Abstract
Recent studies have suggested that neuropilin‐1 (NRP1) may serve as a potential receptor in severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection. However, the biophysical characteristics of interactions between NRP1 and SARS‐CoV‐2 remain unclear. In this study, we examined the interactions between NRP1 and various SARS‐CoV‐2 spike (S) fragments, including the receptor‐binding domain (RBD) and the S protein trimer in a soluble form or expressed on pseudovirions, using atomic force microscopy and structural modeling. Our measurements shows that NRP1 interacts with the RBD and trimer at a higher binding frequency (BF) compared to ACE2. This NRP1‐RBD interaction has also been predicted and simulated via AlphaFold2 and molecular dynamics simulations, and the results indicate that their binding patterns are very similar to RBD‐ACE2 interactions. Additionally, under similar loading rates, the most probable unbinding forces between NRP1 and S trimer (both soluble form and on pseudovirions) are larger than the forces between NRP1 and RBD and between trimer and ACE2. Further analysis indicates that NRP1 has a stronger binding affinity to the SARS‐CoV‐2 S trimer with a dissociation rate of 0.87 s−1, four times lower than the dissociation rate of 3.65 s−1 between NRP1 and RBD. Moreover, additional experiments show that RBD‐neutralizing antibodies can significantly reduce the BF for both ACE2 and NRP1. Together, the study suggests that NRP1 can be an alternative receptor for SARS‐CoV‐2 attachment to human cells, and the neutralizing antibodies targeting SARS‐CoV‐2 RBD can reduce the binding between SARS‐CoV‐2 and NRP1.
Keywords: AlphaFold, force microscopy, MD simulation, neuropilin‐1, SARS‐CoV‐2, virus–host cell interaction
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is a highly transmissible and pathogenic coronavirus (Hu et al., 2021). First identified in Wuhan, China (Gomes, 2020), SARS‐CoV‐2 has rapidly spread globally and is responsible for the COVID‐19 (coronavirus disease 2019) pandemic. According to the World Health Organization (WHO) report, as of August 29, 2023, the global cumulative number of reported cases is now over 770 million, with more than 6.9 million death cases (WHO, 2023). Though several vaccines worldwide have been used for prevention under emergency use authorization (WHO, NaN), it is expected that the transmission of this novel coronaviruses will keep increasing due to more transmissible variants, relaxation of public health social measures, and inequitable vaccine distribution (WHO, 2021). Therefore, the study of SARS‐CoV‐2 is still important for researchers worldwide to provide more insights into prevention and treatments.
Coronaviruses are enveloped, positive‐sense RNA viruses that belong to the family Coronaviridae (Heald‐Sargent and Gallagher, 2012). They are classified into four genera (α, β, γ, and δ) and SARS‐CoV‐2 belongs to the β‐CoV genus. The genome of SARS‐CoV‐2 encodes four structural proteins, spike (S), envelope (E), membrane (M), nucleocapsid (N), and a series of nonstructural proteins that mediate virus replication and inhibit host immune responses (Alanagreh et al., 2020; Naqvi et al., 2020). Among these proteins, the S protein is a heavily N‐linked glycosylated homo‐trimer projecting 200 Å from the surface of SARS‐CoV‐2 (Cao et al., 2021), which is very crucial in mediating receptor binding and hosting cell entry (Bierig et al., 2020; Cao et al., 2021). During SARS‐CoV‐2 entry mediated by S protein, the S protein first binds to its receptor on the host cell membrane, and this binding allows a proteolytic cleavage by host cell proteases, resulting in the exposure of a fusion peptide (FP) (Wrapp et al., 2020). This FP then embeds into the host cell membrane and drives viral‐host membrane fusion. Following membrane fusion, the viral RNA enters the host cell, completing the crucial invasion process (Belouzard et al., 2012; Cao et al., 2021). It is now well‐known that the receptor‐binding domain (RBD) of the SARS‐CoV‐2 S protein binds specifically to ACE2 and initiates the virus‐cell membrane merging (Shang et al., 2020; Yan et al., 2020). However, a recent study by Abebe et al. indicated that the virus also infects tissues with low ACE2 expression or no ACE2 expression (Chekol Abebe et al., 2021). Inspired by this finding, researchers have explored potential receptors that can facilitate virus–cell interactions when the ACE2 level is low.
Neuropilin‐1 (NRP1) is a protein expressed on the surface of certain types of cells in the body, including cells of the immune system and cells of the respiratory tract (Roy et al., 2017; Wild et al., 2012). It is a highly conserved single‐pass membrane protein involved in neuronal development and many other diverse physiological processes, such as cell proliferation, angiogenesis, vascular permeability, and immune functions (Kyrou et al., 2021; Liu et al., 2021). Recent studies indicated that NRP1 could play a role in the infection of cells by SARS‐CoV‐2. A previous study suggested that S protein binding to NRP1 b1 domain will use the same binding site between VEGF ligands and NRP1 b1 domain (Gudowska‐Sawczuk and Mroczko, 2021). Daly et al. (2020) reported that NRP1 was also found to facilitate the virus infection and interact with a polybasic sequence motif in S protein after cleavage directly, and therefore inhibiting NRP1 expression can significantly reduce the infection rate. In the meantime, NRP1 has been found to be upregulated in the respiratory tract during SARS‐CoV‐2 infection according to Cantuti‐Castelvetri et al. (Cantuti‐Castelvetri et al., 2020; Mayi et al., 2021). This may increase the number of receptors on the surface of cells that are available for virus binding, making it easier for the virus to enter and infect the cells. A molecular modeling study under specific docking conditions showed that there are multiple interacting residue pairs between NRP1 b1 domain and SARS‐CoV‐2 RBD (Alnomasy, 2021). Additional interactions were also proposed between NRP1 a2‐b1‐b2 domains and the S protein S1 subunit at multiple locations on both N‐terminal domain and C‐terminal domain including RBD (Li and Buck, 2021). However, it is important to note that the role of NRP1 in SARS‐CoV‐2 infection is not fully understood. In addition, the biomechanical binding strength between NRP1 and SARS‐CoV‐2 S protein fragments that could provide more solid information regarding forces and potential interactions have not been fully studied experimentally.
Here we report our quantification of mechanical strengths and binding affinity between NRP1 and SARS‐CoV‐2 S protein fragments using a custom‐built atomic force microscopy (AFM) based single‐molecule force spectroscopy (SMFS) (Sumbul et al., 2021; Wojcikiewicz et al., 2006), which can directly measure a single bond rupture between two proteins (Xiaohui Zhang et al., 2009; Zhu et al., 2022). In the meantime, AlphaFold2 Multimer and molecular dynamics (MD) simulation were also used for binding prediction to provide additional knowledge regarding the interaction between NRP1 and SARS‐CoV‐2 S protein RBD. We also studied the binding strength between NRP1 and SARS‐CoV‐2 pseudovirions and compared with ACE2 under the same setting to gain a better knowledge of the role of NRP1 in SARS‐CoV‐2 infection. In addition, we tested the impact of antibodies targeting SARS‐CoV‐2 S protein RBD in inhibiting the binding between NRP1 and SARS‐CoV‐2 pseudovirions. Together, our study suggests that NRP1 can be an alternative receptor for SARS‐CoV‐2 attachment to human cells, and the neutralizing antibodies targeting the RBD could still interrupt the binding between SARS‐CoV‐2 and NRP1.
