Abstract
Explaining why some species are disproportionately impacted by the extinction crisis is of critical importance for conservation biology as a science and for proactively protecting species that are likely to become threatened in the future. Using the most current data on threat status, population trends, and threat types for 446 primate species, we advance previous research on the determinants of extinction risk by including a wider array of phenotypic traits as predictors, filling gaps in these trait data using multiple imputation, and investigating the mechanisms that connect organismal traits to extinction risk. Our Bayesian phylogenetically controlled analyses reveal that insular species exhibit higher threat status, while those that are more omnivorous and live in larger groups have lower threat status. The same traits are not linked to risk when repeating our analyses with older IUCN data, which may suggest that the traits influencing species risk are changing as anthropogenic effects continue to transform natural landscapes. We also show that non-insular, larger-bodied, and arboreal species are more susceptible to key threats responsible for primate population declines. Collectively, these results provide new insights to the determinants of primate extinction and identify the mechanisms (i.e. threats) that link traits to extinction risk.
Keywords: biological traits, conservation, extinction risk, IUCN, multiple imputation, primates
1. Introduction
Anthropogenic activity is causing species to disappear at an alarming rate. However, not all species are affected equally. Explaining why some species are more susceptible to extinction than others has become a major goal of conservation biologists as these contributions help to both explain current extinction patterns and allow for proactive protection of species possessing traits that could increase their probability of becoming imperiled. Previous studies have shown that phenotypic traits affect a species' susceptibility to extinction [1]. Physical traits such as large body size and life-history traits such as long generation lengths have been associated with increased risk of extinction in some clades [2–12]. These findings match expectations that lower population densities and increased hunting pressures put larger species disproportionately at risk and expectations that species with longer life histories have less time to adapt to environmental changes [3–5,13]. Behavioural traits have also been linked to increased extinction risk, including small group size and reduced innovativeness [14–16]: large groups are expected to benefit from reduced predation and enhanced foraging while less innovative species are less well-equipped to solve novel environmental challenges.
While much effort has been put toward identifying the traits that covary with extinction risk, important knowledge gaps have limited the effectiveness of these analyses. First, only a handful of studies have incorporated a broad range of traits in a single analysis. Chichorro et al. [1] reviewed studies investigating the correlates of extinction risk and found significant variability in the traits that were investigated (or controlled for). In addition, some traits have only recently been linked to extinction risk, such as behavioural flexibility [16], and thus have not been widely investigated across clades.
Second, the relationships between the actual anthropogenic drivers of environmental change that are responsible for extinction and species traits are understudied in many clades (e.g. in primates [17]), limiting the impact of these comparative studies in applied conservation [18]. Identifying which threats are most impactful to species with different trait types would enable actionable conservation steps (e.g. mitigating key threats in susceptible species’ ranges). Despite the possible benefit of considering specific threats, previous research has mostly focused on connecting species' traits and threat status. Notably, some studies focused primarily on predictors of threat status have attempted to incorporate information about anthropogenic threats into their analyses (e.g. [11,19–25]) while Richards et al. [26] recently explicitly assessed predictors of anthropogenetic threats to seabirds.
Lastly, we lack information on relevant traits for many species, resulting in incomplete data. The species for which we lack data may be systematically biased towards those that are more difficult to study, such as arboreal or nocturnal species. In addition to reducing statistical power, removing these species from analyses has potential to bias observed relationships between variables [27] and can result in a loss of real information when some traits included in an analysis have better data coverage than others. In recent years, improved methods for imputing missing data have become available, creating opportunities to reduce the number of missing data points in analyses of extinction risk (e.g. [12,26]).
Primates have been especially important in studies assessing predictors of extinction risk [3,8,19,28]. Primates are crucial components of tropical biodiversity, core players in the function of ecosystems, and central to many cultures and religions [17]. It is thus an urgent goal to determine which biological and behavioural traits contribute to primate extinction vulnerability and how these traits interact with anthropogenic impacts to contribute to population declines.
Primates are also one of the most threatened animal clades, with approximately 65% of species at risk of extinction [29], yet the last comprehensive assessment of the major determinants of primate extinction risk was published over 20 years ago [3]. The number of recognized primate species has changed dramatically since earlier studies, having more than doubled from 180 to over 500 in the past few decades [30,31]. As a result of these taxonomic changes and limitations of older phylogenies, previous studies focused on a relatively small number of currently recognized primate species. More speciose and up-to-date phylogenies have recently become available [32], coupled with a greater quantity and quality of trait data for many primate species. These contributions create an opportunity for the inclusion of more described primate species in comparative analyses, bringing us closer to capturing the true scope of primate diversity.
Here, we analyse the biological and behavioural determinants of primate extinction risk using a phylogenetic comparative approach. We investigate the relationship between multiple phenotypic traits and two measures of extinction risk reported by the International Union for Conservation of Nature (IUCN): threat status and population trend. We then assess how these same traits covary with vulnerability to the major threats facing primate species. This research addresses the gaps above by including multiple traits in the analysis and using imputation approaches based on phylogeny and phenotypic traits to fill in data for species with missing trait values. In addition, by investigating population trends and specific threats, we improve understanding of the connections between specific traits and the abundance of primates.
We focus on 10 key traits with proposed links to extinction risk (table 1).
Table 1.
The predicted direction of effect of biological and behavioural traits on extinction risk.
