− and Cln3−/− mice, respectively.
(D, E) Untargeted polar metabolite analyses of brain lysosomes and tissues upon CLN3 loss (Supplementary Table 4 and methods). Principal component (PC) analysis comparing the levels of the 189 unique compounds (see text) between whole-tissue or lysosome samples from brains of Cln3+/− and Cln3−/− mice in (D). (E) Volcano plot of lysosomal changes in these compounds. Horizontal line indicates a p-value of 0.05, and vertical ones represent fold changes of 4. n=3 male mice (7 months of age) per genotype. P-values were calculated by ANOVA with Tukey HSD test. The p-values were corrected by the Benjamini-Hochberg method, FDR= 5%.
Because alterations in lysosome-mediated lipid catabolism have been associated with multiple forms of LSDs28,29, we used untargeted lipidomics (see methods) to analyze brain lysosomes from the mice lacking CLN3. Consistent with previous reports30, we detected a significant decrease in tissue and lysosomal levels of bis(monoacylglycero)phosphates (BMPs), a class of lysosomal lipids (Fig. 2B, Extended Data Fig. 2C, Supplementary Table 3). Interestingly, we also found significant increases in various glycerophospholipids (termed phospholipids hereafter) (Supplementary Table 3), including the degradation intermediate lysophosphatidylglycerol (LPG) (Fig. 2B, C, Extended Data Fig. 2C). While these changes could represent a link between CLN3 function and phospholipid catabolism in the lysosome, which was previously noted in yeast models31, it is not clear how they are directly linked to CLN3 loss.
Thus, we turned to untargeted polar metabolomics to search for additional lysosomal metabolites that could be impacted by CLN3 loss (see methods and Extended Data Fig. 3A). We detected 1073 putative compounds in lysosomes after background removal and signal thresholding (Supplementary Table 4). After further filtering for compounds in known compound databases and with reliable fragmentation data we arrived at 189 unique compounds for which we had various degrees of confidence in their identities based on the Metabolomics Standards Initiative (MSI) guidelines (Extended Data Fig. 3A, Supplementary Table 4). Principal component analyses of the 189 compounds revealed that while the metabolic landscapes of the Cln3+/− and Cln3−/− whole-brain samples were similar, those in their respective lysosomes were distinct (Fig. 2D), with four compounds at high levels in lysosomes lacking CLN3 to be the most differentially abundant (Fig. 2E). Critically, these same four compounds were the most significantly affected by CLN3 loss independently of how we filtered the datasets (Supplementary Table 4), indicating that CLN3 function impacts a specific set of metabolites in lysosomes rather than the whole lysosomal metabolome.
CLN3-deficient lysosomes accumulate GPDs
To determine the chemical identity of the accumulating metabolites, we used MS1 as well as MS/MS fragmentation data (Extended Data Fig. 3A). This effort indicated that the four metabolites were likely all glycerophosphodiesters (GPDs), which are formed by the deacylation of phospholipids (Supplementary Table 4, Fig. 3A). We tentatively annotated the compounds as glycerophosphoglycerol (GPG), glycerophosphocholine (GPC), glycerophosphoinositol (GPI), and glycerophosphoethanolamine (GPE), which represent four of the five major GPDs (Fig. 3A). To validate the compounds as GPDs, we showed that commercially available standards for GPC and GPI had retention times and MS/MS spectra matching those in biological samples (Fig. 3B, C, Extended Data Fig. 3B) and (Fig. 3D, E, Extended Data Fig. 3C). For GPE and GPG, because of the lack of commercial standards, we generated these metabolites in house, and validated that the retention times, m/z values and MS/MS spectrum from the biological samples matched those of the standards (Extended Data Fig. 3D–F) and, when available, previously experimentally reported fragments32. While the fifth major GPD, glycerophosphoserine (GPS), was not detected in the original untargeted metabolite profiles, follow-up work showed that its lysosomal levels are also increased upon CLN3 loss (Fig. 3G). Its identity was validated by comparing m/z values and MS/MS spectrum in our samples with those of in silico fragmentation patterns generated with competitive fragmentation modelling-ID (CFM-ID)33 (Extended Data Fig. 3G). Altogether, our approach rigorously identified and annotated the metabolites that accumulate in the lysosome upon CLN3 loss according to MSI guidelines34 (Fig. 3F). We also manually extracted ion chromatograms for GPDs, which confirmed large and significant accumulations of the GPDs in brain lysosomes from the Batten disease model, but also showed almost no or only very modest increases in their corresponding levels in the whole brain samples (Fig. 3G). These data are consistent with lysosomes occupying a minor fraction of total cellular volume and underscore the value of directly measuring metabolites in lysosomes.
Fig. 3: Glycerophosphodiesters (GPDs) accumulate in lysosomes upon CLN3 loss.
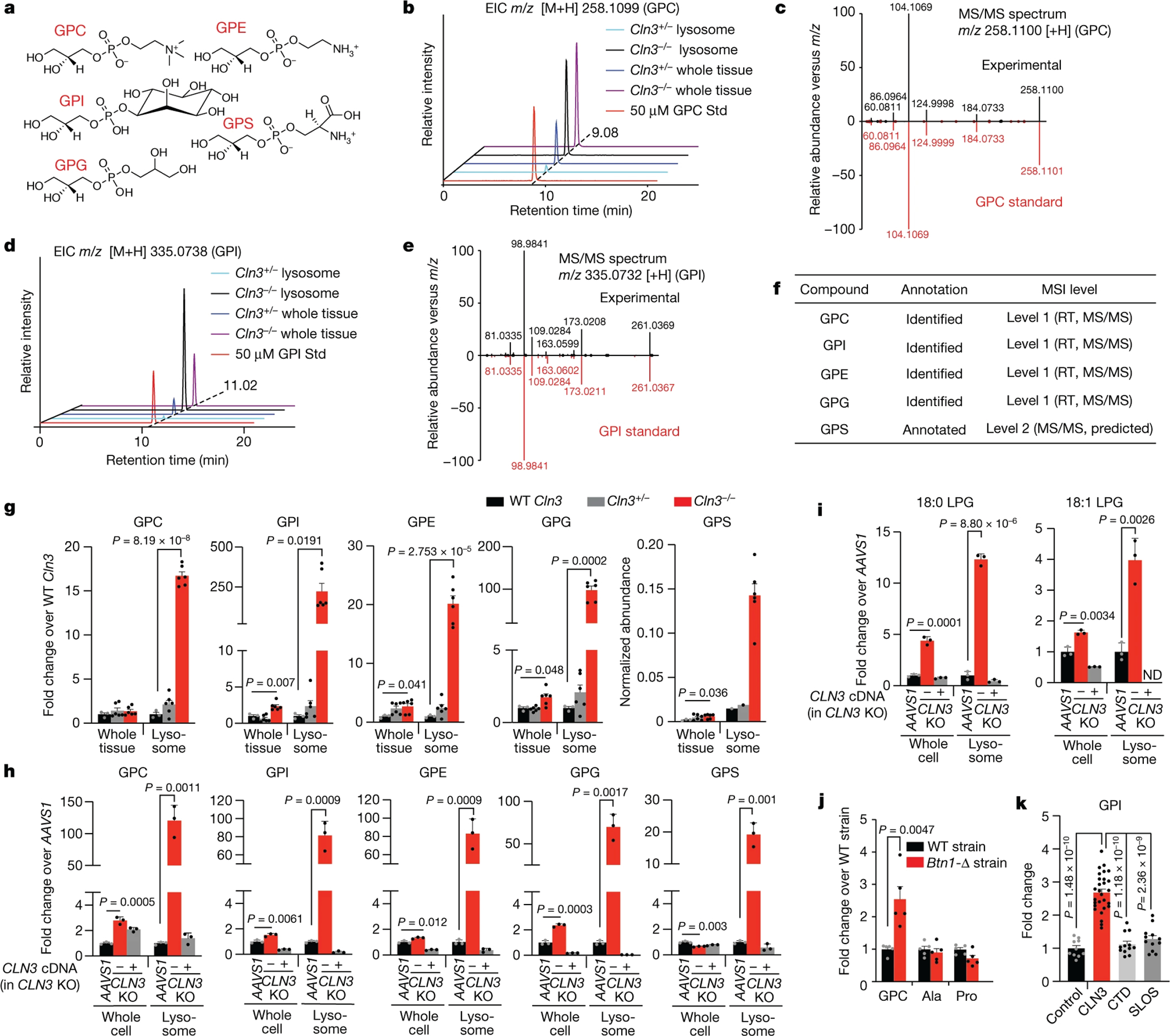
(A) The chemical structure of GPDs in this study.
(B) Extracted ion chromatogram (EIC) for GPC across a range of samples alongside an authentic standard (Std) showing a matching retention time.
(C) Mirror plot for MS/MS spectrum of GPC in a representative sample (lysosomes from Cln3−/− mouse brain) and an authentic standard.
(D) As in B for GPI. (E) As in C for GPI.
(F) The level of confidence in the identification of GPDs in this study according to MSI guidelines.
(G) Targeted analyses of GPDs in brain lysosomes. Fold changes between lysosomes were calculated after subtracting the background signal in control IPs and normalizing to methionine. GPS was only detected in one WT and Cln3+/− lysosomal sample, thus its abundance was reported instead. Data are mean ± SEM; WT Cln3 (n=3), Cln3+/− (n=6) and Cln3−/− (n=6). Male mice from two independent experiments including that in Fig. 2E.
(H) Fold change in the levels of GPDs between HEK-293T cells and their lysosomes upon CLN3 knockout and rescue. sgRNA targeting the AAVS locus was used as control.
(I) Targeted analyses of LPGs from cells as in (H). Data are mean ± SD (H, I); n=3 biologically independent samples (H, I).
(J) Fold change in the levels of GPC, alanine (Ala), and proline (Pro) between wild-type (WT) and Btn1-Δ yeast strains. Data are mean ± SEM (n=5 biologically independent samples).
(K) Fold change in GPI levels in CSF from patients with CLN3 Batten disease (CLN3, n=28), creatine transporter deficiency (CTD, n=12), and Smith-Lemli-Opitz Syndrome (SLOS, n=12) compared to anonymized pediatric controls (Control, n=10). Data are mean ± SEM. See Supplementary Table 5 for the characteristics of study participants. Two-tailed unpaired t-test (G-K).
To rule out that the increase in GPDs in the brain lysosomes of the Batten disease mice is a consequence of neurodevelopment defects, we generated and analyzed HEK-293T cells (Extended Data Fig. 3H) that are null for CLN3 and also express TMEM192–3xHA. In lysosomes from these cells, all five GPDs also accumulated to very high levels and reverted to control amounts or below upon expression of the CLN3 cDNA (Fig. 3H, Extended Data Fig. 3I). Importantly, we found that regardless of the tagging system used to purify lysosomes, similar levels of GPD accumulation are observed upon CLN3 loss (Extended Data Fig. 3J). As in the brain, the HEK-293T lysosomes lacking CLN3 also had increases in LPG levels (Fig. 3I).
Human CLN3 can complement the functions of its yeast homologue Btn1p in multiple lysosomal assays17, indicating that they share the same function. Indeed, yeast lacking Btn1p significantly accumulated GPC, the only detectable GPD in yeast, suggesting that the function of CLN3 in GPD metabolism is conserved (Fig. 3J). Importantly, we found elevated levels of GPI in the cerebrospinal fluid (CSF) of individuals with CLN3 Batten disease compared to that from individuals without CLN3 disease35 (Fig. 3K), suggesting its potential use as a clinical biomarker.
Thus, the use of untargeted metabolomics in combination with the LysoIP approach reveals that in the mouse brain and human cells in culture, loss of CLN3 causes a large lysosomal accumulation of GPDs along with a more modest increase in lysophospholipids, which are the metabolites directly upstream of GPDs in the pathway for phospholipid catabolism. Although the specific lysosomal phospholipases that generate GPDs from phospholipids have not been defined, we note that lysosomes do contain the phospholipase A/B activities required for their release from phospholipids36.
CLN3 is required for the efflux of GPDs
The acidic lysosomal lumen establishes a proton gradient with the cytosol that drives the efflux of several metabolites, including GPDs (Extended Data Fig. 4C, D)12. A plausible explanation for GPD accumulation is that CLN3 loss increases lysosomal pH, which attenuates their egress from the lysosome. However, consistent with previous reports37, CLN3 loss in HEK-293T cells does not increase lysosomal pH but, if anything, leads to a minor decrease (Extended Data Fig. 4A, B). Consistent with this, we observed no lysosomal accumulation, upon CLN3 loss, of amino acids (proline, alanine and glutamate) whose efflux from the lysosome is dependent on pH, which functionally confirms our pH measurements (Extended Data Fig. 4D). Thus, the lysosomal accumulation of GPDs in CLN3-deficient lysosomes is independent of perturbations in lysosomal pH.
Given these data and that CLN3 is a multi-pass transmembrane protein, we hypothesized that CLN3 can be either a lysosomal glycerophosphodiesterase (GDE) required for hydrolyzing GPDs into glycerol 3-phosphate and their corresponding headgroups, an activity that is mostly associated with membrane proteins38, or, alternatively, required for the efflux from lysosomes of GPDs produced by the lysosomal catabolism of phospholipids. To test the former, we purified CLN3 and, as a positive control, the established glycerophosphodiesterase GDE1. While GDE1 readily hydrolyzed deuterated GPC in vitro, we found no such activity for CLN3 (Extended Data Fig. 4E). These results are consistent with previous reports indicating that GPDs are the final products of phospholipid degradation in mammalian lysosomes36.