2. RESULTS
2.1. NRP1 binds to SARS‐CoV‐2 S protein RBD and trimer with mechanical strength comparable to ACE2
The binding strengths between SARS‐CoV‐2 S protein RBD/trimer and NRP1 were characterized by using AFM. These proteins were attached to the cantilever AFM tips and glass surface via an established protocol (Figure 1a). Measurements were made by utilizing a custom‐built AFM, which is specifically designed for operation in the force spectroscopy mode (Krieg et al., 2019; Newton et al., 2017; Wojcikiewicz et al., 2004). During AFM force scanning, the cantilever would firstly be lowered to the surface of the glass substrate, where immobilized soluble NRP1 would interact with the RBD/trimer. Then, the cantilever was retracted from the substrate surface under a set velocity. Any binding between the RBD/trimer and NRP1 would generate an adhesive pull‐off force determined by the cantilever's deflection and detected by a position‐sensitive two‐segment photodiode (Figure 1b).
FIGURE 1.
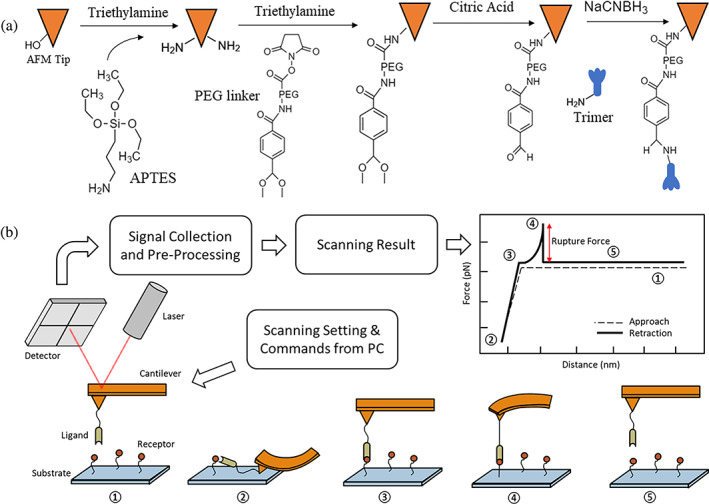
(a) The process of AFM tip functionalization and protein immobilization. The amino functionalized substrate also utilized the same procedure from PEG coating to protein immobilization. (b) Scheme of receptor–ligand interaction measurement using a custom‐built AFM. In our study, S protein fragments or pseudovirions were immobilized on the AFM tip, and receptors were fixed on the functionalized glass substrate. An example of typical force scanning result from AFM is shown. Dashed line in the plot shows the tip approaching and the solid line shows the tip retraction along with a detected unbinding between receptor and ligand. ①–⑤ illustrate the entire procedure of a single force scan, where the tip approaches the substrate from ① to ②, and the receptor‐ligand complex is unfolding from ② to ④, binding is going to break at ④, generating a peak in the retraction line, and the tip is back to an undeformed status and keeps retracting during ⑤. AFM, atomic force microscopy; PEG, polyethylene glycol.
The biophysical characteristics of RBD/trimer‐NRP1 interactions were studied through a dynamic force spectrum (DFS), which is a plot of the most probable unbinding force as a function of the loading rate. All unbinding forces measured for each specific interaction were first divided into at least four groups in the order of their loading rates. Data in each group were analyzed by histograms to determine the most probable unbinding force (Figure 2a,b). Figure 2c and Figure 2d show the unbinding force between the RBD/trimer and NRP1, which increases linearly with the loading rate in logarithm. While ACE2 exhibits a stronger unbinding force with RBD as seen in Figure 2c, the introduction of the trimer instead of the RBD alone reveals that NRP1 has a greater unbinding force with the trimer than ACE2 (Figure 2d). This suggests the presence of additional potential binding sites for NRP1 within the trimer. To examine if the interaction is specific, an adhesion frequency measurement was conducted under the same measurement conditions, and the measurement included a control group, which is the interaction between the trimer and bovine serum albumin (BSA). The result shows the BF between RBD/trimer and NRP1 or ACE2 is about 20%–50% on average, while the BF between RBD/trimer and BSA is around 5% or less, almost no adhesion between the RBD/trimer and BSA (Figure 2e), indicating that most of the interaction detected stemmed from the binding between the receptors and ligands.
FIGURE 2.
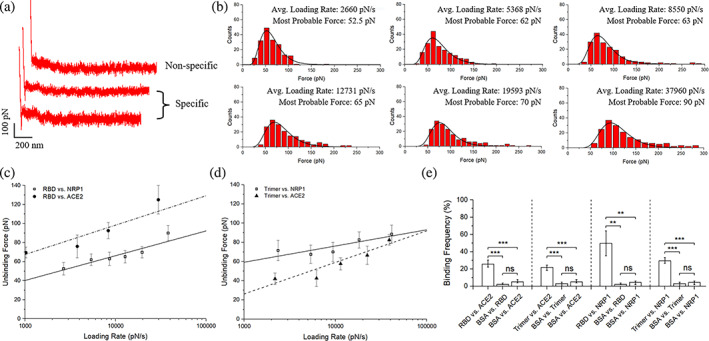
SMFS results among SARS‐CoV‐2 S protein fragments, NRP1, and ACE2. (a) Typical force scans for nonspecific interaction and specific interaction. (b) Exemplary statistical analysis of force scan results from the RBD versus NRP1 experiments to determine most probable unbinding forces. (c) Dynamics force spectrum (DFS) with the linear fit for interactions between RBD and NRP1, compared with interactions between RBD and ACE2. (d) DFS with the linear fit for interactions between trimer and NRP1 with interaction between trimer and ACE2 as a comparison. (e) The interaction specificity is compared by the binding frequency among experiment groups and control groups using bovine serum albumin (BSA). Contact force, dwell time, and retraction speed during the binding frequency measurement were set to 200 pN, 0 s, and 3.7 μm/s, respectively. Error bars are given to show the range. Significance comparison was determined by unpaired t‐test. **<0.01; ***<0.001; ns: not significant. NRP1, neuropilin‐1; RBD, receptor‐binding domain; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; SMFS, single‐molecule force spectroscopy.
A more detailed analysis of the biophysical characteristics of these interactions was proceeded by fitting the DFS data to the Bell–Evans model. According to the model, an external force could distort the intermolecular potential of a receptor‐ligand complex, resulting in the decrease of the activation energy and the increase of the dissociation rate, :
| (1) |
where is the dissociation rate constant in the absence of the external pulling force; is the position of the transition state, indicating the energy barrier location, also called the reaction length; T is the temperature in Kelvin; and is the Boltzmann constant. The probability of protein–protein unbinding as a function of the force can be written as:
| (2) |
The most probable unbinding force () at a given loading rate () can then be given by:
| (3) |
By fitting the DFS data to Equation (3), we calculated the and for these interactions. The results show that the dissociation rate of RBD‐NRP1 is about 3.65 ± 3.33 s−1 with a reaction length of 0.33 ± 0.07 nm, and the dissociation rate of trimer‐NRP1 is about 0.87 ± 0.52 s−1 with a reaction length of 0.39 ± 0.04 nm. Smaller dissociation in the interaction between trimer and NRP1 indicates that they have stronger binding than the interaction between RBD and NRP1, but slightly weaker binding compared to ACE2‐RBD interaction from our previous study (a dissociation rate around 0.047 ± 0.034 s−1 with an activation barrier position of 0.39 ± 0.05 nm) (Cao et al., 2021).