| trait | expected risk high when | reason |
|---|---|---|
| body mass (g) | large body | animals with large bodies have slow life histories, lower population densities, and may be subject to increased hunting pressures [4,5,13] |
| generation length (yrs) | long generations | slow life histories mean fewer generations to adapt to environmental changes [3] |
| home range size (ha) | large home range size | species with individuals that maintain large home ranges are particularly vulnerable to habitat loss, degradation, and edge effects [3,33] |
| group size | small group size | small groups may be more vulnerable to predation and experience foraging disadvantages [14,15] |
| brain volume (cm3) | small brain volume | large relative brain size is a proxy of general intelligence and behavioural flexibility [34,35] which allow animals to solve novel environmental problems [16] |
| omnivory (true or false) | false | animals with a large dietary breadth can rely on a wider range of food types when resources become limited [36] |
| social system | polygynandry | species characterized by complex social organization are hypothesized to have larger critical population sizes (i.e. more individuals must persist to maintain a healthy population) and may therefore go extinct more quickly than species in simpler social systems [37] |
| lifestyle | arboreal | strictly arboreal species are disproportionately affected from losing habitat via deforestation [38] |
| insularity (true or false) | true | island ecosystems are particularly vulnerable to anthropogenic change due to small population sizes, low habitat availability, and low functional redundancy [39–41] |
| nocturnal (true or false) | false | diurnal species are more likely to be disturbed by human activity (e.g. traffic) and diurnal activity has been connected to extinction risk [3] |
2. Methods
(a) . Data
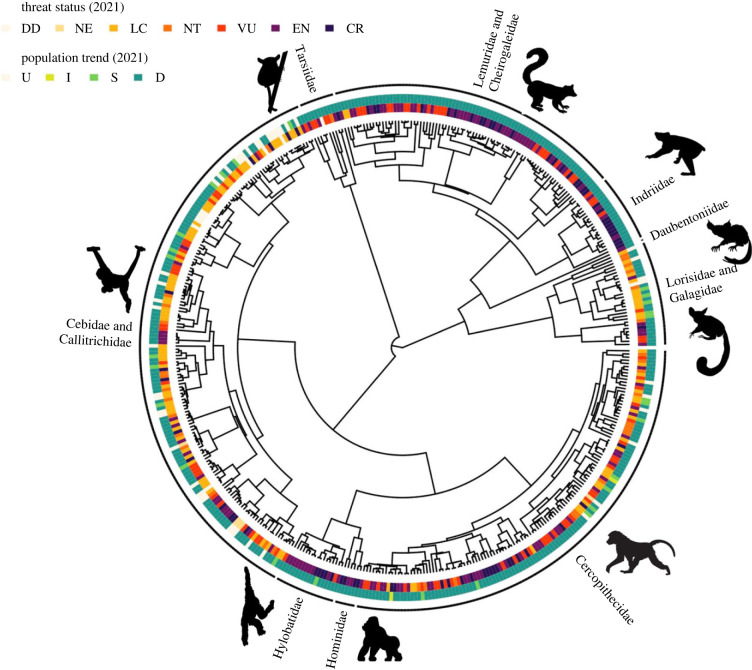
We collected information on threat status (least concern = LC, near threatened = NT, vulnerable = VU, endangered = EN, critically endangered = CR, data deficient = DD, and not evaluated = NE) and population trend (increasing = I, stable = S, decreasing = D, and unknown = U), from the IUCN [29] for 446 primate species present in the ultrametric primate phylogeny published by Upham et al. [32] (figure 1). We also collected a list of active threat types affecting each species in the IUCN [29] as defined by the Salafsky et al. [42] threat classification system: 1 = residential and commercial development, 2 = agriculture and aquaculture, 3 = energy production and mining, 4 = transportation and service corridors, 5 = biological resource use, 6 = human intrusions and disturbance, 7 = natural system modifications, 8 = invasive and other problematic species and genes, 9 = pollution, 10 = geological events, and 11 = climate change and severe weather.
Figure 1.
Phylogenetic distribution of threat status and population trends [29] for 446 primate species in the Upham et al. [32] phylogeny. Images of representative species are presented next to family labels. Codes for threat status: data deficient = DD, not evaluated = NE, least concern = LC, near threatened = NT, vulnerable = VU, endangered = EN, and critically endangered = CR. Codes for population trend: unknown = U, increasing = I, stable = S, and decreasing = D [29].
For each of the 446 species in our dataset, we recorded data on 10 different biological and behavioural traits that have been proposed to be associated with extinction risk from various sources: body mass (g) [43], generation length (yrs) [29], home range size (ha) [43], group size [44], brain volume (cm3) [45], omnivory (true or false), social system (solitary, pair-living, harem polygyny, and polygynandry) ([44,46] and other sources), lifestyle (arboreal, terrestrial, or both) ([44] and other sources), insular (true or false) (inferred from range data available in [44] and [29]), and nocturnal (true or false) [17,29]. The full list of references for trait values is available in the supplementary data [47]. Table 1 summarizes how we expected each trait to be associated with primate extinction risk. Previous studies have included geographic range size as a covariate in similar analyses (e.g. [3,28]). However, a species’ geographic range size is one of the main criteria used by the IUCN to assign threat status: species with small population sizes that have small or restricted geographical ranges are considered to be more imperiled [29] (i.e. threatened species have small geographical ranges by definition). Because we were interested in how biological and behavioural traits contribute to extinction risk, including effects on what geographical ranges they can occupy, we did not include geographic range size in our analysis. Notably, by including insularity in our analysis we controlled for the fact that species on small islands may not be able to maintain geographical ranges large enough to be considered healthy by the IUCN due to geographical barriers.
Following Powell et al. [45], for sexually dimorphic clades (size difference greater than 10%) only brain volume and body mass data from adult females were used in analysis. For all other species, averages for all adults measured in the original source were used. Species found exclusively on Madagascar, Borneo, or Sumatra were not scored as insular since these islands are large enough to support large geographical ranges comparable to many mainland species. Further details on operational definitions and trait coding are provided in the electronic supplementary material, along with a correlation matrix of all traits and response variables (electronic supplementary material, figure S1), and a comparison of trait data from different sources (electronic supplementary material, figures S2, S3, and S4).
(b) . Analysis
(i) . Trait imputation
The availability of data varied across the species in our dataset. Percentages of species with missing trait data were: body mass (6%), omnivory (17%), generation length (23%), home range size (25%), group size (39%), and brain volume (46%). Restricting the analysis to only species with observed data on all traits reduced our sample size of species by over half (e.g. from n = 430 to n = 151 in our analysis of threat status). We thus opted to use multiple imputation for these six traits to avoid losing species from our analysis where one or more traits had missing observations. The other four traits in our analysis (lifestyle, nocturnality, insularity, and social system) had full data coverage and no imputation procedure was necessary for these traits.