To test the possibility that CLN3 is required for the lysosomal efflux of GPDs, we utilized an assay in HEK-293T cells in which we deliver to lysosomes via endocytosis a phospholipid with a labelled head group, which upon lysosomal catabolism produces a labelled GPD whose fate we can monitor by mass spectrometry (Fig. 4A). Specifically, we conjugated phosphatidylglycerol with a deuterium-labeled head group (D5-PG) to bovine serum albumin (BSA) and added it to the medium of serum-starved HEK-293T cells expressing TMEM192–3xHA (Fig. 4A). The relative amounts of the deuterated phosphatidylglycerol were the same in the whole-cell and lysosomal fractions of control and CLN3-null cells, indicating that loss of CLN3 does not impact uptake of the phospholipid into lysosomes (Fig. 4B). Strikingly, we could only detect a small signal for deuterated GPG—the GPD released from the labelled phosphatidylglycerol—in the whole-cell and lysosomal samples from the control cells, consistent with GPG being quickly effluxed from normal lysosomes and further catabolized in the cytosol. In contrast, in the CLN3-null cells deuterated GPG readily accumulated in lysosomes and was also easily detected in whole-cell samples (Fig. 4C). These results indicate that in cells lacking CLN3 the labelled GPG is trapped in lysosomes and so spared from catabolism in the cytosol (Fig. 4C). Consistent with our previous metabolite profiles (Fig 3I), in the CLN3-null cells we also observe a significant accumulation of labeled lysophosphatidylglycerols (Fig. 4D), the metabolites directly upstream of GPD. Thus, we conclude that CLN3 is required for transporting out of the lysosome the GPDs released from phospholipids.
Fig. 4: CLN3 is required for the efflux of GPDs out of lysosomes.
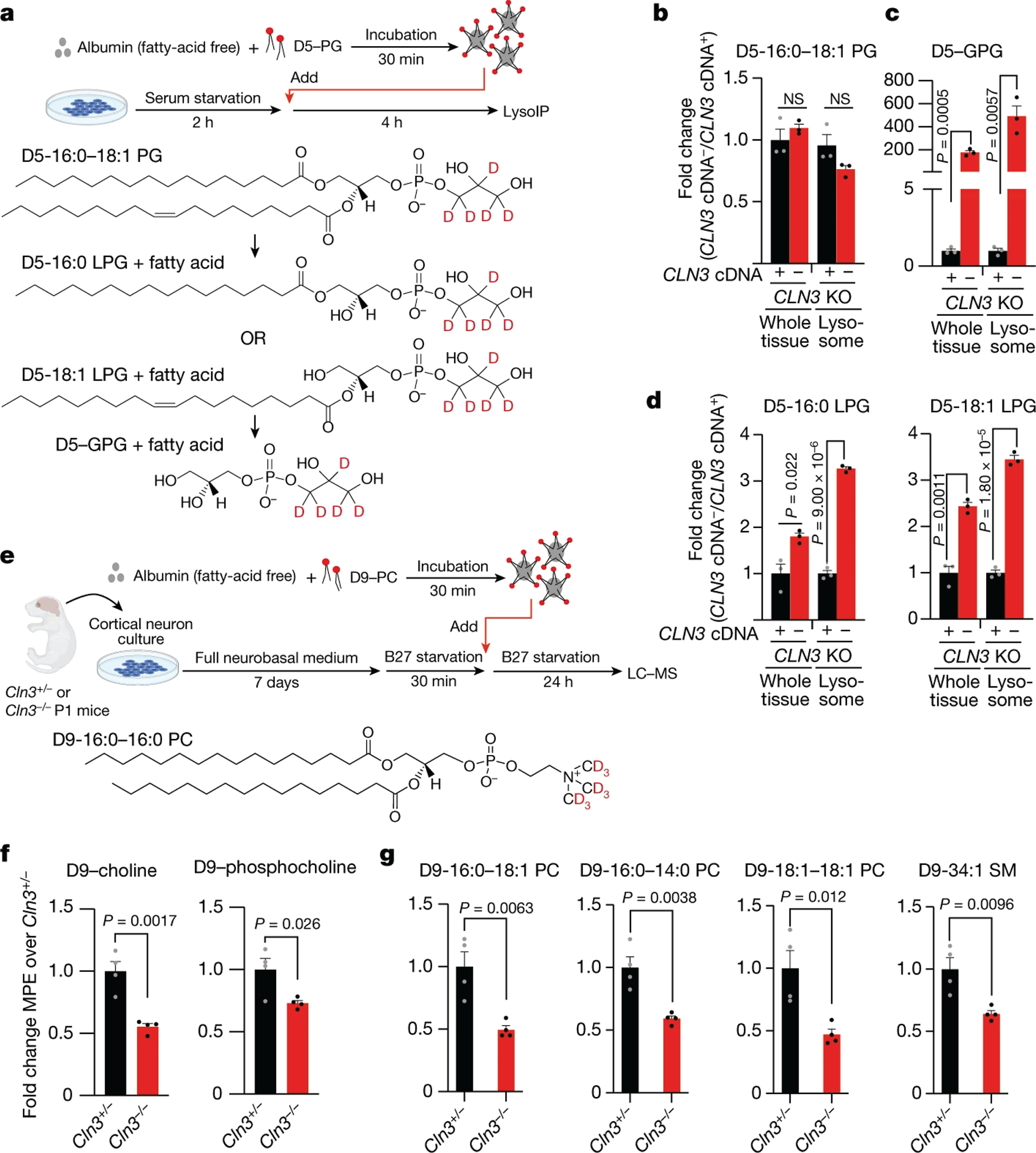
(A) Depiction of an approach to deliver exogenously added deuterated phospholipids to lysosomes. D5-PG: phosphatidylglycerol with 5 deuterium atoms (D) in its headgroup.
(B) CLN3 loss does not affect the cellular uptake of exogenous D5-PG. Fold changes in the whole-cell and lysosomal abundances of D5-PG in CLN3 KO cells relative to those in the CLN3 KO cells rescued with the CLN3 cDNA.
(C) Loss of CLN3 blocks the efflux of GPG from lysosomes. Data are presented as in (B) but for D5-GPG.
(D) LPGs generated in lysosomes accumulate upon CLN3 loss. Data are presented as in (B) but for the two possible D5-LPGs that are generated from the degradation of the exogenously added D5-PG. Data are mean ± SD, n=3 (B-D).
(E) to (G) GPC-derived choline contributes to lipid biosynthesis in a CLN3-dependent manner in primary cortical neurons (E) Depiction of the approach to trace the metabolic fate of the choline moiety of GPC, which is generated from Phosphatidylcholine (PC) degradation in the lysosome. Phosphatidylcholine having 9 deuterium atoms in its choline headgroup (D9–16:0–16:0 PC) was delivered to lysosomes in primary cultured neurons isolated from the cerebral cortex of Cln3+/− (control) or Cln3−/− mice. Mouse drawing from BioRender.
(F) CLN3 loss significantly reduces the cellular levels of deuterated phosphocholine and its precursor choline.
(G) CLN3 loss leads to a reduced contribution of choline derived from lysosomal GPC to the biosynthesis of PC and sphingomyelin (SM) in the cell.
Data are presented as fold changes in the whole-cell molar percent enrichment (MPE) of D9-choline-containing metabolites in cortical neuron cultures prepared from Cln3−/− mice relative to those from Cln3+/− animals. Data are mean ± SEM, n=4 (F, G). Two-tailed unpaired t-test (B-D, F-G). n represents biologically independent samples (B-D, F-G).
To further validate this model and to understand the impact of impaired GPD recycling in neurons, we used phosphatidylcholine with a deuterium-labeled head group (D9-PC) to trace the metabolic fate of its choline moiety in the cell, which is generated by the catabolism of GPC by GDE activity in the cytoplasm (Fig. 4E). CLN3 loss significantly reduced the pools of labeled choline and phosphocholine in primary neuronal cultures (Fig. 4F). Consequently, we observed a significant reduction in the deuterated isotopomers of multiple choline-containing lipid species, including those of PC and sphingomyelin (SM) (Fig. 4G). Of importance, loss of CLN3 did not decrease the uptake of the tracer into neuronal cells (Extended Data Fig. 4F), and did not affect lipid biosynthesis or turnover as indicated by the similar labeling patterns, regardless of CLN3 status, when free D9-choline was used as a tracer (Extended Data Fig. 4G). Altogether, these data further support a critical role for CLN3 in the egress of GPDs from the lysosome for their catabolism and utilization by the rest of the cell.
Discussion
Interestingly, over 60 years ago Christian deDuve discovered that GPDs are terminal products of phospholipid degradation in the lysosome36. This finding implies that a mechanism must exist to efflux GPDs from the lysosome so that they can be degraded by GDEs in the cytosol36,38. However, no transporter for GPDs has been identified in vertebrates, although a plasma membrane GPD transporter has been reported in yeast39. Here, we show that CLN3, whose loss causes a severe neurodegenerative disease in children, is required for the efflux of GPDs from the lysosome. Its membrane localization and predicted atypical solute carrier structure24 suggests that CLN3 is a lysosomal GPD exporter or a key component of one, although further work using purified CLN3 will be needed to test this possibility. Additionally, the massive accumulation of GPDs caused by CLN3 loss might inhibit as yet unidentified enzymes upstream of GPD production to cause the increase we observe in lysophospholipids, which are known to be neurotoxic40. Indeed, GPDs have been previously reported to inhibit lysophospholipase activities in vitro41. Furthermore, our tracing experiments suggest that CLN3 loss might contribute to neurodegeneration by causing the extralysosomal deprivation of nutrients that are essential for brain cell function and membrane integrity such as choline in addition to the potentially toxic effects of storage material in the lysosome2.
Our work provides a framework for future studies into how loss of CLN3 might impact cellular homeostasis, and, more generally, for using the in vivo LysoIP method in combination with untargeted metabolite profiling for understanding the function of disease-associated lysosomal proteins.
Methods
Mouse studies
Mice were maintained on a standard light-dark cycle with access to food and water ad libitum in a room that have a controlled temperature of 22°C, and humidity (around 50%). Cages were cleaned every 4–5 days and supplies of water and food are checked daily. For the generation of Rosa26; lox-stop-lox-TMEM192–3xHA, the TMEM192 coding sequence containing a C-terminal 3xHA epitope tag was inserted into the CTV vector (Addgene plasmid # 15912)13, linearized with AsiSI and electroporated into embryonic stem cells of the 129 background. Embryonic stem (ES) cell colonies were picked, and successfully targeted clones were identified by long-range PCR amplifying regions spanning each homology arm. Positive ES cell clones with proper integration into the Rosa26 locus were then injected into blastocysts and transferred into pseudo-pregnant females to obtain chimeric mice. Chimeras were bred to C57BL/6J mice for at least three generations before maintaining the line in a homozygous form. To generate a constitutive LysoTag expressing line, Rosa26; lox-stop-lox-TMEM192–3xHA mice were crossed with CMV-Cre transgenic mice of the C57BL/6J background, excision of lox-stop-lox was validated using our genotyping PCR method (see details below), and positive progeny were bred to C57BL/6J mice for at least three generations. The conditional LysoTag mouse was deposited at the Jackson Laboratory (Strain #035401) with Common Name: conditional LysoTag (Rosa26; lox-stop-lox-TMEM192–3xHA).
The Batten disease mouse model (Cln3−/−) was purchased from the Jackson Laboratory (stock no. 029471, B6.129S6-Cln3tm1Nbm/J) and maintained in the homozygous state. For LysoIP experiments, Cln3−/− mice were bred with LysoTag mice to generate LysoTag; Cln3+/− (used as controls) and the LysoTag; Cln3−/− experimental group. Statistical methods were not used to predetermine sample size. Sample blinding was not performed for most experiments as they did not involve qualitative scoring (see reporting summary for details).
All procedures involving mice were carried out in accordance with the approved guidelines by the Institutional Animal Care and Use Committee at MIT, the Whitehead Institute for Biomedical Research, and Stanford University.
Mouse genotyping
Genomic DNA was isolated from ear biopsies using QuickExtract DNA Extraction Solution (Lucigen) and polymerase chain reaction (PCR) was performed using GoTaq DNA polymerase (Promega). To check for the presence of lox-stop-lox-TMEM192–3xHA cassette in the Rosa26 locus, two pairs of primers were designed. The non-targeted allele will be amplified by primer pair P1 which produces a PCR product of 588 base pairs (bps), whereas if the lox-stop-lox-TMEM192–3xHA cassette exists it will be amplified by primer pair P2, which gives a 215 bps product. Primer sequences are: P1F: 5′-CAGTAAGGGAGCTGCAGTGG-3′; P1R: 5′-GCAGAAGGAGCGGGAGAAAT-3′; P2F: 5′-TACCTGAAGCGACACAATGC-3′ (within TMEM192 sequence); P2R: 5′-CCGCTCCCTCCAGCATAATC-3′ (within HA sequence). The PCR reaction was performed with four primers in multiplex using the following conditions: 95°C for 3 minutes followed by 35 cycles of 95°C for 30 seconds, 56°C for 30 seconds, and 72°C for 2 minutes, final extension step was added for 72°C for 5 minutes. In order to test for the excision of lox-stop-lox fragment by the Cre-recombinase, two forward primers were designed; one within the loxp stop cassette and another upstream the first loxp site, both having a common reverse primer within the TMEM192 sequence. The non-excised allele will generate a 331 bps product, while successful excision of the loxp-stop-loxp site gives a 193 bps product. Primer sequences were: P3F1: 5′-GGGCAACGTGCTGGTTATTG-3′; P3F2: 5′-CCTCCCCCTGAACCTGAAAC-3′; P3R: 5′GAAGCTGGGCATCCAGAAGT-3. The PCR reaction was performed with three primers using the following conditions: 95°C for 3 minutes followed by 35 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1:20 minutes and a final extension step was added for 72°C for 5 minutes.