2.2. Prediction via AlphaFold2 and MD simulation provide potential RBD‐NRP1 complexes
AlphaFold2 with multimer extension was utilized to predict complex models of NRP1 a1a2b1b2 domains (residues 22–591) and SARS‐CoV‐2 S protein RBD (residues 319–537), considering the efficiency and restrictions on Google Colab and following verification via MD simulations. AlphaFold2 provided several predictions based on the user's setting. We first compared the predicted RBD (p‐RBD) structure in the complex models to the experimental structures (PDB ID: 6VXX for closed state RBD; PDB ID: 6VYB for open state RBD) in RCSB PDB using TM‐score. The higher the TM‐score is, the more similar these two structures are. Statistically, a TM‐score of greater than 0.5 suggests that two proteins are in about the same fold; on the contrary, when the TM‐score is smaller than 0.17, two proteins are in two random structures. However, it should be noted that, when TM‐score is greater than 0.3, the p‐value corresponding to the TM‐score is still significantly small (p < 0.001) (Xu and Zhang, 2010), meaning that two proteins are still likely in about the same fold.
Our comparison results (Figure 3) show that all p‐RBDs are in about the same fold to the experimental structures and are slightly closer to the closed‐state RBD structure. Enabling or disabling Amber force field (Amber FF) during predictions did not have an obvious impact on the structural similarity result. It should be noted that this type of comparison may not be able to align flexible domains such as twists around hinges in some structures. The predicted NRP1 structure was not compared to the experimental structures in PDB since most structures before 2018 have been used for AlphaFold2 training. This structural comparison initially confirmed that all predicted protein structures are reasonable. MD simulations were then conducted to examine the stability of the complex structure and obtain insights on potential interacting residue pairs.
FIGURE 3.
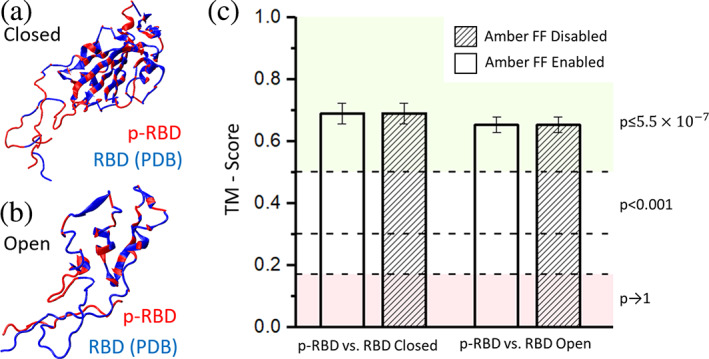
Structural similarity comparison between predicted models and PDB experimental structures using TM‐score. Shown are overlaid structures of the predicted model (p‐RBD) and the reported experimental structures of (a) closed‐state RBD (from PDB 6VXX) and (b) open‐state RBD (from PDB 6VYB). (c) Columns show the mean value of TM‐score and error bar shows the standard deviation. The influence of enabling Amber force field (Amber FF) on model prediction was also considered and no significant difference was found between models with and without the Amber FF. RBD, receptor‐binding domain.
The MD simulation results on Alphafold2‐predicted bound complexes have further narrowed down to two candidates that can form stable interactions, model 2 and 3 (m2, m3). Figure 4a shows an initial structure of m2. From the beginning, the receptor binding motif (RBM, a crucial binding sequence in RBD) is in close contact with b1 domain (residue 254–403, pink) of NRP1. As the simulation progressed, the RBM loop structure reoriented its position, having more pair residue interactions between RBM and b1 domain of NRP1. The representative interacting residue pairs of RBD and of NRP1 are: K458‐D320 (RBD‐NRP1), F456‐Y297, N501‐K351, and T500‐K350 (Figure 4b). Interestingly, most interacting residues in RBM at the RBD‐NRP1 interface are also involved in contacts with ACE2. For example, our previous studies showed that F456, N501, and T500 in RBD exhibited strong interactions with T27, D30, K31, K353, Y41, D355, and R357 in ACE2 at the RBD‐ACE2 interface (Cao et al., 2021; Jawad et al., 2021; Kim et al., 2021, 2023; Lupala et al., 2021). More specifically, we reported that F456, N501, and T500 residues had hydrophobic, charge–charge, and hydrophobic interactions, respectively. Notably, these types of interactions are also found at the RBD‐NRP1 interface resulting from the simulation.
FIGURE 4.
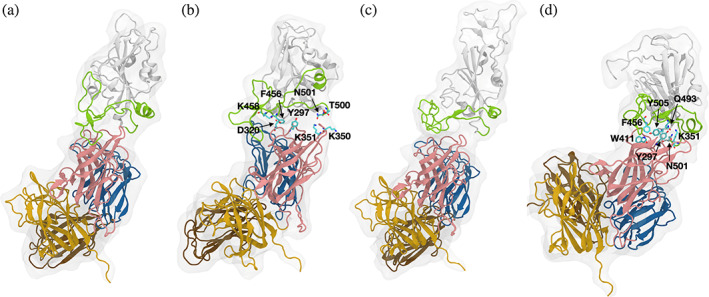
Predicted bound complexes and corresponding MD simulation results. (a and c) Initial structures of m2 and m3 from AlphaFold2 prediction. (b and d) Snapshots of m2 and m3 after 200‐ns MD simulations. The RBD is displayed in white and the RBM is shown in green. The a1, a2, b1, and b2 domains of NRP1 are colored in brown, blue, pink, and yellow, respectively. Interacting residues are represented by stick model with labels. MD, molecular dynamics; NRP1, neuropilin‐1; RBD, receptor‐binding domain; RBM, receptor binding motif.
The m3 originally did not show close contacts between RBM and NRP1 as much as in m2 (Figure 4c). However, during the simulation, NRP1 changed its orientation and the interface gained multiple pair residue interactions. It should be noted that the interacting residues in m3 are slightly different from those in m2. In m3, Q493 and Y505 in RBD are involved in the interface interactions, where Q493 shows a charge–charge interaction with K351 and Y297, and Y505 shows hydrophobic interactions with Y297. NRP1 Y297 also interacts with RBD N501, and RBD F456 has close contact with NRP1 W411 through π–π interactions. Finally, residues that are mainly having contacts with the NRP1 b1 domain are largely located at the beginning and end of the interface, exhibiting a feature of hinge, which was also reported in our prior study (Kim et al., 2023). Overall, at the RBM‐NRP1 interface, interacting residue pairs, nature, and patterns (i.e., hinge motion) are similar to those in the RBD‐ACE2 interface.
2.3. Quantification of SARS‐CoV‐2 pseudovirion–NRP1 interactions using single‐virus force spectroscopy
Pseudovirion is a modified viral particle that is typically not contagious but has a genome that can be used to produce and express specific proteins on its surface. The pseudovirion used here is provided by Dr. Wendy Maury (University of Iowa). The pseudovirus, named rVSV/Spike, has been recently used to study SARS‐CoV‐2–receptor interactions (Bohan et al., 2021). Compared to the S protein trimer we tested, pseudovirion can arrange SARS‐CoV‐2 S proteins in a more realistic conformation expressed on the virion surface. The distribution of the protein and its surrounding environment could be more natural and closer to the authentic SARS‐CoV‐2 viruses. By following the protocol mentioned previously, pseudovirions were immobilized on the AFM tip via the polyethylene glycol (PEG) linker and NRP1 was immobilized on the functionalized glass substrate. We repeated the SMFS using the custom‐built AFM (Figure 5a).
FIGURE 5.