Multiple imputation was accomplished using 100 randomly sampled trees available from Upham et al. [32]. From each tree, we first generated a variance–covariance matrix which we then dissolved into 445 eigenvectors using the ‘PVRdecomp’ function from the R package PVR [48]. To impute body mass (the trait with the best data coverage requiring imputation), we built a linear model where body mass was the response and phylogenetic eigenvectors were predictors. Using forward–backward model selection, we then determined which phylogenetic eigenvectors had the best support for inclusion in models predicting body mass based on Akaike information criterion (AICc) scores using the ‘stepAIC’ function from MASS [49]. We chose how many eigenvectors to include in model selection for the imputation based on model performance in cross validation (electronic supplementary material, table S1). We repeated this process for all traits with missing data. The imputation of traits was ordered so that imputed information could be used to inform subsequent model fits along with phylogenetic information (e.g. once body mass was imputed it was used to inform model fits for the imputation of other traits). Body mass (including both sexes) and female body mass were imputed separately, and female body mass was used thereafter as body mass for sexually dimorphic clades. The top model for each trait was used to impute values for each species with missing data using the ‘predict’ function from the car package [50]. To propagate error, we then used the fits and standard deviations associated with predicted values to take a randomly sampled trait value for each species from the normal or binomial distribution (depending on whether traits were continuous or binary).
The imputation of traits was repeated once for each tree, resulting in 100 imputed datasets. We performed leave-one-out cross-validation for each imputation, where we removed observed datapoints and used our imputation method to predict their values. When comparing these imputed datapoints to the observed datapoints performance proved to be good in all cases (predictive accuracy > 0.8 for continuous variables and area under the ROC curve > 0.8 for binary variables; see electronic supplementary material, table S1).
Different methods of trait imputation can lead to different trait estimates, which may affect conclusions in downstream analyses. To assess the consistency of our results using different methods, we ran a second imputation procedure with the ‘phylopars’ function from Rphylopars [51]. Rphylopars uses a maximum-likelihood approach with a phylogeny and sparse trait matrix to impute missing data [51,52]. Rphylopars provides an advantage over the previously described imputation approach by incorporating the best supported model of trait evolution (in our case the Ornstein–Uhlenbeck model) to impute missing trait values [51], whereas our other imputation method did not subscribe to a particular model of trait evolution. However, Rphylopars also has some limitations including a lack of customizability (e.g. optimizing model inputs based on cross-validation performance) and can still lead to biased trait estimates particularly for traits with a large amount of missing data [52]. Given the advantages and disadvantages of each approach we decided run our main analyses with 100 sets of imputed data from both imputation methods to inform our conclusions about which traits are associated with primate extinction risk.
(ii) . Modelling threat status, population trends, and threat types
We ran multiple models to test predictors of three types of outcomes for primate populations. First, we tested which biological and behavioural traits are associated with the threat status of a species. Second, we tested the effects of species' traits on population trends. Third, we tested which traits were associated with species’ susceptibility to the most prevalent threats faced by primates.
To determine which biological and behavioural traits are associated with threat status, we ran two phylogenetic generalized linear mixed models using Bayesian approximation as implemented in the MCMCglmm R package [53]. The first model had an ordinal error structure, and the response variable was an ordinal measure of threat status scored as follows: LC = 0, NT = 1, VU = 2, EN = 3, and CR = 4 [54]. We ran the second model with a threshold error structure; here the response variable was threat status scored as a binary outcome where species were scored as either being threatened (VU, EN, or CR) (scored as 1) or not threatened (NT or LC) (scored as 0).
To test the effects of traits on population trends we ran a phylogenetic generalized linear mixed model with a threshold error structure [53]. In this model, each species was assigned a binary outcome of either declining (scored as 1) or not declining (i.e. stable or increasing) (scored as 0).
Finally, to determine which biological and behavioural traits were associated with species’ susceptibility to the most important threats that primate species face, we ran five phylogenetic generalized linear mixed models with threshold error structures [53], one for each of the top five threats to primates identified by the IUCN [29]. These top five threats identified for primate species were: 1 = residential and commercial development (35% of species), 2 = agriculture and aquaculture (80%), 3 = energy production and mining (22%), 5 = biological resource use (82%), and 7 = natural system modifications (23%). Here, each of our five models had a binary outcome of 1 (indicating that a species was affected by a particular threat) or 0 (indicating that a species was not affected by the threat).
Each model described above included all 10 traits of interest as fixed effects (i.e. traits were included together as predictors in the same model) and each model was run on 100 imputed datasets and phylogenies to account for uncertainty in phylogeny and trait estimates [55]. Models ran a total of 550 001 iterations, with a thinning interval of 500 and a burn-in of 50 000 to ensure convergence had occurred, which we assessed using trace plots [53]. Species were dropped from analyses when the true value of a response variable was unknown by the IUCN [29] (e.g. if the threat status was DD or NE). Continuous variables were ln-transformed, centred with respect to the mean, and scaled by two standard deviations in all models to make their effect sizes comparable to those reported for binary variables [56]. We used a weakly informative Gelman prior for fixed effects and fixed the residual variance (R) to one [53]. For the phylogenetic component (G), we used parameter expanded priors where V = 1, nu = 1, alpha.mu = 0, and alpha.V = 100 [57]. These analyses were repeated using 100 imputed datasets from both our model selection imputation approach and Rphylopars.
We also tested the hypothesis that some traits previously shown to be associated with primate extinction risk are losing signal as more species become imperiled, for example, if anthropogenic threats are becoming so overwhelming that all species are beginning to suffer regardless of their attributes. This analysis involved repeating our analyses of threat status using an older IUCN threat status dataset and species list obtained from Harcourt & Parks [58].
To interpret the output from our Bayesian analyses, we provide (i) the distribution of posterior means for tests from all 100 imputed datasets in graphical form (figures 2 and 3), (ii) the 89% credible intervals (per [59]) from the full posterior distribution of estimates in graphical form (figures 2 and 3), and (iii) the percentage of iterations from all 100 models that were consistently positive or negative (electronic supplementary material, tables S2 to S20). We focused on results that were most supported based on these outcomes and supported consistently using data from both imputation procedures. For the purposes of providing an estimate of the magnitude of an effect in the main text, posterior means were pooled across datasets using Rubin's rules [55] (hereafter, ‘pooled posterior mean’). Results reported in the main text are from our model selection imputation procedure and were qualitatively consistent using Rphylopars unless otherwise stated.