Antibodies, chemicals and plasmids
Detailed information on the antibodies, chemicals and plasmids used in this study are listed in Supplementary Table 6. Additional reagents include: Secondary antibodies for IF (used at 1:1000): Goat anti-Rat Alexa Fluor 488 (Thermo A-11006), Goat anti-rabbit Alexa Fluor 488 (Thermo A-21206), Goat anti-Mouse Alexa Fluor 647 (ThermoA-21240), Goat anti-Mouse Alexa Fluor 488 (Thermo, A-11001), and Goat anti-rabbit Alexa Fluor 594 (Thermo, A32740). Secondary antibodies for immunoblotting (used at 1:3000): Anti-rabbit HRP (CST, 7074), anti-mouse HRP (CST, 7076), anti-rat HRP (CST, 7077), and Normal Donkey Serum, which were purchased from Cell Signaling Technology (CST); Inactivated fetal calf serum (IFS) from Invitrogen; DMEM high glucose, anti-HA magnetic beads, and ECL2 western blotting substrate from Thermo Scientific; XtremeGene9 DNA Transfection Reagent, Complete Protease Inhibitor Cocktail, and PhosSTOP phosphatase inhibitor from Roche; and QuickExtract DNA Extraction Solution from Lucigen.
Rapid isolation of lysosomes from brain and liver tissues (LysoIP)
Immediately following euthanasia, mouse brains were collected and cerebral hemispheres dissected on an ice-cold plastic dish. For liver samples, we obtained a small round piece using a biopsy punch (4 mm diameter), which helped to reduce variability between samples and minimize processing time prior to the LysoIP. The liver tissues and cerebral hemispheres were used immediately without freezing following similar steps as described in the original LysoIP protocol12.
Protein analyses
4 mm liver biopsy or half cerebral hemisphere samples were gently homogenized in 1 mL ice-cold PBS supplemented with protease and phosphatase inhibitor cocktails (Roche) using a 2 mL dounce homogenizer (douncer). Both liver and brain samples required 25 strokes for optimal homogenization. Homogenates were collected and 25 μL was saved on ice as the input fraction while the remaining was spun down at 1,000×g for 2 minutes at 4°C. The supernatant was then incubated with anti-HA beads that were prewashed 3 times with same buffer, incubated for 20 minutes on a gentle rotator shaker at 4°C, and followed by three rounds of washing in same cold buffer. Washes were done by pipetting the beads three times in each wash, then discarding the supernatant by placing the tube on a magnet to trap the beads. To increase the purity of the lysosomal preparation, we changed the tube during the last wash and always used protein low binding tubes. The input and lysosomal fractions were resuspended in ice-cold Triton lysis buffer (40 mM HEPES pH 7.4, 1% Triton X-100, 10 mM β-glycerol phosphate, 10 mM pyrophosphate, 2.5 mM MgCl2 and protease and phosphatase inhibitor cocktails). Samples were incubated for 10 minutes at 4°C and centrifuged at maximum speed in cold. The supernatant was collected and saved for analysis. The lysosomal fraction was not centrifuged but rather incubated on the magnet and transferred to new tubes twice. For immunoblots, an equivalent of 0.08% and 15% of the whole tissue input and captured lysosomes were run, respectively.
Polar metabolite and lipid analyses
The procedure was similar to the steps above but processing was in ice-cold KPBS (136 mM KCl, 10 mM KH2PO4, pH 7.25 in Optima LC/MS water) with no protease or phosphatase inhibitors. The homogenates were incubated with pre-washed anti-HA beads for 5 minutes for polar metabolites and 20 minutes for lipid experiments. We noticed that incubation for 3–15 minutes still returns similar results for polar metabolites, but 5 minutes incubation was used for convenience. For polar metabolites, the input fractions were resuspended in 225 μL, while the lysosomal fractions were resuspended in 50 μL of ice-cold 80% methanol in LC/MS water containing 500 nM isotope-labeled amino acids used as internal standards (Cambridge Isotope Laboratories). Samples were stored at −80°C until analyzed. On the day of analysis, samples were vortexed for 10 minutes at 4°C and centrifuged at 17,000×g. For lipid extraction, 800 μL of chloroform:methanol at ratio of 2:1 (v/v) containing 750 ng/mL of SPLASH LipidoMIX™ internal standard mix (Avanti) followed by the addition of 160 μl of 0.9% (w/v) NaCl and vortexing for 10 minutes at 4°C. The mixture was centrifuged at 3000×g for 15 minutes at 4°C. The lower phase containing lipids was collected and dried using a SpeedVac. At this stage, samples can be stored at −80°C. On the day of analysis, dried lipid extracts were reconstituted in 50 μL of ACN:IPA:H2O at a ratio of 13:6:1 (v/v/v) and vortexed for 10 minutes at 4°C. Samples were centrifuged for 10 minutes at 4°C and transferred to glass insert vials for LC/MS.
Proteomics
Eluates from LysoIPs and aliquots of the matched starting lysates (corresponding to approximately 50 μg of protein extract) were processed as described in Wyant et al.43. Briefly, proteins were solubilized by addition of SDS to a final concentration of 2% (w/v), followed by sonication in Bioruptor Plus (Diagenode) and heating for 10 minutes at 95°C. Following reduction and alkylation, proteins were precipitated by cold acetone precipitation. The resulting pellets were resuspended in digestion buffer (3M Urea in 100mM HEPES pH8.0) and digested by consecutive addition of LysC (Wako, 3 hours at 37°C) and trypsin (Promega, 16 hours at 37°C). The obtained digested peptides were acidified and desalted with a Waters Oasis® HLB μElution Plate 30 μm (Waters) following the manufacturer instructions. The desalted peptides were dissolved in 5% (v/v) acetonitrile, 0.1% (v/v) formic acid to a peptide concentration of approximatively 1 μg/μL and spiked with iRT peptides (Biognosys AG) prior to analysis by LC-MSMS. Approximatively 1μg of reconstituted peptides were analyzed by Data Independent Acquisition (DIA) using the UltiMate 3000 UPLC system (Thermo Fisher Scientific) fitted with a trapping (Waters nanoEase M/Z Symmetry C18, 5 μm, 180 μm × 20 mm) and an analytical column (Waters nanoEase M/Z Peptide C18, 1.7μm, 75μm × 250mm). The outlet of the analytical column was coupled directly to a QExactive HF (Thermo Fisher Scientific) using the Proxeon nanospray source. Solvent A was water, 0.1% (v/v) FA and solvent B was 80% (v/v) acetonitrile, 0.08% (v/v) FA. Peptides were eluted via a non-linear gradient from 1% to 62.5% B in 131 minutes. Total runtime was 150 minutes, including clean-up and column re-equilibration. The S-lens RF value was set to 60. MS acquisition parameters were set as follows: Full scan MS spectra with a mass range 350–1650 m/z were acquired in profile mode in the Orbitrap with resolution of 120,000 FWHM. The filling time was set at maximum of 60 ms with an AGC target of 3× 106 ions. DIA scans were acquired with 40 mass window segments of differing widths across the MS1 mass range. The default charge state was set to 3+. HCD fragmentation (stepped normalized collision energy; 25.5, 27, 30%) was applied and MS/MS spectra were acquired with a resolution of 30,000 FWHM with a fixed first mass of 200 m/z after accumulation of 3×106 ions or after filling time of 35 ms (whichever occurred first). Data were acquired in profile mode. For data acquisition and processing Tune version 2.9 and Xcalibur 4.1 were employed.
Acquired data were processed using Spectronaut Professional v13.10 (Biognosys AG). Raw files were searched by directDIA search with Pulsar (Biognosys AG) against the mouse UniProt database (Mus musculus, entry only, release 2016_01) with a list of common contaminants appended, using default settings. For quantification, default BGS factory settings were used, except: Proteotypicity Filter = Only Protein Group Specific; Major Group Quantity = Median peptide quantity; Major Group Top N = OFF; Minor Group Quantity = Median precursor quantity; Minor Group Top N = OFF; Data Filtering = Qvalue percentile with Fraction = 0.2 and Imputing Strategy = Global Imputing; Normalization Strategy = Local normalization; Row Selection = Automatic. The candidates and protein report tables were exported from Spectronaut and used for volcano plots generation and Principal Component Analysis (PCA), respectively, using R version 3.4.1 and RStudio server version 1.1.463. Protein groups were considered as significantly enriched in LysoIP samples if they displayed a Q value < 0.01 and average log2 ratio > 1.5. Known lysosomal proteins were defined based on Gene Ontology Cellular Component or UniProt sub-cellular localization annotation. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE44 partner repository with the dataset identifier PXD018624.
Polar metabolomics
The initial metabolite profiling was conducted on a QExactive benchtop orbitrap mass spectrometer equipped with an Ion Max source and a HESI II probe, which was coupled to a Dionex UltiMate 3000 HPLC system (Thermo Fisher Scientific). To prevent any systematic bias from possible column carryover, sample order was randomized in LC/MS runs. External mass calibration was performed using the standard calibration mixture every 7 days. Typically, 5 uL were injected onto a SeQuant® ZIC®-pHILIC 150 × 2.1 mm analytical column equipped with a 2.1 × 20 mm guard column (both 5 mm particle size; EMD Millipore). Buffer A was 20 mM ammonium carbonate, 0.1% ammonium hydroxide; Buffer B was acetonitrile. The column oven and autosampler tray were held at 25°C and 4°C, respectively. The chromatographic gradient was run at a flow rate of 0.150 mL/min as follows: 0–20 minutes: linear gradient from 80–20% B; 20–20.5 minutes: linear gradient from 20–80% B; 20.5–28 minutes: hold at 80% B. The mass spectrometer was operated in full-scan, polarity-switching mode, with the spray voltage set to 3.0 kV, the heated capillary held at 275∘C, and the HESI probe held at 350°C. The sheath gas flow was set to 40 units, the auxiliary gas flow was set to 15 units, and the sweep gas flow was set to 1 unit. MS data acquisition was performed in a range of m/z = 70–1000, with the resolution set at 70,000, the AGC target at 1×106, and the maximum injection time at 20 msec. In order to improve sensitivity of detection of LysoTracker DND-99 and deuterated GPG (D5-GPG) in lipid uptake experiments, additional targeted selected ion monitoring (tSIM) scans were included in positive mode, with m/z centered on 400.21150 and 252.08920, respectively. For these scans, the resolution was set to 70,000, the AGC targeted was 1×105, the max IT was 250 msec, and the isolation window around each target mass was set to 2.0 m/z.
Relative quantitation of polar metabolites was performed by extracting ion chromatograms with XCalibur QuanBrowser 4.1 (Thermo Fisher Scientific) using a 5 ppm mass tolerance and referencing an in-house library of chemical standards. Metabolites and runs were subject to pre-defined quality control parameters: CV (standard deviation/ mean peak area across multiple injections of a representative (pooled) biological sample) below 0.25; R2 (linear correlation across a three-point dilution series of the representative (pooled) biological sample) greater than 0.90.
For a metabolite to be lysosomal in the targeted analysis performed in Figure 1, its abundance in the IP from LysoTag+ mice had to be twice higher than that in a control IP in at least 3 of the 4 samples. Furthermore, all of the fold change calculations in the abundance of lysosomal metabolites, where targeted analyses are presented, were determined after subtracting the abundance of the metabolites in the control IPs. For polar targeted metabolites from cultured cells, we also used Lysotracker detected with LC/MS to account for any gross changes in lysosomal mass or capturing efficiency by normalizing metabolite intensities to the total captured LysoTracker divided by the total cellular LysoTracker signal (capturing efficiency). Whole-cell abundances were normalized to total cell volume.
It is important to note that some metabolite profiling data were acquired on an ID-X tribrid mass spectrometer (Thermo Fisher Scientific) at Stanford University. The same HILIC method and column parts were used for a Vanquish Horizon UPLC system. Different mass spectrometer parameters are as follows: Orbitrap resolution, 120,000; maximum injection time, 80 ms. It is worth noting that due to the change of instrumentation, there is an approximately 1 min retention time shift for those polar metabolite data collected at Stanford University hence the difference in the retention time of GPE and GPG standards from those reported in the experimental samples in Supplementary Table 4. Experimental samples were re-run and metabolite identities were validated using the new instrumentation alongside our standards.