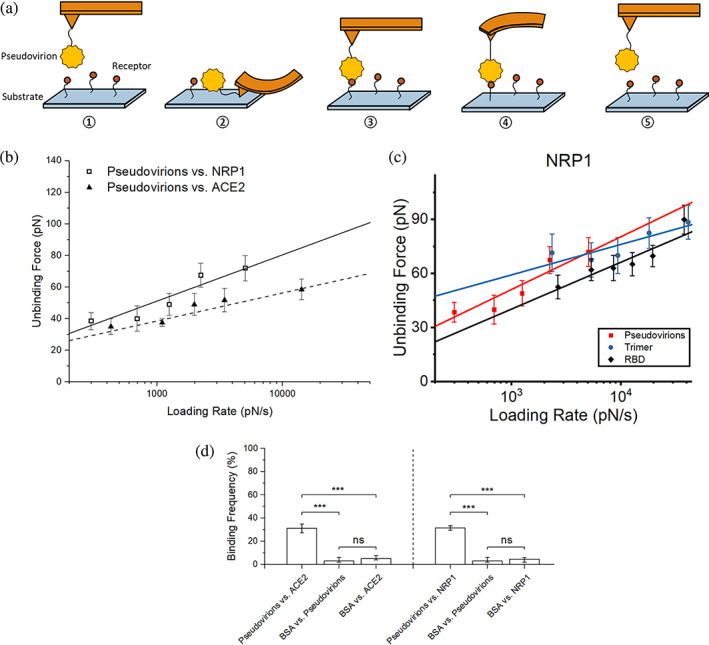
SMFS results among SARS‐CoV‐2 pseudovirions, NRP1, and ACE2. (a) Scheme of receptor–pseudovirion interaction measurement using a custom‐built AFM. In our study, SARS‐CoV‐2 pseudovirions were immobilized on the AFM tip, and receptors were fixed on the functionalized glass substrate. ①–⑤ illustrate the entire procedure of a single force scan, where the tip approaches the substrate (①–②), and the receptor‐pseudovirion complex is unfolding (②–④), binding breaks (④), generating a peak in the retraction line, and the tip is back to an undeformed status and keeps retracting (⑤). (b) Dynamics force spectrum (DFS) with the linear fit for interactions between SARS‐CoV‐2 pseudovirions and NRP1 with ACE2 as comparison. (c) DFS with the linear fit for interactions between SARS‐CoV‐2 S protein fragments or pseudovirions and NRP1. (d) The interaction specificity was compared by the binding frequency among experiment groups and control groups using BSA. Contact force, dwell time, and retraction speed during the binding frequency measurement were set to 200 pN, 0 s, and 3.7 μm/s, respectively. Error bars are given to show the range. Significance comparison was determined by unpaired t‐test. ***<0.001; ns: not significant. AFM, atomic force microscopy; BSA, bovine serum albumin; NRP1, neuropilin‐1; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; SMFS, single‐molecule force spectroscopy.
The result is shown in Figure 5. Similarly, a controlled test of adhesion frequency on receptors and BSA was also conducted to ensure interaction specificity, and a detailed analysis of the biophysical characteristics of interactions was generated by fitting the data to the Bell–Evans model, using Equation (3). The interaction specificity result (Figure 5d) shows that the experimental groups (pseudovirions vs. receptors) have a BF around 30% on average, and the BF of control groups (ligands or receptors against BSA) is around 5% or less, almost no adhesion, indicating that most of the interactions detected are specific. Compared to ACE2, SARS‐CoV‐2 pseudovirions and NRP1 have a slightly higher unbinding force (Figure 5b). Also, the unbinding force between pseudovirions and NRP1 is larger than that between RBD and NRP1 (Figure 5c). Interestingly, measured loading rates under the same settings between pseudovirions and NRP1 are on average lower than the others. The measured dissociation rate between SARS‐CoV‐2 pseudovirions and NRP1 is 0.93 ± 0.40 s−1 which is much smaller than that of RBD‐NRP1 but slightly larger than that of trimer‐NRP1. The results further strengthen our findings that NRP1 has a stronger binding affinity against SARS‐CoV‐2 S protein trimer compared to RBD.
We also estimated the association rate () between pseudovirions and NRP1 using the method developed by the Hinterdorfer group (Dragovich et al., 2019; Rankl et al., 2011). When interactions follow a pseudo first‐order kinetics, the association rate can be calculated using Equation (4):
| (4) |
where gives the effective volume where the interaction can take place; is Avogadro constant; is the number of binding partners; is the interaction time. The effective radius is estimated based on the radius of AFM tip provided by the manufacturer, the length of PEG linker, and the size of pseudovirions. Here all materials and protocols we used are the same as our previous work (Dragovich et al., 2019). Thus, the effective radius is about 130.8 nm. The binding partners can be estimated from unbinding force histogram or probability density curves at the longest contact time (Dragovich et al., 2019). The interaction time can be obtained by fitting the relation between BF and contact time (t):
| (5) |
where A is the maximum BF; is the lag time; is the interaction time. Measurements were made by using our custom‐built AFM and the retraction speed was set to a constant value. In addition, dwell time was set to several different values such as 0/60, 5/60, 10/60, 20/60, 40/60 s, etc. to provide multiple different hold time lengths when the AFM tip contacted the substrate. Time spent on indentation force loading and unloading was also counted for correction.
Fitting on the BF and contact time (Figure 6b) shows that the maximum BF between SARS‐CoV‐2 pseudovirions and NRP1 is about 49.5% and it also gives an interaction time constant of 167.74 ± 20 ms. Binding partners is about 2 based on the histogram of rupture force at the longest contact time length (Figure 6a). The association rate () can then be calculated using Equation (4) and information provided above, which is . The dissociation constant can then be estimated by the ratio between dissociation rate () and association rate (), which is about 110.8 nM.
FIGURE 6.
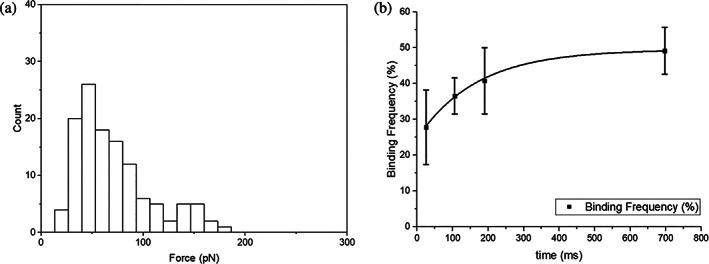
Association rate measurement results of interaction between pseudovirions and NRP1. (a) Histogram of rupture force at the longest contact time to estimate the number of binding partners. According to the histogram, it is likely to have two binding partners since there are two potential possible rupture force peaks, one around 50 pN and the other one around 140 pN. (b) Fitting binding frequency of interactions between SARS‐CoV‐2 pseudovirions and NRP1 to estimate association rate. NRP1, neuropilin‐1; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
2.4. Antibody blocking experiment
A previous study showed that soluble inhibitors for SARS‐CoV‐2 S protein RBD can significantly decrease the interaction between the S proteins and ACE2 (Yang et al., 2020). To explore the blocking efficiency on SARS‐CoV‐2 and NRP1 interaction, the antibodies for RBD (AcroBiosystems, Cat. No. SAD‐S35) were dissolved into PBS with a concentration of 10 μg/mL, which is a saturated concentration for blocking of S protein and ACE2 interactions based on manufacturer technical sheet. Pseudovirions with SARS‐CoV‐2 S proteins were immobilized on the AFM tip, and receptors (NRP1, and ACE2 for comparison) were immobilized on the amino‐functionalized glass substrate using the protocol previously mentioned (Ebner et al., 2007, 2008; Rankl et al., 2008; Riener et al., 2003). Similar SMFS experiments were conducted under the same conditions. We first measured the BF of interactions without antibodies, and the frequency was consistent with the aforementioned results. After adding the antibody into the PBS solution for 15 min, we measured the BF again between pseudovirions and receptors. The result shows that adhesion frequencies for NRP1 and ACE2 both have decreased significantly after treating with the SARS‐CoV‐2 S protein RBD antibody (Figure 7). This result combining with our MD simulation suggests that the interactions between RBD‐NRP1 and between RBD‐ACE2 could be similar and share binding residues, which leads to a competition between RBD‐ACE2 and RBD‐NRP1. Co‐existence of ACE2 and NRP1 on the cell membrane could possibly increase the chance of viral attachment, resulting in high vulnerability of cells, but current vaccines or treatments targeting on RBD could also prevent the infection effectively.