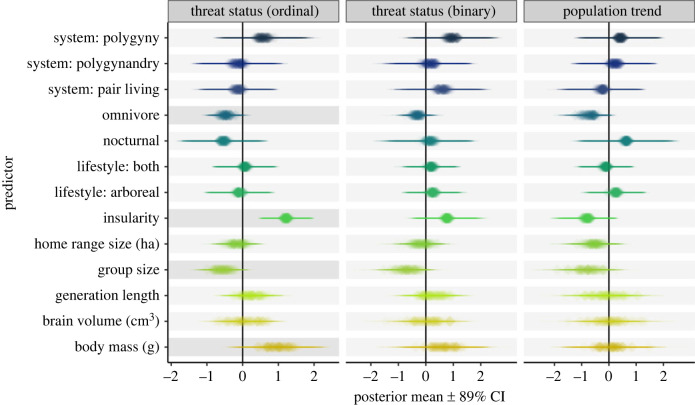
Figure 2.
Outcomes from three sets of models testing the relationship between traits and: threat status scored as an ordinal variable (first panel), threat status scored as a binary variable (second panel), and population trend scored as a binary variable (third panel). Each cell contains 100 posterior means (plotted as translucent diamonds) with their associated 89% credible intervals (plotted as translucent horizontal lines) obtained from 100 MCMCglmm models run with 100 randomly sampled phylogenies across 100 trait datasets with missing datapoints obtained through multiple imputation. Darker shading behind effects interpreted in text. Continuous variables were ln-transformed, centred with respect to the mean, and scaled by two standard deviations.
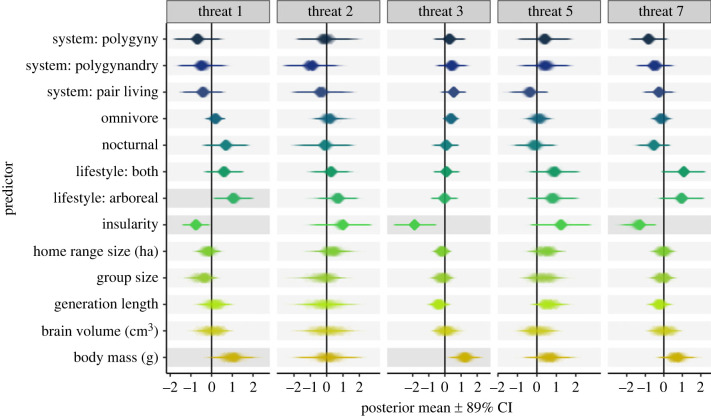
Figure 3.
Outcomes from five sets of models testing the relationship between traits and species susceptibility to: threat 1 = residential and commercial development (first panel), threat 2 = agriculture and aquaculture (second panel), threat 3 = energy production and mining (third panel), threat 5 = biological resource use (fourth panel), and threat 7 = natural system modifications (fifth panel). Each cell contains 100 posterior means (plotted as translucent diamonds) with their associated 89% credible intervals (plotted as translucent horizontal lines) obtained from 100 MCMCglmm models run with 100 randomly sampled phylogenies across 100 trait datasets with missing datapoints obtained through multiple imputation. Darker shading behind effects interpreted in text. Continuous variables were ln-transformed, centred with respect to the mean, and scaled by two standard deviations.
3. Results
(a) . Predictors of threat status and population trends
Our dataset included 430 primate species with known threat statuses. When scored as an ordinal outcome (LC = 0, NT = 1, VU = 2, EN = 3, and CR = 4), primate threat status was positively associated with insularity (pooled posterior mean = 1.214; 100% of 100 100 posterior estimates > 0) (figure 2; electronic supplementary material, table S2). Ordinal threat status was negatively associated with omnivory (pooled posterior mean = −0.474; 95% estimates < 0) and group size (pooled posterior mean = −0.561; 94% estimates < 0) (figure 2; electronic supplementary material, table S2). Body mass, which is often considered a main predictor of extinction risk in many clades, only weakly predicted threat status after controlling for other covariates (pooled posterior mean = 0.980; 93% estimates > 0) (figure 2; electronic supplementary material, table S2) and this weak support disappeared when using imputed data from Rphylopars (pooled posterior mean = 0.517; 74% estimates > 0) (electronic supplementary material, table S8). In our analysis using older IUCN data, we found that only insularity (pooled posterior mean = 2.255; 100% estimates > 0) and home range size (pooled posterior mean = 1.498; 100% estimates > 0) were associated with threat status (ordinal) in the predicted direction (electronic supplementary material, table S3).
Threat status was not strongly associated with any biological or behavioural traits when scored as a binary response (figure 2; electronic supplementary material, table S4). We also ran three separate models with body mass, generation length, and brain volume as sole predictors to determine if correlations among these variables (see electronic supplementary material, figure S1) contributed to the lack of strong associations. However, these additional analyses consistently showed no strong effect of any traits (electronic supplementary material, table S5). In our analysis using older IUCN data, we found that insularity (pooled posterior mean = 2.556; 100% estimates > 0) and home range size (pooled posterior mean = 1.078; 99% estimates > 0) were associated with threat status (binary) in the predicted direction (electronic supplementary material, table S6).
Population trend was not consistently associated with any biological or behavioural traits across 401 species with known population trends (figure 2; electronic supplementary material, table S7).
(b) . Predictors of specific threat types
Our analyses of threat types included 404 species with known threats. Insularity was negatively associated with threat 1 = residential and commercial development (pooled posterior mean = −0.752; 98% estimates < 0; electronic supplementary material, table S11), threat 3 = energy production and mining (pooled posterior mean = −1.807; 100% estimates < 0; electronic supplementary material, table S13), and threat 7 = natural system modifications (pooled posterior mean = −1.312; 100% estimates < 0; electronic supplementary material, table S15) (figure 3). Species living a strictly arboreal lifestyle were more likely to be affected by threat 1 = residential and commercial development than strictly terrestrial species (i.e. the baseline) (pooled posterior mean = 1.045; 98% estimates > 0; electronic supplementary material, table S11) (figure 3). Species with larger body masses were more likely to be affected by threat 3 = energy production and mining (pooled posterior mean = 1.232; 99% estimates > 0; electronic supplementary material, table S13) (figure 3). Large bodied species were also more likely to be affected by threat 1 = residential and commercial development (pooled posterior mean = 1.004; 96% estimates > 0; electronic supplementary material, table S11), but like our analysis of ordinal threat status, this effect was no longer compelling using imputed data from Rphylopars (pooled posterior mean = 0.741; 83% estimates > 0; electronic supplementary material, table S16) which instead supported a negative relationship between threat 1 and group size (pooled posterior mean = −0.720; 97% estimates < 0; electronic supplementary material, table S16).