Untargeted polar metabolomics
Data were acquired as described above, with additional data-dependent (dd) MS/MS collected on pooled samples to aid with unknown metabolite identification. For ddMS/MS, the top 10 ions in each full scan were isolated with a 1.0-Da window, fragmented with a step-wise collision energy of 15, 30, and 45 units, and analyzed at a resolution of 17,500 with an AGC target of 2×105 and a maximum injection time of 100 msec. The underfill ratio was set to 0. The selection of the top 10 ions was set to isotopic exclusion, a dynamic exclusion window of 5.0 sec, and an exclusion list of background ions based on a solvent blank. Data were analyzed using Compound Discoverer 3.1 (Thermo Fisher Scientific) and by including an in-house mass-list. Normalization was performed using the default total ion count (TIC) parameter. Identification/ annotation of metabolite species was confirmed by matching retention times and MS/MS data to authentic standards. In cases where there was no authentic standard available, MS/MS data were matched first to published experimental MS/MS data (i.e. through the Thermo Fisher mzCloud spectral library), or to predicted MS/MS data produced using CFM-ID33. See Supplementary Table 4 and Figure 3 for details for each metabolite annotated/ identified in this study.
Compound Discoverer (CD) workflow:
Features were extracted in the m/z 70–1000 mass range, from 0.1–16 minutes of the pHILIC chromatographic runs. The signal to noise threshold for feature detection was set to 1.5. Compounds were detected using the following parameter settings: mass tolerance was set to 10 ppm, retention time tolerance was set to 0.4 minutes, minimum peak intensity was set to 100,000 AU, and signal to noise threshold for compound detection was set to 3. Isotopes and adducts were grouped using the default ions and base ions lists. Missing peaks were gap filled and mock extraction blank samples were used to determine background compounds for subsequent subtraction. Areas were normalized by constant median excluding the blank samples.
To predict elemental compositions of the compounds, the relative intensity tolerance was set to 30% for isotope pattern matching. For the mzCloud database search, both the precursor and fragment mass tolerance were set to 10 ppm. The databases used for matching compounds in ChemSpider for structural search were BioCyc, Human Metabolome Database and KEGG, and the mass tolerance in ChemSpider Search was set to 5 ppm. The mass tolerance for matching compounds in Metabolika pathways was set to 5 ppm. Compounds were also matched to our in-house mass list, with mass tolerance set to 5 ppm and RT tolerance set to 1min. mzLogic was applied to score structure candidates based on MS/MS, with fragment mass tolerance set to 10 ppm and match factor threshold set to 30. Compounds were assigned by comparing annotations using the following nodes in order of priority: #1: Mass List Search; #2: mzCloud search; #3: Predict Compositions; #4 ChemSpider Search; #5: Metabolika Search.
The differential analysis was performed on log10 transformed values using defined sample ratios (Cln3+/− vs Cln3−/−, both in LysoIP fraction and whole cell fraction), and p-values were calculated. A results table of 1073 detected compounds was generated from this workflow (Supplementary Table 4) after (1) background compound subtraction and (2) applying a filter of maximum area intensity greater than 500,000 AU to filter out low signal peaks.
The 1073 detected compounds include possible artifacts as well as redundant or mis-annotated compounds. To focus on unique compounds with good peak quality and sufficient information for validation studies, we performed additional filtering, which included the following: (1) at least 1 compound match in the ChemSpider search; (2) at least 1 compound match in the mzCloud search. This resulted in 237 compounds. We then manually removed unreliable compounds: those with poorly aligned chromatograms and/or those that were obviously redundant. The final filtered compound list contained 189 compounds with assigned chemical formula and fragmentation results to help with compound identification (Supplementary Table 4). Among the 189 compounds after filtering, 3.2% had at least 1 log2 fold difference between the Cln3+/− vs Cln3−/− LysoIP samples, with adjusted p-values less than 0.05. The top compounds are structurally related and can be grouped as a class of compounds termed glycerophosphodiesters (GPDs). Principal Component Analysis in Fig. 2D was performed using PC1 and PC3, because PC3 includes the metabolites that separate the lysosomes of different genotypes. Whole tissues are not separated regardless of the PC used. P-values were calculated by running the Tukey HSD test (post-hoc) after ANOVA (for two group comparison, the ANOVA is simplified to unpaired t-test). The p-values were corrected by the Benjamini-Hochberg method for the false discovery rate. These analyses were done using the differential analysis node in compound discoverer v3.1.
For targeted analyses, Thermo Scientific TraceFinder software v5.0 was used to help in processing targeted metabolites screening and obtaining quantitative data from LC-MS.
In the final result table with 189 detected compounds, compound identification was performed based on accurate mass, retention time, and MS/MS spectra comparisons. We assigned to each compound one of four different levels of identification/annotation according to guidelines issue by the Metabolomics Standards Initiative (MSI)45. Level 1 defines compounds identified with both experimental retention time and MS/MS fragmentation patterned matched to authentic chemical standards that were run under the sams chromatographic conditions. Level 2 defined putatively annotated compounds, with only 1 of the orthogonal properties (e.g. RT) matched with an authentic chemical standard, or an MS2 match to only online in-silico spectra library or the Thermo Fisher mzCloud database. Level 3 defined putatively characterized compound classes, and finally level 4 defined unknown compounds.
Lipidomics
Lipids were separated on an Ascentis Express C18 2.1 × 150 mm 2.7 um column (Sigma-Aldrich) connected to a Vanquish Horizon UPLC system and an ID-X tribrid mass spectrometer (Thermo Fisher Scientific) equipped with a heated electrospray ionization (HESI) probe. External mass calibration was performed using the standard calibration mixture every seven days. Between 2 and 5 uL of sample were injected onto the column, with separate injections for positive and negative ionization modes. Mobile phase A in the chromatographic method consisted of 60:40 water: acetonitrile with 10 mM ammonium formate and 0.1% formic acid, and mobile phase B consisted of 90:10 isopropanol: acetonitrile, with 10 mM ammonium formate and 0.1% formic acid. The chromatographic gradient was adapted from46. Briefly, the elution was performed with a gradient of 40 minutes; during 0−1.5 minutes isocratic elution with 32% B; from 1.5 to 4 minutes increase to 45% B, from 4 to 5 minutes increase to 52% B, from 5 to 8 minutes to 58% B, from 8 to 11 minutes to 66% B, from 11 to 14 minutes to 70% B, from 14 to 18 minutes to 75% B, from 18 to 21 minutes to 97% B, during 21 to 35 minutes 97% B is maintained; from 35−35.1 minutes solvent B was decreased to 32% and then maintained for another 4.9 minutes for column re-equilibration. The flow rate was set to 0.260 mL/minutes. The column oven and autosampler were held at 55°C and 15°C, respectively. The mass spectrometer parameters were as follows: The spray voltage was set to 3.25 kV in positive mode and 3.0 kV in negative mode, and the heated capillary and the HESI were held at 300°C and 375°C, respectively. The S-lens RF level was set to 45, and the sheath and auxillary gas were set to 40 and 10 units, respectively. These conditions were held constant for both positive and negative ionization mode acquisitions.
The mass spectrometer was operated in full-scan-ddMS/MS mode with an orbitrap resolution of 120,000 (MS1) and 30,000 (MS/MS). Internal calibration using Easy IC was enabled. Quadrupole isolation was enabled, the AGC target was 1×105, the maximum injection time was 50 msec, and the scan range was m/z = 200–2000. For ddMS/MS, the cycle time as 1.5 sec, the isolation window was 1, and an intensity threshold of 1×103 was used. HCD fragmentation was achieved using a step-wise collision energy of 15, 25, and 35 units, and detected in the orbitrap with an AGC target of 5×104 and a maximum injection time of 54 ms. Isotopic exclusion was on, a dynamic exclusion window of 2.5 seconds was used, and a top 100 exclusion list was generated using a solvent bank.
High-throughput annotation and relative quantification of lipids was performed using LipidSearch v4.2.21 (Thermo Fisher Scientific/ Mitsui Knowledge Industries)47,48 using the HCD and ‘Labelled’ databases (the latter was searched to detect the Avanti Splash LipidoMix that was used as an internal standard). Precursor ion tolerance was set to 5 ppm and product ion tolerance was set to 8 ppm. LipidSearch nomenclature uses underscores to separate the fatty acyl chains to indicate the lack of sn positional information (for example, PC(16:0_18:1)) and not (PC(16:0/18:1)). Raw peak areas for all annotated lipids were exported to Microsoft Excel and filtered according to the following predetermined quality control criteria: Rej (reject parameter calculated by LipidSearch) equal to 0; PQ (peak quality parameter calculated by LipidSearch software) greater than 0.75; CV (s.d./mean peak area across triplicate injections of a represented (pooled) biological sample) below 0.4. Raw peak areas of the filtered lipids were normalized to well-detected deuterated standards found in the Avanti Splash LipidoMix as a control for lipid extraction and efficiency, as well as sample loading. Individual lipid species shown in figures were validated by manually checking the peak alignment and matching the MS/MS spectra to the characteristic fragmentation patterns found in the LIPIDMAPS database (www.lipidmaps.org). Notably, BMP lipid class is not included in the Lipidsearch target database—the PG lipid species from Lipidsearch analysis might be misannotated because BMP and PG are structural isomers. To distinguish between PG and BMP lipid species, annotation for those two lipid classes was performed manually based on the retention time and positive mode MS2 fragmentation. The lipid standards (18:1 (Δ9-Cis) PG and 18:1 BMP (S,R)) were purchased from Avanti Polar Lipids and found to be separable under our lipid chromatographic conditions. The BMP standard elutes before its PG isomer, either when the pure standards were prepared in isopropanol:methanol (1:1) solvent or when spiked into a bovine liver lipid extract. Although BMP and PG have better sensitivity in negative-ionization mode, their positive ammonium adducts were useful precursors for generating distinct neutral loss fragments critical for annotation. Consistent with a previous by Hankin et al49, the PG standard generated loss of the polar head group leading to a diglyceride-like fragment. In contrast, the BMP standard generated loss of the monoacylglycerolphosphate moiety leading to monoglyceride-like fragments. When MS2 fragmentation was not acquired, the species was annotated as BMP/PG.
Protein extractions from mouse tissues
Immediately following euthanasia, tissues were collected and snap-frozen in liquid nitrogen. Prior to protein extraction, tissues were powdered in liquid nitrogen using a mortar and a pestle. RIPA lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease and phosphatase inhibitors was added to 25 mg of tissue powder and samples were incubated on shaker for 1 hour at 4°C. Lysates were centrifugated for 30 minutes at maximum speed and protein concentrations were determined using the Pierce BCA Protein Assay kit (Thermo Fisher Scientific).
Immunoblotting
Lysates were resolved by SDS-PAGE (Thermo Fisher Scientific) at 120 V. Resolved proteins were transferred for 2 hours at 40 V to PVDF membranes. Membranes were blocked with 5% nonfat dry milk in TBST (Tris-buffered saline with Tween 20) for 1 hour, then incubated overnight with primary antibodies in 5% bovine serum albumin (BSA) in TBST at 4°C. All primary antibody dilutions are reported in supplementary table 6. Following incubation, membranes were washed with TBST for 3 times for 5 minutes per wash and then incubated with the appropriate secondary antibodies diluted 1:3000 in 5% BSA for 1 hour at room temperature. Membranes were then washed 3 times with TBST and visualized using ECL2 western blotting substrate (Thermo Fisher Scientific).
Electron Microscopy
For electron microscopy studies, mice were perfused with 2% paraformaldehyde and 2.5 % glutaraldehyde. Brains and livers were then isolated and postfixed in the same fixing solution. Coronal 200 μm sections were cut on a vibratome Leica VT-1000 (Leica, Heidelberg, Germany). Sections were post-fixed with 2% osmium, rinsed, dehydrated, and embedded in Durcupan resin (Sigma-Aldrich, St. Louis, USA). Ultra-thin sections (0.07 – 0.09 μm) were cut with a diamond knife with an ultramicrotome (Leica EM UC6, Leica microsystems, Wetzlar, Germany), stained with lead citrate (Reynolds solution) and examined with a transmission electron microscope FEI Tecnai G2 Spirit BioTwin (Thermo Fisher Scientific company, Oregon, USA), using a digital camera (Morada, Olympus Soft Image Solutions GmbH, Münster, Germany).
Cathepsin B Activity Assay
Cathepsin B activity was assayed fluorometrically using 5 μM of the fluorogenic CTSB substrate, Z-RR-AMC (Sigma) in 100 μl of the cathepsin B activation buffer (KH2PO4 (87.7 mM), Na2HPO4 (12.3 mM), EDTA (4 mM), DTT (2.6 mM) and cysteine (8 mm) in a pH=6.0). The plate was incubated in a Tecan Spark multimode microplate reader (Tecan Trading AG, Switzerland) at 37°C and read at excitation of 380 nm and emission of 460 nm every 5 minutes. The values of Relative fluorescence units (RFU) of all samples were corrected by subtracting the value of a blank sample.
β-hexosaminidase Activity Assay
β-hexosaminidase activity was assayed using a N-acetyl-β-D-glucosaminide (sugar-substrate) conjugated to the fluorophore 4-methylumbelliferone (4-MU). 0.6 mM of 4-MU N-acetyl-β-D-glucosaminide in 50 mM sodium acetate buffer, pH 5.4 was used with 5 μg of brain or liver homogenates and incubated at 37 °C for several time points. The reaction was stopped by adding 0.1 M glycine buffer, pH 10. The fluorescence of released (4-MU) was measured in a Tecan Spark multimode microplate reader (Tecan Trading AG, Switzerland), at an emission wavelength of 448 nm and excitation at 362 nm. The values of relative fluorescence units (RFU) of all samples were corrected by subtracting the value of a blank sample.