FIGURE 7.
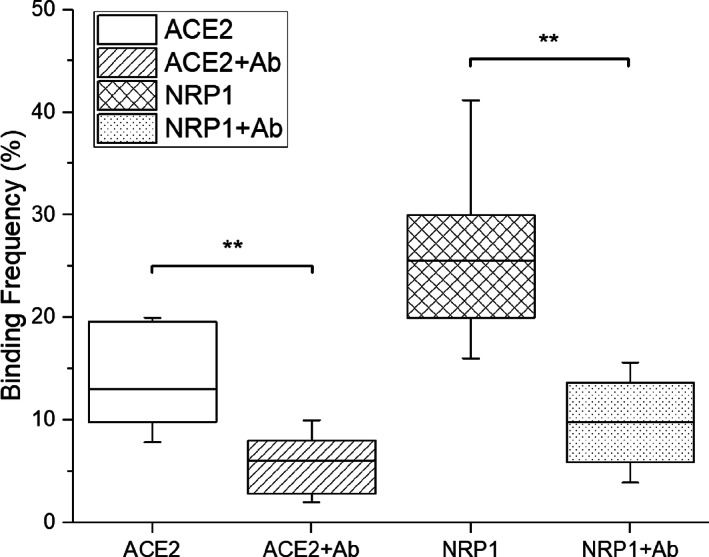
Binding frequencies between pseudovirions and ACE2 or NRP1 before and after adding antibodies for S protein RBD. Contact force, dwell time, and retraction speed during the measurement were set to 200 pN, 0 s, and 3.7 μm/s, respectively. A statistical analysis indicates that the impact of antibodies is significant for both ACE2 and NRP1. Significance comparison was determined by paired t‐test. **<0.01; ns: not significant. NRP1, neuropilin‐1; RBD, receptor‐binding domain.
3. DISCUSSION AND CONCLUSIONS
Our study reveals that SARS‐CoV‐2 S protein trimer/pseudovirions carrying S proteins and NRP1 exhibit a higher unbinding force compared to those with ACE2, and the fitted dissociation rates also indicate their potential high binding affinity. Though SARS‐CoV‐2 S protein RBD and ACE2 still possess strong binding affinity compared to NRP1, our force spectroscopy and MD simulation results suggest that the NRP1 could potentially work as an alternative receptor given the considerable unbinding force and similarity in interaction interface between NRP1 and RBD. In addition, antibodies specifically targeting S protein RBD are also found to be effective in reducing the binding probability to NRP1, which correlates well with our findings and provides additional evidence. In the meantime, it is also worth to note that, compared to ACE2, NRP1 may interact with SARS‐CoV‐2 S protein at multiple locations according to the significant difference in our measured dissociation rates for RBD‐NRP1 and trimer‐NRP1. This implies that not only can the binding possibility be increased, but also the viral docking on the cell surface can be improved when the NRP1 is present. Previous studies (Daly et al., 2020; Li and Buck, 2021) demonstrated that the NRP1 could interact with S protein at some other locations. For instance, a MD simulation study by Li and Buck showed NRP1 interacting with RRAR motif in S protein S1 subunit, where NRP1 collaborated together with ACE2 to facilitate S1‐S2 separation and enhance SARS‐CoV‐2 invasion (Li and Buck, 2021). Therefore, it is likely that NRP1 could be another crucial receptor and play multiple roles in SARS‐CoV‐2 infection. Future studies using more powerful computing platform or advanced imaging techniques such as cryo‐EM may help us reveal these multiple binding locations clearly. In general, our findings demonstrate that NRP1 could be a potent alternative receptor especially when the cell membrane is lack of ACE2.
The dissociation rate determined by Equation (3) and AFM data is a dynamic rate constant that describes ligand dissociating from a protein. It can reflect the binding affinity between ligands and receptors. Here, we measured and compared dissociation rates between NRP1 and S protein RBD, trimer, or pseudovirions: 3.65 ± 3.33 s−1 (RBD‐NRP1), 0.87 ± 0.52 s−1 (trimer‐NRP1), and 0.93 ± 0.40 s−1 (pseudovirions‐NRP1). The significant difference in dissociation rates between RBD‐NRP1 and trimer‐NRP1 suggests that in addition to RBD, the trimer offers more potential interaction sites for NRP1. Compared to traditional techniques for binding affinity measurement, our custom‐built AFM established a 2D membrane‐membrane contact environment to study SARS‐CoV‐2's affinity to NRP1, where ligands and receptors are all immobilized. Traditional measurement of binding affinity via methods including BLI, ELISA, etc. has either ligands or receptors in soluble form, which allows more degrees of freedom for studying the interaction between receptor and ligand. Such an experimental condition will provide more potential binding sites and may present a higher binding affinity in the end, which may not be able to exhibit in vivo situation.
It is well‐known that NRP1 is highly expressed in olfactory epithelium and some other neuronal cells. The strong binding affinity between SARS‐CoV‐2 S protein and NRP1 that we detected without any cleavage on S protein also implies that the binding between SARS‐CoV‐2 and NRP1 may play a role in many SARS‐CoV‐2 symptoms related to nervous systems such as loss of smell or neuronal pain, since their binding may change or interrupt signal regulations via NRP1. Previous study by Cantuti‐Castelvetri et al. (2020) concluded that SARS‐CoV‐2 may not infect cells and initiate virus entry by binding with NRP1. However, Moutal et al. (2021) observed that S protein can interfere with VEGF‐A/NRP‐1 pathway, leading to the suppression of spinal activity and reduced neuronal signal, causing analgesia. A recent study by Zazhytska et al. (2022) revealed that SARS‐CoV‐2 can indirectly affect signaling genes without infection, causing COVID‐19‐induced anosmia. Chapoval et al. summarized that a pain‐lowering effect caused by interactions between NRP1 and S protein could explain the disease transmission in asymptomatic individuals, compared to the pain‐promoting effect caused by the interaction between S protein and ACE2 (Chapoval and Keegan, 2021). In addition, researchers have hypothesized that one potential pathway to the central nervous system during SARS‐CoV‐2 infection is through the retrograde transport along axons of olfactory sensory neurons, delivering from olfactory epithelium to the olfactory bulb, then toward the olfactory nucleus in the pyriform cortex (Cardona et al., 2020; Davies et al., 2020). Considering the relatively high expression of NRP1 and its essential role in the nervous system, NRP1 may contribute to the invasion of SARS‐CoV‐2 to the central nervous system which could cause pathological complications and may enhance the deterioration of brain tumors or glioblastoma (Zalpoor et al., 2022). Thus, further research regarding the role of NRP1 in COVID‐19 could promote our understanding in relations between SARS‐CoV‐2 infection and a series of symptoms in COVID‐19 and post‐COVID‐19, which may provide potential solutions to improve patients' recovery.