Some weaker trait associations were also detected (e.g. arboreality or partial arboreality being positively associated with threat 7; electronic supplementary material, table S15). However, given the number of models where credible intervals around these estimates overlapped with zero, we chose to interpret associations with stronger support.
4. Discussion
We investigated the correlates of extinction risk and threat susceptibility in primates using phylogenetic comparative methods to analyse the most complete and up-to-date set of trait data and IUCN data. One novelty of our approach was the use of phylogenetic and trait-based imputation of missing data. In our analysis using threat status as an ordinal outcome—with five ranked categories from least concern to critically endangered—higher threat status was associated with insularity, absence of omnivory, and small group size, consistent with our predictions for these traits. Therefore, primate species that are most imperiled, and thus score highest in ordinal threat status, do tend to be those with biological and behavioural predispositions to extinction. When looking at specific threats, we found that larger-bodied and arboreal species are more vulnerable to key threats, while insular species are less vulnerable to these threats.
Although we did find that some traits were powerful predictors of threat status when scored according to ranked categories, the traits we investigated were not strong predictors of binary extinction risk outcomes (i.e. threat status scored as threatened versus non-threatened and population trend scored as declining versus not declining). This is contrary to findings in other taxonomic groups, such as birds, where multiple traits have been linked to binary measures of extinction risk [7]. Notably, the relative number of threatened and declining species is much higher for primates than for birds. For instance, 93% of primate species with known population trends are in decline while approximately 50% of birds are in decline [7]. This may explain why no predictors emerge for binary outcomes in primates: most species are threatened or becoming threatened, and traits are mostly deterministic of severity.
Some traits that predict threat status in other clades did not emerge as powerful predictors for primates, even in our analysis of ordinal threat status. For example, in a recent study of birds, Ducatez et al. [16] provided evidence that innovativeness—a known measure of behavioural flexibility associated with general intelligence [34,60]—buffers against extinction. However, we did not find an effect of brain size, another known measure of behavioural flexibility [34,61] and a correlate of innovativeness, in our analyses. The caveats associated with each of these measures are discussed in Creighton et al. [61]; however, we suggest that these differences in results could arise because the links between extinction risk and flexibility are complex for a clade like primates, where innovations are frequent and human conflict is common. Certain behavioural innovations may reduce conflict with humans and increase resilience to habitat degradation (e.g. novel approaches for accessing food [62]). However, other innovative behaviours can increase human–wildlife conflict. In many primate species, crop-raiding and garbage eating are innovative behaviours that have become common practice in the context of anthropogenic change (e.g. chimpanzees and baboons [63–65]). These behaviours bring animals in direct conflict with humans and, in some cases, attract them to lower quality habitats [66]. Future contributions should further address the paradox of how flexibility both helps and hinders species' persistence.
When repeating our analyses of extinction risk with an older IUCN dataset documented in Harcourt & Parks [58], we found that insularity and home range size shared a positive relationship with binary and ordinal threat status, while other traits were not powerful predictors. This pattern of results using newer versus older data indicates that some traits (i.e. home range size) have become less powerful predictors of extinction in the past 20 or so years. Meanwhile, some traits identified in our analysis of ordinal 2021 threat status (i.e. group size and omnivory) do not emerge in analyses with older data indicating these traits may be beginning to have a larger signal over time. However, it is also possible that the larger number of species in our 2021 dataset (a consequence of taxonomic reevaluations in many clades [31]) and general improvements in the thoroughness and accuracy of IUCN assessments since Harcourt & Parks [58] provided the statistical power to detect the effects of these traits.
In analyses assessing predictors of direct threats, we found that strictly arboreal species were more likely to be threatened with residential and commercial development—a major driver of deforestation in many regions. We also found that insular species were less likely to be vulnerable to multiple threats (residential and commercial development, energy production and mining, and natural system modifications) despite being more likely to be highly threatened. This indicates that the high threat statuses of insular species may not be driven by anthropogenic activity. Instead, their small geographical ranges enforced by geographical barriers could simply make it impossible to maintain healthy population sizes.
While our results were largely consistent using two methods of imputation, we identified a few discrepancies where support for trait associations changed meaningfully depending on the imputation procedure: one method better supported an effect of body mass on ordinal threat status and threat type 1, while the other better supported an effect of group size on threat 1. One of the main benefits of imputation in multivariable analyses is the ability to preserve real data for well represented traits when another trait has poor coverage. Given that all methods of imputation are imperfect, using multiple methods of imputation with different associated biases may help reveal cases where the imputation is influential in driving an observed relationship between two variables and may help with interpretation of findings, especially when credible intervals are wide or close to overlapping with zero.
One limitation of our analysis was that body mass, generation length, and brain volume were all highly correlated in our dataset (correlation coefficients range between 0.6 and 0.9). These traits each have independent predicted associations with extinction risk in conflicting directions, meaning that reducing these variables to fewer terms (e.g. via principal component analysis) would reduce our ability to draw conclusions about their independent effects. We instead included these predictors in the same models to identify how they independently contributed to extinction risk and threat vulnerability [67]. However, this creates the possibility of collinearity in model estimates for these variables as suggested by the increased uncertainty (i.e. wide credible intervals) around estimates for these three traits. Notably, when we did not detect an effect of these traits, we tested them as predictors in separate models and found that associations among these traits were not strong enough to affect our conclusions. We were also limited in that, like many previous studies, response variables in our analysis come from IUCN assessments. While the IUCN maintains the largest global dataset on species extinction risk and threats useful for comparative analyses, these measures are vulnerable to errors in empirical data and in models used to estimate population declines and extinction risk [68]. As a result, there is likely to be uncaptured uncertainty associated with the measures of extinction risk used in our analyses.
Cardillo & Meijaard [18] identified the limitations of comparative studies of extinction risk when it comes to conservation action, including the difficulty of translating results to policy and on the ground conservation activities. We therefore offer some connections between our findings and real-world conservation questions. First, understanding the biological and behavioural predictors of threat susceptibility in a broader range of taxa could help organizations like the IUCN to identify which threats pose the most imminent harm to species with shared characteristics. When it comes to conservation action, this information can be used to identify which populations are most likely to become threatened by anthropogenic activity in the near future based on a combination of imminent threats, traits, and species’ distributions. For instance, overlaying geographical information about the expansion of threats described above on primate species' distributions would allow us to forecast which populations are most likely to be impacted based on their traits (e.g. arboreal species living in proximity to residential development may be at particularly high risk) and in turn would help identify where to allocate limited funds for conservation and surveillance efforts. This approach would require close collaboration among modellers, policy makers, and on the ground conservationists, and represents an interdisciplinary avenue for future conservation efforts.