Immunofluorescence assays in cells and tissues
Cells:
50,000 cells were plated on fibronectin-coated glass cover slips, fixed with 4% paraformaldehyde and blocked with 5% donkey serum in PBS for 1 hour at room temperature. This was followed by overnight incubation at 4°C with primary antibodies. Primary antibodies used in this study are mouse anti-Lamp2 (1:500) and rabbit anti-Flag (1:500). Secondary antibodies (Alexa-Fluor) were then applied in 1% BSA, 0.3% Triton X-100 for 2 hours. Images were acquired in z-series on a spinning disk confocal system and maximum intensity projections were processed using ImageJ v1.52.
Tissues:
Immediately following euthanasia, brains and livers were removed and fixed by immersion in 4% paraformaldehyde overnight, and embedded in paraffin. For experiments including Cln3-knockout mice, after fixation, tissues were transferred to 30% sucrose overnight, embedded in OCT (TissueTek) and frozen. For immunofluorescence staining of paraffin-embedded tissues, 5-μm sections were deparaffinized. Heat-induced antigen retrieval was performed in Borg Decloaker (Biocare Medical) buffer using a pressure cooker (Dako). For cryo-sections, antigen retrieval was omitted. Sections were blocked in 5% donkey serum, 0.3% Triton X-100 for 2 hours. Primary antibodies (rat anti-Lamp1, mouse anti-HA, and rabbit anti-NeuN) were applied in 1% BSA, 0.3% Triton X-100 for 48 hours. Secondary antibodies were then applied in 1% BSA, 0.3% Triton X-100 for 2 hours. Sections were covered by cover slip with Vectashield (Vector Laboratories). Z-series on a spinning disk confocal system, using software for acquisition.
Images were acquired with Airyscan2 LSM980 microscope or Zeiss AxioVert200M microscope using ZEN 3.2 imaging software or MetaMorph 7, respectively.
and maximum intensity projections were processed using ImageJ v1.52.
Cell lines and plasmids
The pMXs-IRES-Bsd vector was obtained from Cell Biolabs. HEK-293T cells were purchased from ATCC and authenticated by STR profiling. The sequence of the human CLN3 cDNA was mutated to remove the proto-spacer adjacent motif (PAM) and within the seed region of the sgRNA used in this study. Other mutations were also introduced to the sequence targeting another sgRNA site that was not included in the current study.
Cell culture
HEK-293T cells and their derivatives were cultured in DMEM base media with 10% inactivated fetal calf serum (Thermo Fisher Scientific) supplemented with 2 mM glutamine, penicillin, and streptomycin (Thermo Fisher Scientific). All cell lines were maintained at 37°C and 5% CO2. For all LysoIP experiments using HEK-293T cell lines, cells were cultured in RPMI base media supplemented with 4 nM of LysoTracker Red DND-99 for one hour before processing as described in Abu-Remaileh et al., 201712.
Virus production and transduction
Lentiviruses were produced by transfecting HEK-293T cells with lentiviral plasmids of the lysosomal tags in combination with the packaging plasmids VSV-G env elope and ΔVPR. For retrovirus production, pMXs FLAG-CLN3 plasmid and retroviral packaging plasmids Gag-Pol and VSV-G were co-transfected using XtremeGene9 transfection reagent. The culture medium was changed to DMEM supplemented with 30% inactivated fetal calf serum 16 hours post transfection. The virus-containing supernatant was collected 48 hours post transfection and spun for 5 minutes at 400xg to remove cells and then frozen at −80°C. To establish cell lines with stable expression, 500,000 wild-type or CLN3 knock-out HEK-293T cells were plated in 6-well plates in DMEM with 10% inactivated fetal calf serum and 8 μg/mL polybrene and 100–250 μL of virus-containing media. Spin infection was performed at 2,200 RPM for 45 minutes at 37°C. Cells with the virus were then incubated for 16 hours before adding fresh culture medium containing blasticidin (InvivoGen) for at least 72 hours selection. Stable cell lines were tested for proper localization of the lysosomal tag or Flag-CLN3 using immunofluorescence.
Generation of knock-out cell lines using CRISPR-Cas9 technology
Human CLN3 was depleted using the pLentiCRISPRv1 system. The following sense (S) and antisense (AS) oligo-nucleotides were cloned into pLentiCRISPRv1:
sgCLN3 (S): caccgGTCACGCTCAGCAATACCGC
sgCLN3 (AS): aaacGCGGTATTGCTGAGCGTGACc
sgAAVS (S): caccgTCCCCTCCACCCCACAGTG
sgAAVS (AS): aaacCACTGTGGGGTGGAGGGGAc
To generate knock-out cells, HEK-293T cells were transduced as described above and after puromycin selection, cells were single-cell sorted into 96-well plates containing 200 μl of DMEM supplemented with 30% inactivated fetal calf serum. Cell clones with the desired knockouts were identified by Illumina amplicon deep sequencing at Massachusetts General Hospital (Boston, MA). Control cells were generated by targeting the non-coding AAVS1 locus as described before50.
Generation and purification of GPG and GPE
To obtain GPG and GPE, phosphatidylglycerol (16:0 PG) and phosphatidylethanolamine (16:0 PE) (Avanti) were saponified as follows: 25 mg of each was dissolved in a 3 mL 2:1 chloroform:methanol solution in a clean 20 mL scintillation vial provided with a magnetic stirring bar. 1 mL 2 M NaOH was added to the mixture and reaction was stirred at room temperature for 2 hours. 1 mL 2 M HCl was used to neutralize the reaction and glycerophosphodiesters were extracted with 2 mL water twice into separate vessels. Aqueous phases were combined, washed with 2 mL chloroform, frozen in liquid nitrogen, and lyophilized. To separate the crude product from inorganic salts, the lyophilates were dissolved in a minimum amount of methanol, and the desired products were separated from precipitants by paper filtration.
Metabolite extraction from yeast
Saccharomyces cerevisiae (WT strain BY6741 and Btn1Δ strain P35H10) were kindly obtained by the Jarosz lab at Stanford University. Yeast strains were grown in Yeast Nitrogen Base (with amino acids and 2% glucose) and harvested at the early growth phase (OD600 = 0.5–1.0). Cells were centrifugated for 3 minutes at 2700xg at lowest temperature and the cell pellets were washed twice with cold PBS. Cells were then resuspended in 1 mL of pre-chilled quenching solution (2:2:1 mixture of ACN:MeOH:H2O) and transferred to 2 mL vials half-full with 0.5 mm glass beads. The cells were homogenized by 10 pulses of 10–20 seconds each at 3,800–5000 rpm with 2 minutes rest on ice between each pulse. Homogenates were removed and centrifuged at 2700×g at in cold for 2 minutes. Supernatants were removed to pre-chilled tube and centrifuged for 30 minutes at 17000×g in cold. Supernatant were transferred to new tubes and stored at −80°C for subsequent analysis using LC-MS. This protocol was adapted from Krink-Koutsoubelis et al.51
Glycerophosphodiester phosphodiesterase activity assay
HEK 293T cells stably expressing 3xFLAG-GDE or 3xFLAG-CLN3 were harvested and homogenized on ice. The resulting supernatant was immunoprecipitated with anti-FLAG conjugated agarose beads for 4 hours at 4°C. Tagged proteins were competitively eluted using FLAG peptide. The activity assay was carried out in 50 μl containing buffer of 100 mM Tris (pH 7.5), 10 mM MgCl2, 2 μg fatty acid free BSA, 50 μM labelled D9-GPC, and 2 μg of 3xFLAG-GDE or 3xFLAG-CLN3 protein (as described in Zheng et al., 2003)52. The incubation was at 37°C and terminated by addition of 80% cold methanol. GDE activity was assessed by the change in the level of D9-GPC and the generation of D9-Choline using mass spectrometry.
Measurement of lysosomal pH
Lysosomal pH was measured using the LysoSensor™ Yellow/Blue DND-160 indicator dye. In brief, the cells were incubated with 5 μM LysoSensor™ Yellow/Blue DND-160 in complete cell growth medium for 10 min at 37°C. The cells were then washed three times with PBS. The cells were subsequently analyzed using microplate reader and fluorescence emissions were collected at 440 and 540 nm for excitations at 340 and 380 nm, respectively. LysoSensor™ Yellow/Blue is ratiometric, so the ratio of light excited at 340 nm to 380 nm is proportional with lysosomal pH that is measured using a calibration curve. To create a standard curve, the cells with LysoSensor were equalized by incubating them in a series of MES calibration buffer (20 mM MES (2-(N-morpholino) ethanesulfonic acid), 110 mM KCl, 20 mM NaCl, 10 μM monesin, 20 μM nigericin) at pH values ranging from 3.5 to 7.5 for 10 minutes at 37°C.
In vitro lipid uptake assay
1.25% fatty acid-free BSA (FAF-BSA) was dissolved in DMEM medium at 37 °C and subsequently filtered. 0.18 mM of 16:0–18:1 D5-PG lipid (Avanti) was incubated with FAF-BSA at room temperature for 30 minutes on a rotating shaker. The molar ratio of D5-PG to FAF-BSA was 1:1. This mixture of D5-PG conjugated to FAF-BSA was considered as 10X stock and experiments were performed at a 1X working concentration. Prior to supplementation with D5-PG:FAF-BSA, cells were serum starved for 2 hours to enhance endocytosis. Cells were then incubated with the mixture for 4 hours. Treated and untreated cells were subjected to LysoIP followed by LC/MS analysis as described above.
D9-dipalmitoylphosphatidylcholine (16:0–16:0) and D9-choline labeling in primary neurons
Cln3+/− and Cln3−/− postnatal (day 1) mouse pups were euthanized by decapitation, and the cortices were collected by microdissection. Fresh cortices were then washed by dissection medium (Thermo Fisher Scientific 14170161) prior to tissue dissociation by scissors, 0.25% trypsin and pipette trituration. Cell strainers (70 μm) were used to remove tissue chunks from digested cortices. Dissociated neurons were plated with minimum essential medium (Fisher Scientific 21010046) containing 10% inactivated fetal calf serum (Thermo Fisher Scientific 10438026) supplemented with 2 mM glutamine, penicillin, and streptomycin (Thermo Fisher Scientific SV30010) in 24-well plates with coverslips coated by poly-L-lysine (Newcomer Supply 1339A). A complete media change to neurobasal media (Thermo Fisher Scientific 21103049) supplemented with B-27 (Invitrogen 17504044) and 2 mM glutamax (Fisher Scientific 35050061) was performed 24 hours after initial seeding. Primary neuronal cells were maintained at 37°C and 5% CO2. Neurons (day 7) were first starved in neurobasal media without B-27 for 30 minutes, and then were labeled by 37.6 μM d9-dipalmitoylphosphatidylcholine (16:0–16:0) conjugated with 0.25% fatty acid-free bovine serum albumin (BSA) or 37.6 μM d9-choline chloride in neurobasal media and B-27 for 24 hours. Molar percent enrichment (MPE) for trimethyl-d9 labeling is calculated based on the following equation:
where 𝑛 is the number of hydrogen atoms, the abundance of a mass isotopomer and the labeling state (M+i). As a choline-containing metabolite can only be present as unlabeled (M+0) or d9-labeled (M+9), the equation can be simplified as follows:
Human studies
Ethics Statement
The Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health IRB approved the natural history studies. Individuals of any age, sex, and race who have at least one variant in CLN3 and clinical symptoms suggestive of CLN3 disease were enrolled following appropriate provision of consent and assent. For minors, parents or legally authorized representatives provided permission and consent.
CSF Sample Collection
Collection of CSF samples used in this study was as previously described (Dang Do et al., 2020)35. We performed lumbar puncture under sedation, in participants unable to cooperate with the procedure when awake, to collect CSF from individuals with CLN3 [Investigations of Juvenile Neuronal Ceroid Lipofuscinosis (CLN3); NCT033007304], creatine transporter deficiency (NCT02931682), or Smith-Lemli-Opitz syndrome (NCT00001721). Anonymized residual CSF samples from outside clinical laboratories were also collected (NCT00344331). Participants in the CLN3 natural history study have a consistent identifier (SP_._._) across publications to facilitate data comparisons. These are included in Supplementary Table 5.
Data preparation and statistics
Displays of quantitative data were prepared in Microsoft Excel v16.61 and 16.62 and GraphPad Prism v9.0. Statistical comparisons were done as two-tailed unpaired t-test in Prism or Excel, unless stated otherwise in the legend. All displayed measurements represent samples generated independently, or biological replicates. Immunoblot and immunofluorescence data are representative experiments and the number of independent experiments performed is indicated in the legend. Drawings used in creating illustrations in Fig. 1A, Fig. 1C, Extended Data Fig. 2A, and Extended Data Fig. 4A and 4E are from BioRender.com and were used with permission.
Extended Data
Extended Data Fig. 1: The effects of LysoTag expression on lysosomes in vivo.