4. MATERIALS AND METHODS
4.1. Protein constructs
SARS‐CoV‐2 S protein RBD (wildtype) was purchased from Acrobiosystems (Newark, DE, catalog #SPD‐C52H3). It is expressed from human 293 cells (HEK293) with a polyhistidine tag at the C‐terminus and contains AA Arg 319–Lys 537 (accession # QHD43416.1). According to the manufacturer, the purity of RBD is >95% as determined by SDS‐PAGE.
SARS‐CoV‐2 S protein trimer (wildtype) was purchased from Acrobiosystems (Newark, DE, catalog #SPN‐C52H9). It is the ectodomain of SARS‐CoV‐2 S protein which contains AA Val 16–Pro 1213 of the wild‐type (Wuhan‐Hu‐1) strain (accession # QHD43416.1). The recombinant protein is expressed from human 293 cells (HEK293) with a polyhistidine tag at the C‐terminus. Proline substitutions (F817P, A892P, A899P, A942P, K986P, V987P) and alanine substitutions (R683A and R685A) are introduced to stabilize the trimeric prefusion state of SARS‐CoV‐2 S protein and abolish the furin cleavage site. According to the manufacturer, the purity of the protein was >95% as determined by SDS‐PAGE. Trimerization of the protein has been verified by MALS and negative stained electron microscopy.
The ACE2 protein was purchased from Acrobiosystems (Newark, DE, catalog #AC2‐H52H8). It is a recombinant N‐terminal His‐tagged protein consisting of Gln18‐Ser740 of human ACE2 (GenBank accession number: AF291820.1), expressed from HEK293 cells. According to the manufacturer, the protein has been shown to bind the SARS‐CoV‐2 S protein with high affinity.
The human neuropilin‐1 protein was purchased from Acrobiosystems (Newark, DE, catalog #NR1‐H5228). It is expressed from human 293 cells (HEK293) carrying a polyhistidine tag at the C‐terminus and contains AA Phe 22–Lys 644 (accession # AAH07533).
The anti‐SARS‐CoV‐2 RBD neutralizing antibody was purchased from the Acrobiosystems (Newark, DE, catalog #SAD‐S35). The antibody is recombinantly produced from human 293 cells (HEK293) based on antibodies isolated from a SARS‐CoV‐2 infected patient. According to the manufacturer, the antibody can recognize and inhibit the interaction between SARS‐CoV‐2 RBD and ACE2 effectively. No cross‐reactivity is detected with S protein RBD domain of other CoVs.
4.2. Pseudovirus
The pseudovirus, named rVSV/Spike, was a kind gift from Dr. Wendy Maury (University of Iowa). As described earlier, the pseudovirus created stocks as a recombinant VSV virus with the spike protein carrying the D614G mutation in Vero E6 cells. For purification, the supernatants containing the pseudovirions were concentrated by centrifugation, resuspended in PBS, and layered over a 20% sucrose/PBS cushion. After additional centrifugation, the virus pellet was resuspended in PBS (Bohan et al., 2021).
4.3. Cantilever preparation/coverslip preparation
To functionalize AFM cantilevers (MLCT‐BIO‐DC, Bruker, USA) with SARS‐CoV‐2 S protein RBD, trimer, and SARS‐CoV‐2 pseudovirions, the cantilever was first silanized with (3‐aminopropyl)‐triethoxysilane (APTES). SARS‐CoV‐2 S protein RBD, trimer, and SARS‐CoV‐2 pseudovirions at 1 μM were immobilized onto the silanized cantilever using a heterobifunctional PEG crosslinker, Acetal‐PEG‐NHS (2000 MW, Creative PEGworks, USA), according to the detailed protocol developed by Dr. Hermann J. Gruber, Johannes Kepler University (Ebner et al., 2007, 2008; Rankl et al., 2008; Riener et al., 2003). Our previous work on SARS‐CoV‐2 versus ACE2 has shown that the molecular weight of Acetal‐PEG‐NHS does not influence significantly on the measurement results (Cao et al., 2021). Soluble recombinant NRP1 or ACE2 (1 μM) were attached to the amino‐functionalized glass substrates (NANOCS, USA) using the same crosslinking approach. Functionalized cantilevers and glass surfaces were stored in PBS and used for the AFM experiment within 8 h.
4.4. Single‐molecule force spectroscopy (SMFS)
SMFS was conducted using a custom‐designed AFM apparatus. AFM measurements were collected at cantilever retraction speeds ranging from 0.19 to 3.7 μm/s to achieve the desired loading rate (200–2000 pN/s). All measurements were conducted at 25°C in phosphate‐buffered saline (PBS). The contact time and indentation force between the cantilever and the sample were minimized to obtain measurements of the unitary unbinding force.
Calibration of the cantilevers (lever C of MLCT) was done by first measuring the inverse optical lever sensitivity (InvOLS) via recording force curves on a hard glass surface, followed by obtaining the spring constant of the cantilever via thermally‐induced fluctuations (Levy and Maaloum, 2001). The spring constants (9.6 ± 2.8 pN/nm, mean ± SD) of the calibrated cantilevers agreed with the values specified by the manufacturer (10 pN/nm). All the unbinding rupture forces have been corrected for viscous drag force (Franz et al., 2007), which was obtained by multiplying the tip movement velocity by the viscous drag coefficient. The viscous drag coefficient was measured by moving the cantilever at varying velocities near the substrate (Cao et al., 2021; Franz et al., 2007) and is 5.05 pN s/μm for the lever C of MLCT.
To enable the measurement of a single molecular interaction, the contact between the cantilever tip and the substrate was minimized by reducing both the contact duration (<50 ms) and the compression force (~200 pN). The brief contact duration was chosen to ensure that, for the majority of contacts (67% or greater), no adhesion (rupture force) was observed between the AFM tip and surface. Assuming the adhesion bond formation obeyed Poisson statistics, an adhesion frequency of ~33% in the force measurements implies that among the observed unbinding events, the probabilities of forming a single, double, and triple adhesion bond between AFM tip and surface were 81%, 16%, and 2%, respectively (Chesla et al., 1998). Therefore, our experimental condition ensured there was a >80% probability that the adhesion event was mediated by a single bond (Evans, 2001).
4.5. Interaction prediction via AlphaFold2 and MD simulation
NRP1 and SARS‐CoV‐2 S protein RBD complex structures were predicted by AlphaFold2 coupled with Google Colab. Compared to other structural prediction methods, AlphaFold2 is a high‐accuracy prediction tool based on deep learning that can generate the closest computational predicted structures to experiment determined structures (Mirdita et al., 2022; Skolnick et al., 2021). The latest version has included an extension, called Multimer, to predict protein–protein complexes specifically. The sequence information of human NRP1 (residues 22–644) and SARS‐CoV‐2 S protein RBD (residues 319–537) were retrieved from UniProt database (NRP1: O14786; SARS‐CoV‐2 spike: P0DTC2). These sequences were properly trimmed to fit with the sequence information of the human NRP1 and SARS‐CoV‐2 S protein RBD purchased for our experiments. The long free tail of human NRP1 (residues 592–644) was removed to improve computing efficiency. The Amber FF was enabled, the multiple sequence alignment (MSA) mode was selected as “MMseqs2,” the pair mode was set as “unpaired+paired,” and the model type was set to “AlphaFold2‐multimer‐v2” or “auto.”