In summary, by applying new statistical approaches for dealing with missing data to investigate the drivers of extinction and by considering the activities that influence threat status, we have shown that multiple traits contribute to primate extinction risk. Our findings suggest that the effects of some traits, such as home range size, have weakened over the past 20 years, indicating that the traits that influence a species’ threat status are changing as anthropogenic effects continue to transform natural landscapes. Other characteristics shown to affect extinction risk in other clades, such as behavioural flexibility, do not appear to affect primate extinction risk, suggesting that different processes likely govern extinction in different clades. Focusing on mitigating key threats, as identified here, from susceptible species' geographical ranges will be an important and necessary step for future recovery.
Acknowledgements
We thank Dan Greenberg, Mark Janko, and Jarrod Hadfield for statistical advice and Cecile Ane for informative conversation about Rphylopars. We also thank Susan Alberts, members of the Alberts Lab at Duke University, and Brian Lerch for their comments on previous drafts of the manuscript.
Ethics
This work did not require ethical approval from a human subject or animal welfare committee.
Data accessibility
The final data and code used for all analyses can be found on Zenodo: https://doi.org/10.5281/zenodo.8200120 [47].
Supplementary analyses and results tables are provided in electronic supplementary material [69].
Declaration of AI use
We have not used AI-assisted technologies in creating this article.
Authors' contributions
M.J.A.C.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, visualization, writing—original draft, writing—review and editing; C.L.N.: conceptualization, methodology, supervision, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This work was funded by Duke University.
References
- 1.Chichorro F, Juslén A, Cardoso P. 2019. A review of the relation between species traits and extinction risk. Biol. Conserv. 237, 220-229. ( 10.1016/j.biocon.2019.07.001) [DOI] [Google Scholar]
- 2.Bennett PM, Owens IP. 1997. Variation in extinction risk among birds: chance or evolutionary predisposition? Proc. R. Soc. Lond. B 264, 401-408. ( 10.1098/rspb.1997.0057) [DOI] [Google Scholar]
- 3.Purvis A, Gittleman JL, Cowlishaw G, Mace GM. 2000. Predicting extinction risk in declining species. Proc. R. Soc. B 267, 1947-1952. ( 10.1098/rspb.2000.1234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardillo M, Bromham L. 2001. Body size and risk of extinction in Australian mammals. Conserv. Biol. 15, 1435-1440. ( 10.1111/j.1523-1739.2001.00286.x) [DOI] [Google Scholar]
- 5.Cardillo M, Mace GM, Jones KE, Bielby J, Bininda-Emonds OR, Sechrest W, Orme CD, Purvis A. 2005. Multiple causes of high extinction risk in large mammal species. Science 309, 1239-1241. ( 10.1126/science.1116030) [DOI] [PubMed] [Google Scholar]
- 6.Fritz SA, Bininda-Emonds OR, Purvis A. 2009. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol. Lett. 12, 538-549. ( 10.1111/j.1461-0248.2009.01307.x) [DOI] [PubMed] [Google Scholar]
- 7.Lee TM, Jetz W. 2011. Unravelling the structure of species extinction risk for predictive conservation science. Proc. R. Soc. B 278, 1329-1338. ( 10.1098/rspb.2010.1877) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matthews LJ, Arnold C, Machanda Z, Nunn CL. 2011. Primate extinction risk and historical patterns of speciation and extinction in relation to body mass. Proc. R. Soc. B 278, 1256-1263. ( 10.1098/rspb.2010.1489) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ripple WJ, Wolf C, Newsome TM, Hoffmann M, Wirsing AJ, McCauley DJ. 2017. Extinction risk is most acute for the world's largest and smallest vertebrates. Proc. Natl Acad. Sci. USA 114, 10 678-10 683. ( 10.1073/pnas.1702078114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nolte D, Boutaud E, Kotze DJ, Schuldt A, Assmann T. 2019. Habitat specialization, distribution range size and body size drive extinction risk in carabid beetles. Biodiversity Conserv. 28, 1267-1283. ( 10.1007/s10531-019-01724-9) [DOI] [Google Scholar]
- 11.Chichorro F, Correia L, Cardoso P. 2022. Biological traits interact with human threats to drive extinctions: a modelling study. Ecol. Inform. 69, 101604. ( 10.1016/j.ecoinf.2022.101604) [DOI] [Google Scholar]
- 12.Chichorro F, et al. 2022. Trait-based prediction of extinction risk across terrestrial taxa. Biol. Conserv. 274, 109738. ( 10.1016/j.biocon.2022.109738) [DOI] [Google Scholar]
- 13.Cardillo M. 2021. Clarifying the relationship between body size and extinction risk in amphibians by complete mapping of model space. Proc. R. Soc. B 288, 20203011. ( 10.1098/rspb.2020.3011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davidson AD, Hamilton MJ, Boyer AG, Brown JH, Ceballos G. 2009. Multiple ecological pathways to extinction in mammals. Proc. Natl Acad. Sci. USA 106, 10 702-10 705. ( 10.1073/pnas.0901956106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidson AD, Boyer AG, Kim H, Pompa-Mansilla S, Hamilton MJ, Costa DP, Ceballos G, Brown JH. 2012. Drivers and hotspots of extinction risk in marine mammals. Proc. Natl Acad. Sci. USA 109, 3395-3400. ( 10.1073/pnas.1121469109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ducatez S, Sol D, Sayol F, Lefebvre L. 2020. Behavioural plasticity is associated with reduced extinction risk in birds. Nat. Ecol. Evol. 4, 788-793. ( 10.1038/s41559-020-1168-8) [DOI] [PubMed] [Google Scholar]