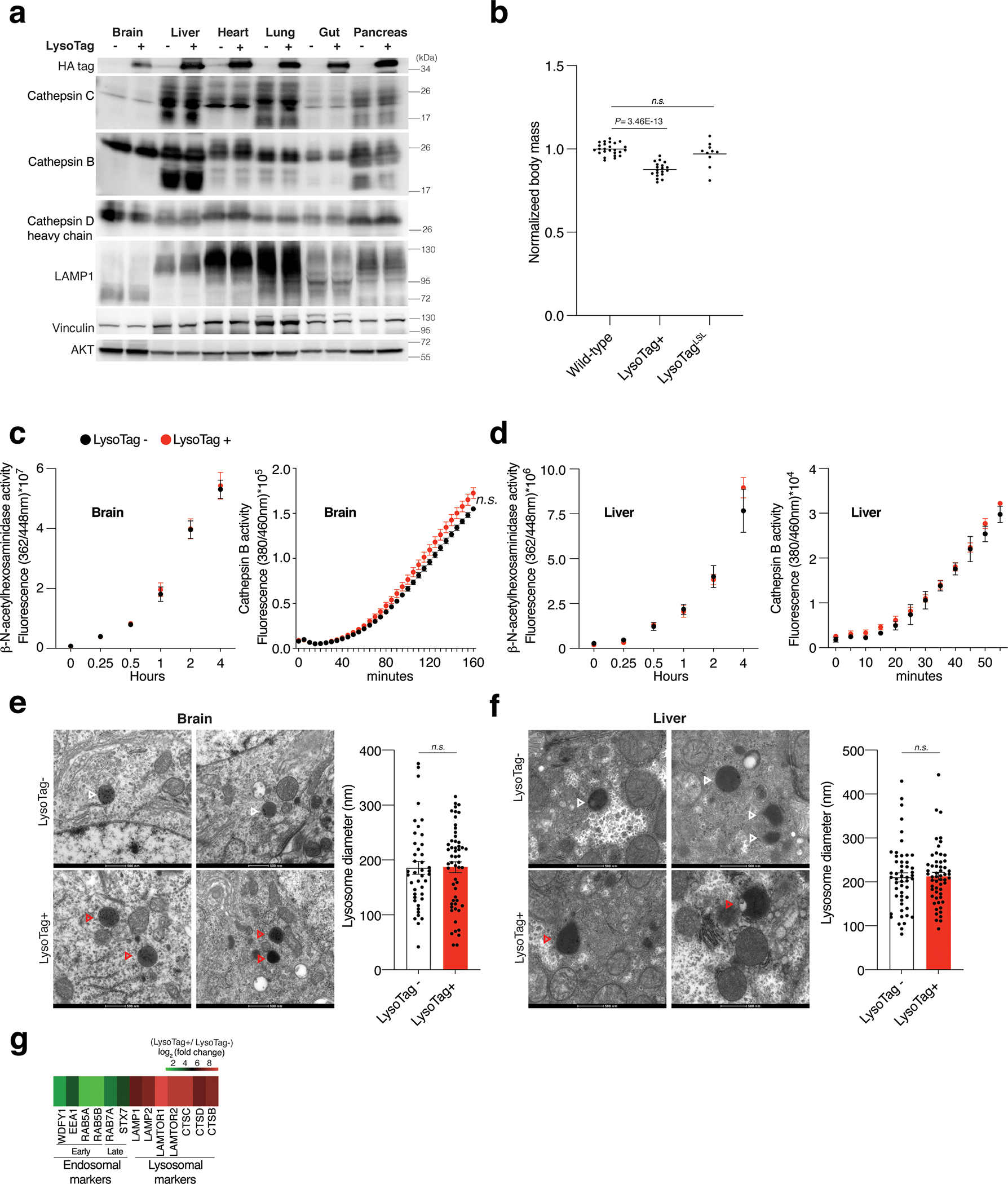
(A) In the constitutive LysoTag mice, TMEM192–3xHA is expressed across all tissues examined as determined by immunoblotting using antibodies to the HA epitope. No difference in the expression of lysosomal markers Lamp-1, Cathepsin B, C, and D is observed upon the expression of TMEM192–3xHA. Vinculin and AKT were used as loading controls. Numbers indicate molecular weights in kDa according to protein standards run on the same gel. Immunoblot is representative of two independent experiments.
(B) Normalized body weights of mice with the indicated genotypes at 5–9 weeks of age. Mice were weighed individually. The body mass of each mouse was normalized to the mean mass of sex-matched wild-type littermates before pooling by genotype for statistical analyses using a two-tailed unpaired t-test. (n=25, 19, and 10 for Wild type, LysoTag+, and LysoTagLSL mice, respectively.).
(C) and (D) Measurement of lysosomal hydrolase activity in brain and liver tissues of the constitutive LysoTag mice shows no effect of TMEM192–3xHA expression on lysosomal function. For measuring Cathepsin B activity, its fluorogenic substrate was incubated with homogenates of the corresponding tissues (see methods). For determining β-hexosaminidase activity, fluorogenic 4-methylumbelliferyl (MUF)- N-acetyl-β-Glucosaminide was used as a substrate (see methods). Fluorescence was measured at 37°C and data were shown following the subtraction of background fluorescence observed in the absence of homogenate (mean ± SEM; n=4).
(E) and (F) Representative transmission electron micrographs showing lysosomes from brain and liver tissues of LysoTag− (control) and LysoTag+ mice. Lysosomes were identified by the presence of a single membrane and granular, electron-dense appearance. White and Red arrowheads indicate lysosomes in cells from LysoTag− and LysoTag+ mice, respectively. n=2 mice per group and scale bar, 500 nm. The graph showing the mean diameter of lysosomes in multiple quantified cells. The diameter of lysosome in each group was generated by measuring lysosomes using ImageJ v1.52. (For brain, n=42 and n=55 measured lysosomes from LysoTag− and LysoTag+ mice, respectively. For liver, n=51 and n=56 measured lysosomes LysoTag− and LysoTag+ mice, respectively. Data presented as mean ± SEM. n.s: non-significant; Two-tailed unpaired t-test (E, F).
(G) Heatmap presentation of the enrichment of early- and late-endosomal markers as well as those for lysosome in the LysoIP performed from mouse liver tissue. Enrichment values are derived from Supplementary Table 1.
Extended Data Fig. 2: Generation and validation of LysoTag mouse model for Batten disease studies.
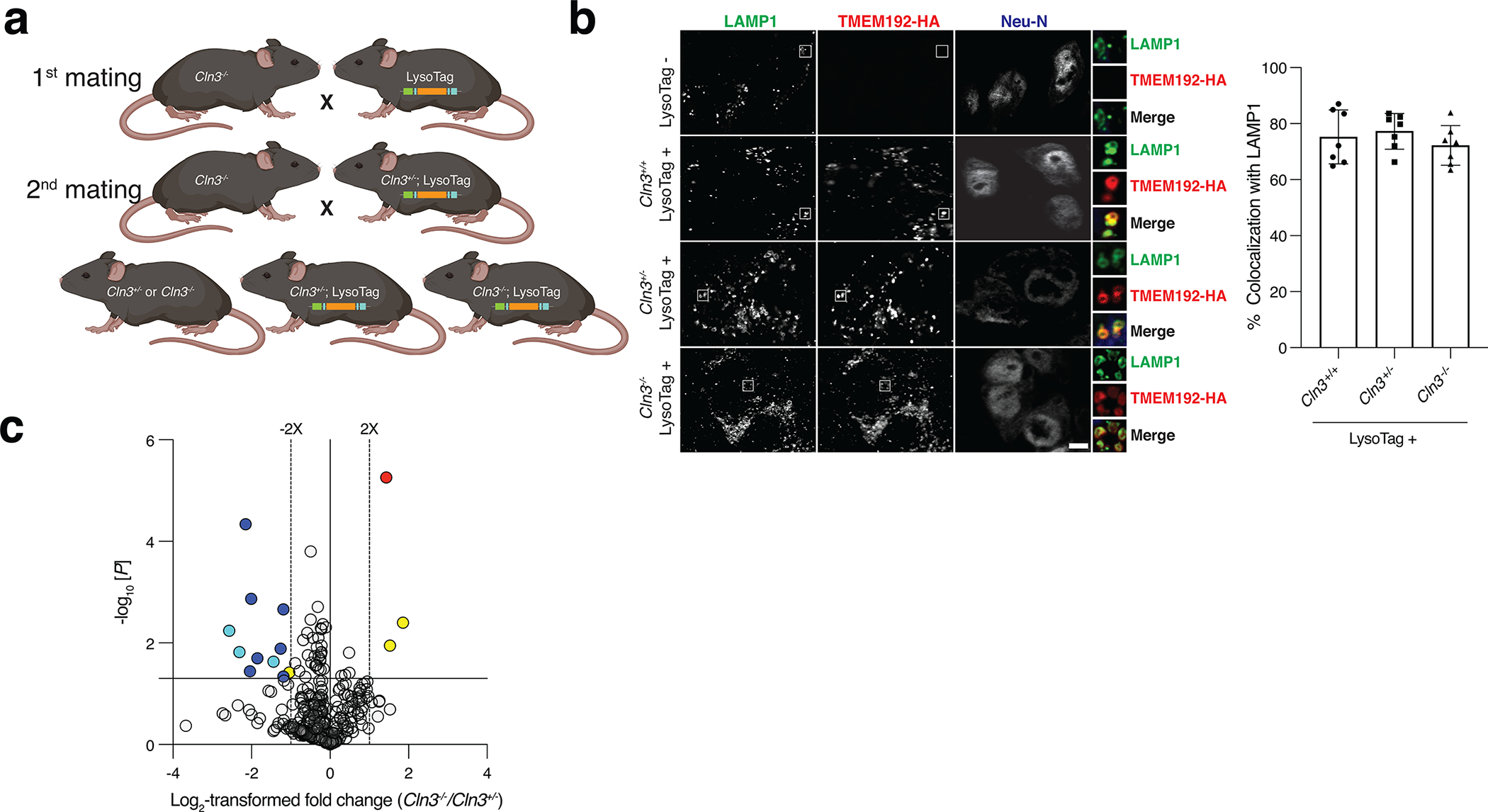
(A) Schematic depicting the breeding strategy to generate LysoTag models for Batten disease studies. Cln3−/− mice were crossed with heterozygous constitutive LysoTag mice and their progeny were then crossed back to Cln3−/− animals to generate the indicated experimental mouse genotypes. Mouse drawing was created with BioRender.
(B) Expression and localization of the TMEM192–3xHA fusion protein to lysosomes in brain neurons of Cln3+/+, Cln3+/− and Cln3−/− LysoTag mice. TMEM192–3xHA, lysosomes and neurons were detected in an immunofluorescence assay using antibodies to the HA epitope the lysosomal marker, Lamp-1 and the neuronal marker Neu-N, respectively. Scale bar = 5 μm, and magnified insets are labeled with white boxes in the main image. Percentage of colocalization between TMEM192–3xHA and Lamp1 shown in the right panel was measured in n=7 cells per genotype. Data are shown as mean ± SEM. Micrographs are representative of at least three independent experiments.
(C) Volcano plot comparing the lipidomic data from whole brain homogenates of Cln3+/− and Cln3−/− mice shows that only few lipids change significantly and they belong to phospholipids (yellow) and lysophosphatidylglycerol (LPG) in red. Loss of CLN3 also led to a decrease in the brain of bis(monoacylglycero)phosphate (BMP), a class of lysosomal lipids (blue). BMP/PG (cyan) annotation was used when MS/MS fragmentation was not acquired (see Supplementary Table 3 and methods for details). Horizontal line indicates a p-value of 0.05, and vertical dotted lines a fold change of 2. Each dot represents a lipid species. Data were acquired in negative ion mode and normalized to internal lipid standards for the best-matched lipid class (n = 5 and 4 for Cln3+/− and Cln3−/− mice, respectively). Female mice with an average age of 7 months were used. Data are in Supplementary Table 3. Two-tailed unpaired t-test was used.
Extended Data Fig. 3: Validation of glycerophosphodiesters (GPDs) as the metabolites that accumulate in lysosomes upon CLN3 loss.