Two predicted models from AlphaFold2 prediction results (i.e., m2 and m3) were used to run MD simulations, as other models from AlphaFold2 showed that RBD residues that are supposed to be imbedded toward S protein (Wrobel et al., 2020) had unrealistic interactions with b1 domain. All systems were prepared with CHARMM‐GUI Solution Builder (Jo et al., 2008; Lee et al., 2016). The initial system composed of approximately 400,000 atoms had a box size of about 160 × 160 × 160 Å3. The CHARMM36(m) FF were used for proteins (Hatcher et al., 2009); 0.15 M KCl was incorporated with TIP3P water model for the solvation (Jorgensen et al., 1983). By employing the force‐based switching function (Dion et al., 2004), the van der Waals interactions switched off smoothly between 10 and 12 Å, and for the long‐range electrostatic interactions, the particle‐mesh Eward method (Essmann et al., 1995) with a mesh size of ~1 Å was utilized. The SHAKE algorithm (Ryckaert et al., 1977) was applied to constrain bond lengths including hydrogen atom. Temperature and pressure were set to 303.15 K and 1 bar, respectively, where Langevin dynamics with a friction coefficient of 1 ps−1 and Monte Carlo barostat were applied to maintain temperature and pressure, respectively (Åqvist et al., 2004; Chow and Ferguson, 1995). The NVT simulations with positional and dihedral restraints were conducted for equilibration simulation. Then, NPT simulations were performed for production simulation without any restraints. The 4 fs of time‐step was used together with the hydrogen mass repartitioning method (Gao et al., 2021; Hopkins et al., 2015). Periodic boundary condition was applied for all simulations. All simulations were conducted at least 200 ns using OpenMM simulation package (Eastman et al., 2017).
4.6. Statistical analysis
Three hundred to 500 force curves were typically recorded for each pulling speed from different sites on the substrate. Recorded force curves were further analyzed by IGOR Pro to collect unbinding forces and loading rates for Bell–Evans model fitting. The fitting was performed using IGOR Pro or Origin software by minimizing the chi‐square statistic for the optimal fit. Unless otherwise stated, the data is reported as the mean and the standard error of the estimate. Statistical analyses between groups were performed using an unpaired or paired t‐test by R Studio based on each specific situation, with a p‐value less than 0.05 considered statistically significant.
ACKNOWLEDGMENTS
This work is supported in part by National Institutes of Health grants AI133634 and AI163708, and a Lehigh University CORE grant. We thank Dr. Wendy Maury for providing the SARS‐CoV‐2 pseudovirus.
Hou D, Cao W, Kim S, Cui X, Ziarnik M, Im W, et al. Biophysical investigation of interactions between SARS‐CoV‐2 spike protein and neuropilin‐1. Protein Science. 2023;32(10):e4773. 10.1002/pro.4773
Review Editor: Aitziber L. Cortajarena
Contributor Information
Wonpil Im, Email: woi216@lehigh.edu.
X. Frank Zhang, Email: frank.zhang@umass.edu.
REFERENCES
- Alanagreh L a, Alzoughool F, Atoum M. The human coronavirus disease COVID‐19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020;9(5):331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnomasy SF. Virus‐receptor interactions of SARS‐CoV‐2 spikereceptor‐binding domain and human neuropilin‐1 b1 domain. Saudi J Biol Sci. 2021;28(7):3926–3928. 10.1016/j.sjbs.2021.03.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åqvist J, Wennerström P, Nervall M, Bjelic S, Brandsdal BO. Molecular dynamics simulations of water and biomolecules with a Monte Carlo constant pressure algorithm. Chem Phys Lett. 2004;384(4–6):288–294. [Google Scholar]
- Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. 10.3390/v4061011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierig T, Collu G, Blanc A, Poghosyan E, Benoit RM. Design, expression, purification, and characterization of a YFP‐tagged 2019‐nCoV spike receptor‐binding domain construct. Front Bioeng Biotechnol. 2020;8:618615. 10.3389/fbioe.2020.618615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohan D, Van Ert H, Ruggio N, Rogers KJ, Badreddine M, Aguilar Briseno JA, et al. Phosphatidylserine receptors enhance SARS‐CoV‐2 infection. PLoS Pathog. 2021;17(11):e1009743. 10.1371/journal.ppat.1009743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti‐Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, et al. Neuropilin‐1 facilitates SARS‐CoV‐2 cell entry and infectivity. Science. 2020;370(6518):856–860. 10.1126/science.abd2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Dong C, Kim S, Hou D, Tai W, Du L, et al. Biomechanical characterization of SARS‐CoV‐2 spike RBD and human ACE2 protein‐protein interaction. Biophys J. 2021;120(6):1011–1019. 10.1016/j.bpj.2021.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona GC, Pájaro LDQ, Marzola IDQ, Villegas YR, Salazar LRM. Neurotropism of SARS‐CoV 2: mechanisms and manifestations. J Neurol Sci. 2020;412:116824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapoval SP, Keegan AD. Perspectives and potential approaches for targeting neuropilin 1 in SARS‐CoV‐2 infection. Mol Med. 2021;27(1):162. 10.1186/s10020-021-00423-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekol Abebe E, Mengie Ayele T, Tilahun Muche Z, Asmamaw Dejenie T. Neuropilin 1: a novel entry factor for SARS‐CoV‐2 infection and a potential therapeutic target. Biol Theory. 2021;15:143–152. 10.2147/BTT.S307352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesla SE, Selvaraj P, Zhu C. Measuring two‐dimensional receptor‐ligand binding kinetics by micropipette. Biophys J. 1998;75(3):1553–1572. 10.1016/S0006-3495(98)74074-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K‐H, Ferguson DM. Isothermal‐isobaric molecular dynamics simulations with Monte Carlo volume sampling. Comput Phys Commun. 1995;91(1–3):283–289. [Google Scholar]
- Daly JL, Simonetti B, Klein K, Chen K‐E, Williamson MK, Antón‐Plágaro C, et al. Neuropilin‐1 is a host factor for SARS‐CoV‐2 infection. Science. 2020;370(6518):861–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Randeva HS, Chatha K, Hall M, Spandidos DA, Karteris E, et al. Neuropilin‐1 as a new potential SARS‐CoV‐2 infection mediator implicated in the neurologic features and central nervous system involvement of COVID‐19. Mol Med Rep. 2020;22(5):4221–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion M, Rydberg H, Schroder E, Langreth DC, van der Lundqvist BI. Waals density functional for general geometries. Phys Rev Lett. 2004;92(24):246401. 10.1103/PhysRevLett.92.246401 [DOI] [PubMed] [Google Scholar]
- Dragovich MA, Fortoul N, Jagota A, Zhang W, Schutt K, Xu Y, et al. Biomechanical characterization of TIM protein‐mediated Ebola virus‐host cell adhesion. Sci Rep. 2019;9(1):267. 10.1038/s41598-018-36449-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman P, Swails J, Chodera JD, McGibbon RT, Zhao Y, Beauchamp KA, et al. OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comput Biol. 2017;13(7):e1005659. 10.1371/journal.pcbi.1005659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner A, Hinterdorfer P, Gruber HJ. Comparison of different aminofunctionalization strategies for attachment of single antibodies to AFM cantilevers. Ultramicroscopy. 2007;107(10–11):922–927. 10.1016/j.ultramic.2007.02.035 [DOI] [PubMed] [Google Scholar]