- 17.Estrada A, et al. 2017. Impending extinction crisis of the world's primates: why primates matter. Sci. Adv. 3, e1600946. ( 10.1126/sciadv.1600946) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardillo M, Meijaard E. 2012. Are comparative studies of extinction risk useful for conservation? Trends Ecol. Evol. 27, 167-171. ( 10.1016/j.tree.2011.09.013) [DOI] [PubMed] [Google Scholar]
- 19.Purvis A, Cardillo M, Grenyer R, Collen B. 2005. Correlates of extinction risk: phylogeny, biology, threat and scale. In Phylogeny and conservation (eds Purvis A, Gittleman JL, Brooks T), pp. 295-316. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 20.Cardillo M, Mace GM, Gittleman JL, Jones KE, Bielby J, Purvis A. 2008. The predictability of extinction: biological and external correlates of decline in mammals. Proc. R. Soc. B 275, 1441-1448. ( 10.1098/rspb.2008.0179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González-Suárez M, Gómez A, Revilla E. 2013. Which intrinsic traits predict vulnerability to extinction depends on the actual threatening processes. Ecosphere 4, 1-16. ( 10.1890/ES12-00380.1) [DOI] [Google Scholar]
- 22.Murray KA, Verde Arregoitia LD, Davidson A, Di Marco M, Di Fonzo MM. 2014. Threat to the point: improving the value of comparative extinction risk analysis for conservation action. Glob. Change Biol. 20, 483-494. ( 10.1111/gcb.12366) [DOI] [PubMed] [Google Scholar]
- 23.Di Marco M, Collen B, Rondinini C, Mace GM. 2015. Historical drivers of extinction risk: using past evidence to direct future monitoring. Proc. R. Soc. B 282, 20150928. ( 10.1098/rspb.2015.0928) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruland F, Jeschke JM. 2017. Threat-dependent traits of endangered frogs. Biol. Conserv. 206, 310-313. ( 10.1016/j.biocon.2016.11.027) [DOI] [Google Scholar]
- 25.Atwood TB, Valentine SA, Hammill E, McCauley DJ, Madin EM, Beard KH, Pearse WD. 2020. Herbivores at the highest risk of extinction among mammals, birds, and reptiles. Sci. Adv. 6, eabb8458. ( 10.1126/sciadv.abb8458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards C, Cooke RS, Bates AE. 2021. Biological traits of seabirds predict extinction risk and vulnerability to anthropogenic threats. Global Ecol. Biogeogr. 30, 973-986. ( 10.1111/geb.13279) [DOI] [Google Scholar]
- 27.Nakagawa S, Freckleton RP. 2008. Missing inaction: the dangers of ignoring missing data. Trends Ecol. Evol. 23, 592-596. ( 10.1016/j.tree.2008.06.014) [DOI] [PubMed] [Google Scholar]
- 28.Machado FF, Jardim L, Dinnage R, Brito D, Cardillo M. 2022. Diet disparity and diversity predict extinction risk in primates. Anim. Conserv. 26, 331-339. ( 10.1111/acv.12823) [DOI] [Google Scholar]
- 29.IUCN. 2021. The IUCN Red List of Threatened Species Version 2021-1. See https://www.iucnredlist.org/en.
- 30.Rylands AB, Mittermeier RA. 2014. Primate taxonomy: species and conservation. Evol. Anthropol. 23, 8-10. ( 10.1002/evan.21387) [DOI] [PubMed] [Google Scholar]
- 31.Creighton MJA, Luo AQ, Reader SM, Mooers AØ. 2022. Predictors of taxonomic inflation and its role in primate conservation. Anim. Conserv. 26, 355-364. ( 10.1111/acv.12825) [DOI] [Google Scholar]
- 32.Upham NS, Esselstyn JA, Jetz W. 2019. Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 17, e3000494. ( 10.1371/journal.pbio.3000494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodroffe R, Ginsberg JR. 1998. Edge effects and the extinction of populations inside protected areas. Science 280, 2126-2128. ( 10.1126/science.280.5372.2126) [DOI] [PubMed] [Google Scholar]
- 34.Reader SM, Hager Y, Laland KN. 2011. The evolution of primate general and cultural intelligence. Phil. Trans. R. Soc. B 366, 1017-1027. ( 10.1098/rstb.2010.0342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarrete AF, Reader SM, Street SE, Whalen A, Laland KN. 2016. The coevolution of innovation and technical intelligence in primates. Phil. Trans. R. Soc. B 371, 20150186. ( 10.1098/rstb.2015.0186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyles JG, Storm JJ. 2007. The perils of picky eating: dietary breadth is related to extinction risk in insectivorous bats. PLoS ONE 2, e672. ( 10.1371/journal.pone.0000672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Höglund J. 1996. Can mating systems affect local extinction risks? Two examples of lek-breeding waders. Oikos 77, 184-188. ( 10.2307/3546056) [DOI] [Google Scholar]
- 38.Munstermann MJ, Heim NA, McCauley DJ, Payne JL, Upham NS, Wang SC, Knope ML. 2022. A global ecological signal of extinction risk in terrestrial vertebrates. Conserv. Biol. 36, e13852. ( 10.1111/cobi.13852) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biber E. 2002. Patterns of endemic extinctions among island bird species. Ecography 25, 661-676. ( 10.1034/j.1600-0587.2002.t01-1-250603.x) [DOI] [Google Scholar]
- 40.Blackburn TM, Cassey P, Duncan RP, Evans KL, Gaston KJ. 2004. Avian extinction and mammalian introductions on oceanic islands. Science 305, 1955-1958. ( 10.1126/science.1101617) [DOI] [PubMed] [Google Scholar]
- 41.Leclerc C, Courchamp F, Bellard C. 2018. Insular threat associations within taxa worldwide. Sci. Rep. 8, 6393. ( 10.1038/s41598-018-24733-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salafsky N, et al. 2008. A standard lexicon for biodiversity conservation: unified classifications of threats and actions. Conserv. Biol. 22, 897-911. ( 10.1111/j.1523-1739.2008.00937.x) [DOI] [PubMed] [Google Scholar]
- 43.Galán-Acedo C, Arroyo-Rodríguez V, Andresen E, Arasa-Gisbert R. 2019. Ecological traits of the world's primates. Sci. Data 6, 1-5. ( 10.1038/s41597-019-0059-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowe N, Myers M. 2011. All the world’s primates. Primate Conservation, Inc. See https://www.alltheworldsprimates.org.