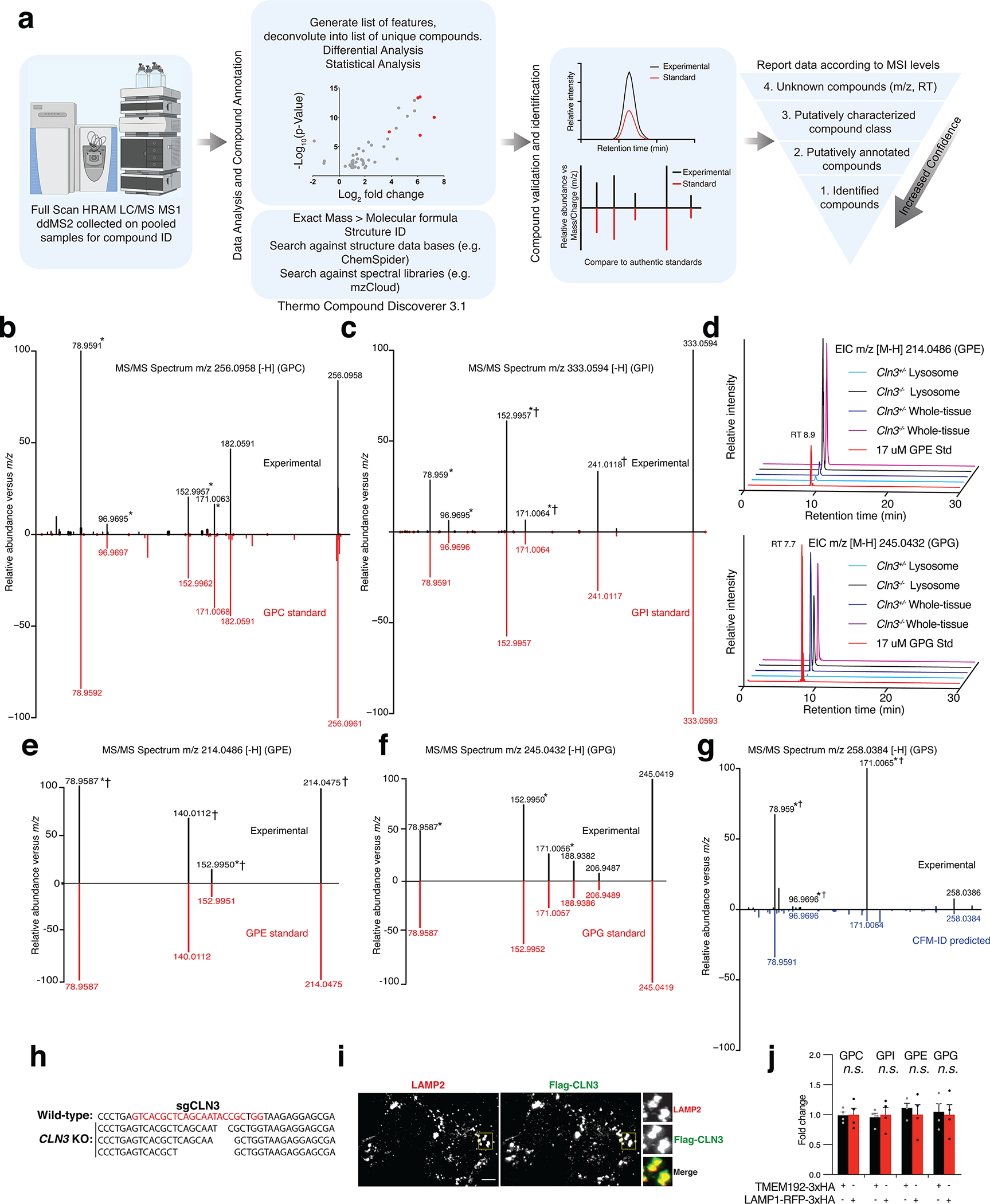
(A) Identification and annotation of metabolites using untargeted metabolomics analyses. Overview of our untargeted metabolomics experimental workflow. MS1 data were collected for every sample using full scan mode on a high-resolution accurate-mass instrument. MS/MS data were collected from pooled samples only to aid with compound identification. Data were then analyzed using Thermo Compound Discoverer v3.1 to generate a list of unique features which were deconvoluted into a list of unique compounds. Differential and statistical analyses were performed. Compounds which were statistically significant in the differential analyses were validated and identified using authentic standards, when available. If no authentic standard was available, compounds were annotated by comparing MS/MS data to a database, or in silico fragmentation prediction (competitive fragmentation modelling-ID, CFM-ID). Data were then reported according to the Metabolomics Standards Initiative (MSI) guidelines.
(B) and (C) Mirror plots for GPC and GPI in negative ion mode, respectively. Fragments common to the glycerol-phosphate group are indicated with an asterisk*. Fragments common to those found in the MS/MS spectra reported by Kopp et al.32 are indicated with †.
(D) EICs for GPE and GPG across a range of samples were analyzed alongside in-house generated standards (Std) and showed a matching retention time (RT).
(E) and (F) Mirror plots for GPE and GPG, respectively. Fragments common to the glycerol-phosphate group are indicated with an asterisk*. Fragments common to those found in the MS/MS spectra reported by Kopp et al.32 are indicated with †.
(G) Mirror plots showing MS/MS spectra for GPS from experimental samples compared to MS/MS spectra generated in silico using CFM-ID. Fragments common to the glycerol-phosphate group are indicated with an asterisk*. Fragments common to those found in the MS/MS spectra reported in Kopp et al.32 are indicated with †.
(H) Amplicon sequencing was used to validate CLN3 knock out cells generated using CRISPR-Cas9. Three mutant alleles were identified and all were found to generate frameshift deletions in the CLN3 coding sequence as compared to the wild-type sequence.
(I) Localization of Flag-CLN3 protein to lysosomes. Flag-CLN3 and lysosomes were detected in an immunofluorescence assay using antibodies to the Flag epitope and the lysosomal marker Lamp-2, respectively. Scale bar = 5 μm. Micrographs are representative images of three experiments.
(J) Changes in GPD levels in CLN3-deficient cells expressing TMEM192–3xHA tag or Lamp1–3xHA tag. Data presented as a comparison between the increase in the lysosomal abundance of GPDs upon CLN3 loss in Lamp1–3xHA tagged lysosomes relative to that in TMEM192–3xHA tagged lysosomes (mean ± SEM; n=4 biologically independent samples). n.s: non-significant; Two-tailed unpaired t-test.
Extended Data Fig. 4: Testing the effects of CLN3 loss on lysosomal pH and cellular lipid metabolism.
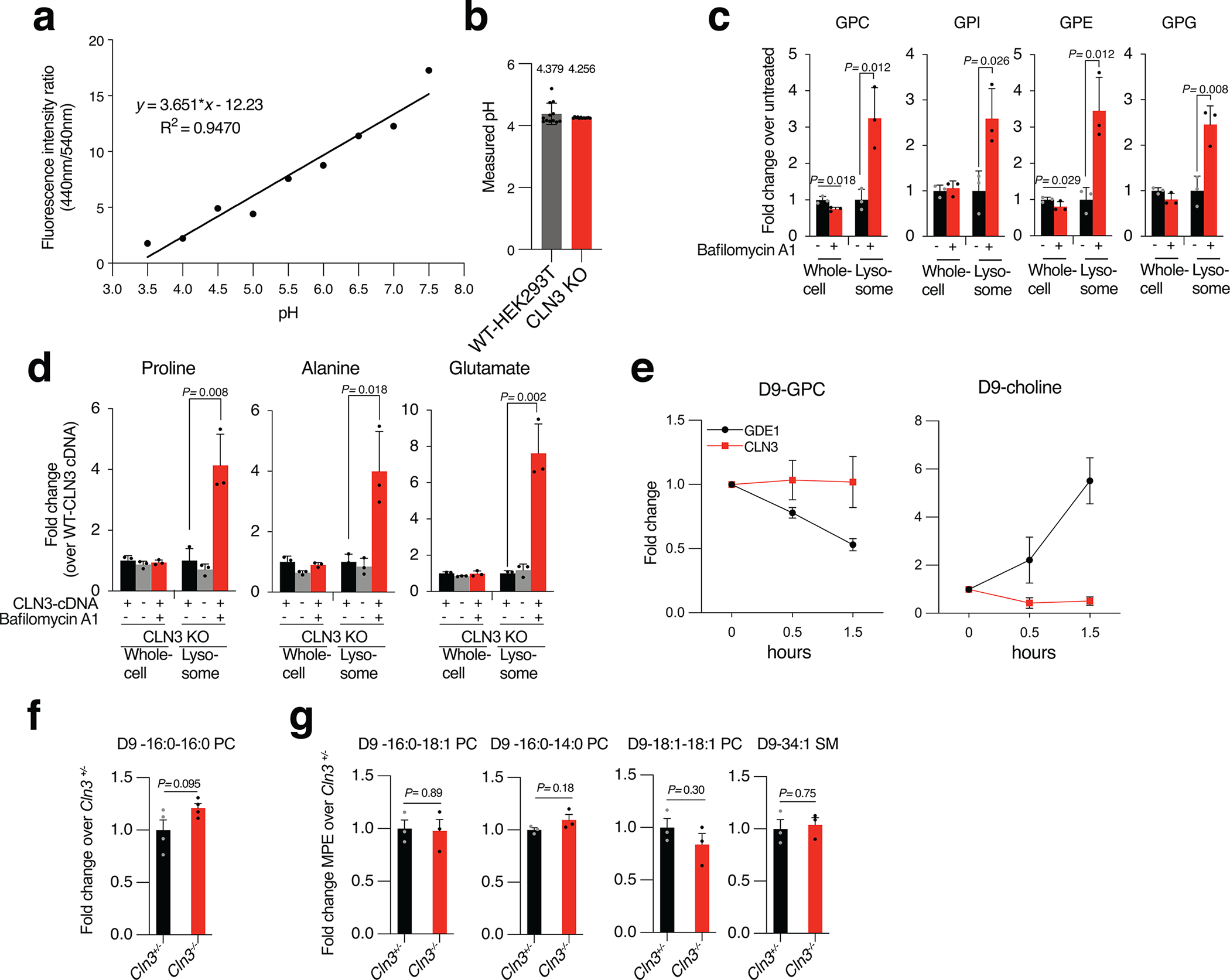
(A) to (D) CLN3 loss does not increase lysosomal pH. (A) A standard calibration curve of the ratiometric dye LysoSensor Yellow/Blue DND-160. See methods for experimental details.
(B) Lysosomal pH in wild-type and CLN3-KO HEK293T cells as calculated using the standard curve in A. Data are presented as mean ± SD, n=12 biologically independent samples.
(C) Targeted analyses of the fold changes in whole-cell and lysosomal levels of GPDs upon treatment with 500 nM Bafilomycin A1 for 6 hours in CLN3 expressing HEK-293T cells. Data are presented as mean ± SD, n=3 biologically independent samples, (Two-tailed unpaired t-test).
(D) The levels of several amino acids whose egress from lysosomes is sensitive to the proton gradient across the lysosomal membrane are not affected by CLN3 loss. The levels of proline, alanine, and glutamate in whole cells and lysosomes were compared between CLN3-KO cells with and without CLN3 cDNA addback and upon treatment with 500 nM Bafilomycin A1 (BafA1) for 6 hours. Data are presented as mean ± SD, n=3 biologically independent samples, (Two-tailed unpaired t-test).
(E) The recombinant CLN3 protein does not have glycerophosphodiesterase activity. GPC-d9 was incubated with recombinant 3xFLAG-CLN3 or the positive control glycerophosphodiesterase1 (3x-FLAG-GDE-1) for the indicated time points. The extent of GPC-d9 hydrolysis was determined by measuring the decline in its level and the increase in the levels of the product D9-choline. Data presented as mean ± SEM of n=3 biologically independent samples.
(F) CLN3 loss does not decrease tracer uptake. Fold change in tracer levels (D9–16:0–16:0 PC) normalized to total protein for samples measured in Figures 4F and G. Data presented as mean ± SEM of n=4 biologically independent samples.
(G) CLN3 loss does not affect the biosynthesis or turnover of PC and sphingomyelin (SM) in cells. Free D9-choline was used as a tracer. Data are presented as fold changes in the whole-cell molar percent enrichment (MPE) of D9-choline-containing lipids in cortical neuron cultures prepared from Cln3−/− mice relative to those from Cln3+/− animals (mean ± SEM, n=3 biologically independent samples, (Two-tailed unpaired t-test)).
Supplementary Material
Table 6: Reagents used in this study.
Table 5: Characteristics of study participants.
Table 4: Untargeted metabolomics of lysosomes derived from brains of Cln3+/− and Cln3−/− mice.P values were calculated by ANOVA with Tukey HSD test. The p values were corrected by the Benjamini-Hochberg method, FDR = 5%. n=3 per genotype.
Table 2: Targeted metabolomics of liver-derived lysosomes. Mean of log2-transformed values are included. Two-tailed unpaired t-test per raw was performed on fold changes between ad libitum and fasted animals. Ad libitum (n=4) and fasted (n=3). Male mice with an average age of 5.5 weeks were used.
Table 3: Lipidomics analysis of whole tissue and lysosomes derived from brains of Cln3+/− and Cln3−/− mice. P values were calculated using two-tailed unpaired t-test. n=5 and 4 female mice with an average age of 7 months for Cln3+/− and Cln3−/− mice, respectively.
Table 1: Proteomic analysis of liver-derived lysosomes. Paired t-test performed at the peptide precursor level. P values were corrected for multiple testing as described by Storey1. n=3 male mice with average age of 6.5 weeks.
Acknowledgements
The authors would like to thank the members of the Abu-Remaileh and Sabatini Labs for helpful insights. We also thank Joanna M. Kirkpatrick and the FLI Core Proteomics. This work was supported by grants from the NIH (DP2-CA271386), Stanford Alzheimer’s Disease Research Center (ADRC), Charles King fellowship, BeatBatten (Netherlands) and NCL-Stiftung (Germany) foundations to M.A-R and from the NIH (R01 CA103866 and R01 CA129105) to D.M.S. N.N.L., W.D. were also supported by BeatBatten (Netherlands) and NCL-Stiftung (Germany) foundations. U.N.M. is supported by Stanford ChEM-H Chemistry/Biology Interface Program, O’Leary-Thiry Graduate Fellowship and NIH T32 training grant (T32GM120007). A.L.C. is supported by F31 predoctoral NRSA (5F31DK113665) and C.G. and R.T. were partially supported by funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)-Projektnummer 239283807 - TRR 152 and the NCL-Stiftung (Germany). A.O. is supported by DFG via the Research Training Group ProMoAge (GRK 2155), the Else Kröner Fresenius Stiftung (award number: 2019_A79), the Fritz-Thyssen foundation (award number: 10.20.1.022MN) and the Chan Zuckerberg Initiative Neurodegeneration Challenge Network (award numbers: 2020-221617 and 2021-230967). The FLI is a member of the Leibniz Association and is financially supported by the Federal Government of Germany and the State of Thuringia. The CLN3 natural history study is funded by an NIH Clinical Center Bench to Bedside award and the NICHD intramural research program (ZIA HD008989). We thank participants and families in this study for their invaluable contributions. M.A-R. is a Terman Faculty Fellow at Stanford University.
Footnotes
Competing Interests
M.A-R. is a scientific advisory board member of Lycia Therapeutics. A.D.D. and F.D.P. have a collaborative research agreement with Amicus Therapeutics, Inc. All other authors declare no competing interests.
Storey, J. D. A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 64, 479–498, doi:https://doi.org/10.1111/1467-9868.00346 (2002).
Data availability
The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018624. Known lysosomal proteins in our proteomics analysis were defined based on Gene Ontology Cellular Component or UniProt sub-cellular localization annotation (https://www.uniprot.org/). The conditional LysoTag mouse was deposited at the Jackson Laboratory (Strain #035401). Other unique biological materials in the form of plasmids or cell lines are available from corresponding author upon request. Detailed lipidomics and metabolomics data are provided in Supplementary Table 3 and 4, respectively. Other data generated are available from the corresponding author upon request.
References
- 1.Ballabio A & Bonifacino JS Lysosomes as dynamic regulators of cell and organismal homeostasis. Nature reviews. Molecular cell biology 21, 101–118, doi: 10.1038/s41580-019-0185-4 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Medoh UN, Chen JY & Abu-Remaileh M Lessons from metabolic perturbations in lysosomal storage disorders for neurodegeneration. Current Opinion in Systems Biology 29, 100408, doi: 10.1016/j.coisb.2021.100408 (2022). [DOI] [Google Scholar]
- 3.Platt FM, d’Azzo A, Davidson BL, Neufeld EF & Tifft CJ Lysosomal storage diseases. Nat Rev Dis Primers 4, 27, doi: 10.1038/s41572-018-0025-4 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Ferguson SM Neuronal lysosomes. Neurosci Lett 697, 1–9, doi: 10.1016/j.neulet.2018.04.005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perera RM & Zoncu R The Lysosome as a Regulatory Hub. Annual review of cell and developmental biology 32, 223–253, doi: 10.1146/annurev-cellbio-111315-125125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savini M, Zhao Q & Wang MC Lysosomes: Signaling Hubs for Metabolic Sensing and Longevity. Trends Cell Biol 29, 876–887, doi: 10.1016/j.tcb.2019.08.008 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballabio A & Gieselmann V Lysosomal disorders: from storage to cellular damage. Biochimica et biophysica acta 1793, 684–696, doi: 10.1016/j.bbamcr.2008.12.001 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Boustany RM Lysosomal storage diseases--the horizon expands. Nature reviews. Neurology 9, 583–598, doi: 10.1038/nrneurol.2013.163 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Marques ARA & Saftig P Lysosomal storage disorders - challenges, concepts and avenues for therapy: beyond rare diseases. J Cell Sci 132, doi: 10.1242/jcs.221739 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Wallings RL, Humble SW, Ward ME & Wade-Martins R Lysosomal Dysfunction at the Centre of Parkinson’s Disease and Frontotemporal Dementia/Amyotrophic Lateral Sclerosis. Trends Neurosci 42, 899–912, doi: 10.1016/j.tins.2019.10.002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, Telpoukhovskaia MA, Bahr BA, Chen X & Gan L Endo-lysosomal dysfunction: a converging mechanism in neurodegenerative diseases. Curr Opin Neurobiol 48, 52–58, doi: 10.1016/j.conb.2017.09.005 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Abu-Remaileh M et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813, doi: 10.1126/science.aan6298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thai TH et al. Regulation of the germinal center response by microRNA-155. Science 316, 604–608, doi: 10.1126/science.1141229 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Pisoni RL, Acker TL, Lisowski KM, Lemons RM & Thoene JG A cysteine-specific lysosomal transport system provides a major route for the delivery of thiol to human fibroblast lysosomes: possible role in supporting lysosomal proteolysis. The Journal of cell biology 110, 327–335 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eiberg H, Gardiner RM & Mohr J Batten disease (Spielmeyer-Sjogren disease) and haptoglobins (HP): indication of linkage and assignment to chr. 16. Clin Genet 36, 217–218, doi: 10.1111/j.1399-0004.1989.tb03193.x (1989). [DOI] [PubMed] [Google Scholar]
- 16.Lerner TJ et al. Isolation of a novel gene underlying Batten disease, CLN3. The International Batten Disease Consortium. Cell 82, 949–957, doi: 10.1016/0092-8674(95)90274-0 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Mirza M et al. The CLN3 gene and protein: What we know. Mol Genet Genomic Med 7, e859, doi: 10.1002/mgg3.859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butz ES, Chandrachud U, Mole SE & Cotman SL Moving towards a new era of genomics in the neuronal ceroid lipofuscinoses. Biochim Biophys Acta Mol Basis Dis, 165571, doi: 10.1016/j.bbadis.2019.165571 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Jarvela I et al. Biosynthesis and intracellular targeting of the CLN3 protein defective in Batten disease. Hum Mol Genet 7, 85–90, doi: 10.1093/hmg/7.1.85 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Storch S, Pohl S & Braulke T A dileucine motif and a cluster of acidic amino acids in the second cytoplasmic domain of the batten disease-related CLN3 protein are required for efficient lysosomal targeting. The Journal of biological chemistry 279, 53625–53634, doi: 10.1074/jbc.M410930200 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Mao Q, Foster BJ, Xia H & Davidson BL Membrane topology of CLN3, the protein underlying Batten disease. FEBS Lett 541, 40–46, doi: 10.1016/s0014-5793(03)00284-9 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Ezaki J et al. Characterization of Cln3p, the gene product responsible for juvenile neuronal ceroid lipofuscinosis, as a lysosomal integral membrane glycoprotein. J Neurochem 87, 1296–1308, doi: 10.1046/j.1471-4159.2003.02132.x (2003). [DOI] [PubMed] [Google Scholar]
- 23.Oetjen S, Kuhl D & Hermey G Revisiting the neuronal localization and trafficking of CLN3 in juvenile neuronal ceroid lipofuscinosis. J Neurochem 139, 456–470, doi: 10.1111/jnc.13744 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Perland E, Bagchi S, Klaesson A & Fredriksson R Characteristics of 29 novel atypical solute carriers of major facilitator superfamily type: evolutionary conservation, predicted structure and neuronal co-expression. Open Biol 7, doi: 10.1098/rsob.170142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchison HM et al. Targeted disruption of the Cln3 gene provides a mouse model for Batten disease. The Batten Mouse Model Consortium [corrected]. Neurobiol Dis 6, 321–334, doi: 10.1006/nbdi.1999.0267 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Kovacs AD & Pearce DA Finding the most appropriate mouse model of juvenile CLN3 (Batten) disease for therapeutic studies: the importance of genetic background and gender. Dis Model Mech 8, 351–361, doi: 10.1242/dmm.018804 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lojewski X et al. Human iPSC models of neuronal ceroid lipofuscinosis capture distinct effects of TPP1 and CLN3 mutations on the endocytic pathway. Hum Mol Genet 23, 2005–2022, doi: 10.1093/hmg/ddt596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platt FM Sphingolipid lysosomal storage disorders. Nature 510, 68–75, doi: 10.1038/nature13476 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Fuller M & Futerman AH The brain lipidome in neurodegenerative lysosomal storage disorders. Biochem Biophys Res Commun 504, 623–628, doi: 10.1016/j.bbrc.2018.03.042 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Hobert JA & Dawson G A novel role of the Batten disease gene CLN3: association with BMP synthesis. Biochem Biophys Res Commun 358, 111–116, doi: 10.1016/j.bbrc.2007.04.064 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Padilla-Lopez S, Langager D, Chan CH & Pearce DA BTN1, the Saccharomyces cerevisiae homolog to the human Batten disease gene, is involved in phospholipid distribution. Dis Model Mech 5, 191–199, doi: 10.1242/dmm.008490 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopp F et al. The glycerophospho metabolome and its influence on amino acid homeostasis revealed by brain metabolomics of GDE1(−/−) mice. Chem Biol 17, 831–840, doi: 10.1016/j.chembiol.2010.06.009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen F, Pon A, Wilson M, Greiner R & Wishart D CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res 42, W94–99, doi: 10.1093/nar/gku436 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sumner LW et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221, doi: 10.1007/s11306-007-0082-2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dang Do AN et al. Neurofilament light chain levels correlate with clinical measures in CLN3 disease. Genet Med 23, 751–757, doi: 10.1038/s41436-020-01035-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fowler S & De Duve C Digestive activity of lysosomes. 3. The digestion of lipids by extracts of rat liver lysosomes. The Journal of biological chemistry 244, 471–481 (1969). [PubMed] [Google Scholar]
- 37.Schmidtke C et al. Lysosomal proteome analysis reveals that CLN3-defective cells have multiple enzyme deficiencies associated with changes in intracellular trafficking. The Journal of biological chemistry 294, 9592–9604, doi: 10.1074/jbc.RA119.008852 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corda D et al. The emerging physiological roles of the glycerophosphodiesterase family. The FEBS journal 281, 998–1016, doi: 10.1111/febs.12699 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Patton-Vogt J Transport and metabolism of glycerophosphodiesters produced through phospholipid deacylation. Biochimica et biophysica acta 1771, 337–342, doi: 10.1016/j.bbalip.2006.04.013 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Rigoni M et al. Equivalent effects of snake PLA2 neurotoxins and lysophospholipid-fatty acid mixtures. Science 310, 1678–1680, doi: 10.1126/science.1120640 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Fallbrook A, Turenne SD, Mamalias N, Kish SJ & Ross BM Phosphatidylcholine and phosphatidylethanolamine metabolites may regulate brain phospholipid catabolism via inhibition of lysophospholipase activity. Brain Res 834, 207–210, doi: 10.1016/s0006-8993(99)01570-x (1999). [DOI] [PubMed] [Google Scholar]
- 42.Storey JD A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 64, 479–498, doi: 10.1111/1467-9868.00346 (2002). [DOI] [Google Scholar]
- 43.Wyant GA et al. NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 360, 751–758, doi: 10.1126/science.aar2663 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez-Riverol Y et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450, doi: 10.1093/nar/gky1106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salek RM, Steinbeck C, Viant MR, Goodacre R & Dunn WB The role of reporting standards for metabolite annotation and identification in metabolomic studies. Gigascience 2, 13, doi: 10.1186/2047-217X-2-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bird SS, Marur VR, Sniatynski MJ, Greenberg HK & Kristal BS Serum lipidomics profiling using LC-MS and high-energy collisional dissociation fragmentation: focus on triglyceride detection and characterization. Anal Chem 83, 6648–6657, doi: 10.1021/ac201195d (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taguchi R & Ishikawa M Precise and global identification of phospholipid molecular species by an Orbitrap mass spectrometer and automated search engine Lipid Search. J Chromatogr A 1217, 4229–4239, doi: 10.1016/j.chroma.2010.04.034 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Yamada T et al. Development of a lipid profiling system using reverse-phase liquid chromatography coupled to high-resolution mass spectrometry with rapid polarity switching and an automated lipid identification software. J Chromatogr A 1292, 211–218, doi: 10.1016/j.chroma.2013.01.078 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Hankin JA, Murphy RC, Barkley RM & Gijon MA Ion Mobility and Tandem Mass Spectrometry of Phosphatidylglycerol and Bis(monoacylglycerol)phosphate (BMP). Int J Mass Spectrom 378, 255–263, doi: 10.1016/j.ijms.2014.08.026 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang T, Wei JJ, Sabatini DM & Lander ES Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84, doi: 10.1126/science.1246981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krink-Koutsoubelis N et al. Engineered Production of Short-Chain Acyl-Coenzyme A Esters in Saccharomyces cerevisiae. ACS Synth Biol 7, 1105–1115, doi: 10.1021/acssynbio.7b00466 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Zheng B, Berrie CP, Corda D & Farquhar MG GDE1/MIR16 is a glycerophosphoinositol phosphodiesterase regulated by stimulation of G protein-coupled receptors. Proceedings of the National Academy of Sciences of the United States of America 100, 1745–1750, doi: 10.1073/pnas.0337605100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 6: Reagents used in this study.
Table 5: Characteristics of study participants.
Table 4: Untargeted metabolomics of lysosomes derived from brains of Cln3+/− and Cln3−/− mice.P values were calculated by ANOVA with Tukey HSD test. The p values were corrected by the Benjamini-Hochberg method, FDR = 5%. n=3 per genotype.
Table 2: Targeted metabolomics of liver-derived lysosomes. Mean of log2-transformed values are included. Two-tailed unpaired t-test per raw was performed on fold changes between ad libitum and fasted animals. Ad libitum (n=4) and fasted (n=3). Male mice with an average age of 5.5 weeks were used.
Table 3: Lipidomics analysis of whole tissue and lysosomes derived from brains of Cln3+/− and Cln3−/− mice. P values were calculated using two-tailed unpaired t-test. n=5 and 4 female mice with an average age of 7 months for Cln3+/− and Cln3−/− mice, respectively.
Table 1: Proteomic analysis of liver-derived lysosomes. Paired t-test performed at the peptide precursor level. P values were corrected for multiple testing as described by Storey1. n=3 male mice with average age of 6.5 weeks.
Data Availability Statement
The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018624. Known lysosomal proteins in our proteomics analysis were defined based on Gene Ontology Cellular Component or UniProt sub-cellular localization annotation (https://www.uniprot.org/). The conditional LysoTag mouse was deposited at the Jackson Laboratory (Strain #035401). Other unique biological materials in the form of plasmids or cell lines are available from corresponding author upon request. Detailed lipidomics and metabolomics data are provided in Supplementary Table 3 and 4, respectively. Other data generated are available from the corresponding author upon request.