- 45.Powell LE, Isler K, Barton RA. 2017. Re-evaluating the link between brain size and behavioural ecology in primates. Proc. R. Soc. B 284, 20171765. ( 10.1098/rspb.2017.1765) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeCasien AR, Williams SA, Higham JP. 2017. Primate brain size is predicted by diet but not sociality. Nat. Ecol. Evol. 1, 0112. ( 10.1038/s41559-017-0112) [DOI] [PubMed] [Google Scholar]
- 47.Creighton MJA, Nunn CL. 2023. Data and code from: Explaining the primate extinction crisis: predictors of extinction risk and active threats (Version 1). Zenodo. ( 10.5281/zenodo.8200120) [DOI] [PMC free article] [PubMed]
- 48.Santos T, Diniz-Filho JA, e Luis TR, Bini M, Santos MT. 2018. Package ‘PVR’. See https://cran.r-project.org/web/packages/PVR/index.html.
- 49.Ripley B, Venables B, Bates DM, Hornik K, Gebhardt A, Firth D, Ripley MB. 2013. Package ‘MASS’. CRAN Repository. See https://cran.r-project.org/web/packages/MASS/index.html.
- 50.Fox J, et al. 2012. Package ‘car’. See https://cran.r-project.org/web/packages/car/index.html.
- 51.Goolsby EW, Bruggeman J, Ané C. 2017. Rphylopars: fast multivariate phylogenetic comparative methods for missing data and within-species variation. Methods Ecol. Evol. 8, 22-27. ( 10.1111/2041-210X.12612) [DOI] [Google Scholar]
- 52.Johnson TF, Isaac NJ, Paviolo A, González-Suárez M. 2021. Handling missing values in trait data. Global Ecol. Biogeogr. 30, 51-62. ( 10.1111/geb.13185) [DOI] [Google Scholar]
- 53.Hadfield JD. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 33, 1-22. ( 10.18637/jss.v033.i02)20808728 [DOI] [Google Scholar]
- 54.Butchart SH, et al. 2007. Improvements to the red list index. PLoS ONE 2, e140. ( 10.1371/journal.pone.0000140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakagawa S, De Villemereuil P. 2019. A general method for simultaneously accounting for phylogenetic and species sampling uncertainty via Rubin's rules in comparative analysis. Syst. Biol. 68, 632-641. ( 10.1093/sysbio/syy089) [DOI] [PubMed] [Google Scholar]
- 56.Gelman A. 2008. Scaling regression inputs by dividing by two standard deviations. Stat. Med. 27, 2865-2873. ( 10.1002/sim.3107) [DOI] [PubMed] [Google Scholar]
- 57.Hadfield JD. 2018. MCMCglmm course notes. See http://cran.nexr.com/web/packages/MCMCglmm/vignettes/CourseNotes.pdf.
- 58.Harcourt AH, Parks SA. 2003. Threatened primates experience high human densities: adding an index of threat to the IUCN Red List criteria. Biol. Conserv. 109, 137-149. ( 10.1016/S0006-3207(02)00146-5) [DOI] [Google Scholar]
- 59.McElreath R. 2018. Statistical rethinking: a Bayesian course with examples in R and Stan. New York: NY: Chapman and Hall/CRC. [Google Scholar]
- 60.Reader SM, Laland KN. 2002. Social intelligence, innovation, and enhanced brain size in primates. Proc. Natl Acad. Sci. USA 99, 4436-4441. ( 10.1073/pnas.062041299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Creighton MJA, Greenberg DA, Reader SM, Mooers AØ. 2021. The role of behavioural flexibility in primate diversification. Anim. Behav. 180, 269-290. ( 10.1016/j.anbehav.2021.07.013) [DOI] [Google Scholar]
- 62.Beever EA, Hall LE, Varner J, Loosen AE, Dunham JB, Gahl MK, Smith FA, Lawler JJ. 2017. Behavioral flexibility as a mechanism for coping with climate change. Front. Ecol. Environ. 15, 299-308. ( 10.1002/fee.1502) [DOI] [Google Scholar]
- 63.Maples WR, Maples MK, Greenhood WF, Walek ML. 1976. Adaptations of crop-raiding baboons in Kenya. Am. J. Phys. Anthropol. 45, 309-315. ( 10.1002/ajpa.1330450216) [DOI] [Google Scholar]
- 64.Hahn NE, Proulx D, Muruthi PM, Alberts S, Altmann J. 2003. Gastrointestinal parasites in free-ranging Kenyan baboons (Papio cynocephalus and P. anubis). Int. J. Primatol. 24, 271-279. ( 10.1023/A:1023092915171) [DOI] [Google Scholar]
- 65.Hockings KJ, Anderson JR, Matsuzawa T. 2009. Use of wild and cultivated foods by chimpanzees at Bossou, Republic of Guinea: feeding dynamics in a human-influenced environment. Am. J. Primatol. 71, 636-646. ( 10.1002/ajp.20698) [DOI] [PubMed] [Google Scholar]
- 66.McLennan MR, Spagnoletti N, Hockings KJ. 2017. The implications of primate behavioral flexibility for sustainable human–primate coexistence in anthropogenic habitats. Int. J. Primatol. 38, 105-121. ( 10.1007/s10764-017-9962-0) [DOI] [Google Scholar]
- 67.Freckleton RP. 2002. On the misuse of residuals in ecology: regression of residuals vs. multiple regression. J. Anim. Ecol. 71, 542-545. ( 10.1046/j.1365-2656.2002.00618.x) [DOI] [Google Scholar]
- 68.Rueda-Cediel P, Anderson KE, Regan TJ, Regan HM. 2018. Effects of uncertainty and variability on population declines and IUCN Red List classifications. Conserv. Biol. 32, 916-925. ( 10.1111/cobi.13081) [DOI] [PubMed] [Google Scholar]
- 69.Creighton MJA, Nunn CL. 2023. Explaining the primate extinction crisis: predictors of extinction risk and active threats. Figshare. ( 10.6084/m9.figshare.c.6791601) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Creighton MJA, Nunn CL. 2023. Data and code from: Explaining the primate extinction crisis: predictors of extinction risk and active threats (Version 1). Zenodo. ( 10.5281/zenodo.8200120) [DOI] [PMC free article] [PubMed]
Data Availability Statement
The final data and code used for all analyses can be found on Zenodo: https://doi.org/10.5281/zenodo.8200120 [47].
Supplementary analyses and results tables are provided in electronic supplementary material [69].