- Ebner A, Wildling L, Zhu R, Rankl C, Haselgrubler T, Hinterdorfer P, et al. Functionalization of probe tips and supports for single‐molecule recognition force microscopy. Top Curr Chem. 2008;285:29–76. 10.1007/128_2007_24 [DOI] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
- Evans E. Probing the relation between force—lifetime—and chemistry in single molecular bonds. Annu Rev Biophys Biomol Struct. 2001;30(1):105–128. [DOI] [PubMed] [Google Scholar]
- Franz CM, Taubenberger A, Puech P‐H, Muller DJ. Studying integrin‐mediated cell adhesion at the single‐molecule level using AFM force spectroscopy. Sci STKE. 2007;2007(406):pl5. [DOI] [PubMed] [Google Scholar]
- Gao Y, Lee J, Smith IPS, Lee H, Kim S, Qi Y, et al. CHARMM‐GUI supports hydrogen mass repartitioning and different protonation states of phosphates in lipopolysaccharides. J Chem Inf Model. 2021;61(2):831–839. 10.1021/acs.jcim.0c01360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes C. Report of the WHO‐China joint mission on coronavirus disease 2019 (COVID‐19). Braz J Implantol Health Sci. 2020;2(3). [Google Scholar]
- Gudowska‐Sawczuk M, Mroczko B. The role of neuropilin‐1 (NRP‐1) in SARS‐CoV‐2 infection: review. J Clin Med. 2021;10(13):2772. 10.3390/jcm10132772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatcher E, Guvench O, Mackerell AD. CHARMM additive all‐atom force field for aldopentofuranoses, methyl‐aldopentofuranosides, and fructofuranose. J Phys Chem B. 2009;113(37):12466–12476. 10.1021/jp905496e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald‐Sargent T, Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4(4):557–580. 10.3390/v4040557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins CW, Le Grand S, Walker RC, Roitberg AE. Long‐time‐step molecular dynamics through hydrogen mass repartitioning. J Chem Theory Comput. 2015;11(4):1864–1874. [DOI] [PubMed] [Google Scholar]
- Hu B, Guo H, Zhou P, Shi Z‐L. Characteristics of SARS‐CoV‐2 and COVID‐19. Nat Rev Microbiol. 2021;19(3):141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawad B, Adhikari P, Podgornik R, Ching W‐Y. Key interacting residues between RBD of SARS‐CoV‐2 and ACE2 receptor: combination of molecular dynamics simulation and density functional calculation. J Chem Inf Model. 2021;61(9):4425–4441. [DOI] [PubMed] [Google Scholar]
- Jo S, Kim T, Iyer VG, Im W. CHARMM‐GUI: a web‐based graphical user interface for CHARMM. J Comput Chem. 2008;29(11):1859–1865. 10.1002/jcc.20945 [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- Kim S, Liu Y, Lei Z, Dicker J, Cao Y, Zhang XF, et al. Differential interactions between human ACE2 and spike RBD of SARS‐CoV‐2 variants of concern. J Chem Theory Comput. 2021;17(12):7972–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Liu Y, Ziarnik M, Seo S, Cao Y, Zhang XF, et al. Binding of human ACE2 and RBD of omicron enhanced by unique interaction patterns among SARS‐CoV‐2 variants of concern. J Comput Chem. 2023;44(4):594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg M, Fläschner G, Alsteens D, Gaub BM, Roos WH, Wuite GJ, et al. Atomic force microscopy‐based mechanobiology. Nat Rev Phys. 2019;1(1):41–57. [Google Scholar]
- Kyrou I, Randeva HS, Spandidos DA, Karteris E. Not only ACE2—the quest for additional host cell mediators of SARS‐CoV‐2 infection: neuropilin‐1 (NRP1) as a novel SARS‐CoV‐2 host cell entry mediator implicated in COVID‐19. Signal Transduct Target Ther. 2021;6(1):1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, et al. CHARMM‐GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput. 2016;12(1):405–413. 10.1021/acs.jctc.5b00935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R, Maaloum M. Measuring the spring constant of atomic force microscope cantilevers: thermal fluctuations and other methods. Nanotechnology. 2001;13(1):33–37. [Google Scholar]
- Li Z‐L, Buck M. Neuropilin‐1 assists SARS‐CoV‐2 infection by stimulating the separation of spike protein S1 and S2. Biophys J. 2021;120(14):2828–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S‐D, Zhong L‐P, He J, Zhao Y‐X. Targeting neuropilin‐1 interactions is a promising anti‐tumor strategy. Chin Med J. 2021;134(5):508–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupala CS, Li X, Lei J, Chen H, Qi J, Liu H, et al. Computational simulations reveal the binding dynamics between human ACE2 and the receptor binding domain of SARS‐CoV‐2 spike protein. Quant Biol. 2021;9(1):61–72. [Google Scholar]
- Mayi BS, Leibowitz JA, Woods AT, Ammon KA, Liu AE, Raja A. The role of neuropilin‐1 in COVID‐19. PLoS Pathog. 2021;17(1):e1009153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19(6):679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutal A, Martin LF, Boinon L, Gomez K, Ran D, Zhou Y, et al. SARS‐CoV‐2 spike protein co‐opts VEGF‐A/neuropilin‐1 receptor signaling to induce analgesia. Pain. 2021;162(1):243–252. 10.1097/j.pain.0000000000002097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A, et al. Insights into SARS‐CoV‐2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020;1866(10):165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton R, Delguste M, Koehler M, Dumitru AC, Laskowski PR, Muller DJ, et al. Combining confocal and atomic force microscopy to quantify single‐virus binding to mammalian cell surfaces. Nat Protoc. 2017;12(11):2275–2292. 10.1038/nprot.2017.112 [DOI] [PubMed] [Google Scholar]
- Rankl C, Kienberger F, Wildling L, Wruss J, Gruber HJ, Blaas D, et al. Multiple receptors involved in human rhinovirus attachment to live cells. Proc Natl Acad Sci U S A. 2008;105(46):17778–17783. 10.1073/pnas.0806451105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankl C, Wildling L, Neundlinger I, Kienberger F, Gruber H, Blaas D, et al. Determination of the kinetic on‐and off‐rate of single virus–cell interactions. Atomic force microscopy in biomedical research. Totowa, New Jersey: Humana Press; 2011. p. 197–210. [DOI] [PubMed] [Google Scholar]
- Riener CK, Stroh CM, Ebner A, Klampfl C, Gall AA, Romanin C, et al. Simple test system for single molecule recognition force microscopy. Anal Chim Acta. 2003;479(1):59–75. [Google Scholar]
- Roy S, Bag AK, Singh RK, Talmadge JE, Batra SK, Datta K. Multifaceted role of neuropilins in the immune system: potential targets for immunotherapy. Front Immunol. 2017;8:1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert J‐P, Ciccotti G, Berendsen HJ. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n‐alkanes. J Comput Phys. 1977;23(3):327–341. [Google Scholar]
- Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS‐CoV‐2. Nature. 2020;581(7807):221–224. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnick J, Gao M, Zhou H, Singh S. AlphaFold 2: why it works and its implications for understanding the relationships of protein sequence, structure, and function. J Chem Inf Model. 2021;61(10):4827–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumbul F, Valotteau C, Fernandez I, Meola A, Baquero E, Kostrz D, et al. Dynamics and binding strength of the spike protein of SARS‐Cov‐2 probed by high‐speed atomic force microscopy. Biophys J. 2021;120(3):3a. [Google Scholar]
- WHO . COVID‐19 weekly epidemiological update, edition 49, 20 Jul 2021. 2021.
- WHO . COVID‐19 weekly epidemiological update, edition 132, 1 Mar 2023. 2023.
- WHO . WHO lists additional COVID‐19 vaccine for emergency use and issues interim policy recommendations [Internet]. WHO, Last Modified (2021), 7.
- Wild JR, Staton CA, Chapple K, Corfe BM. Neuropilins: expression and roles in the epithelium. Int J Exp Pathol. 2012;93(2):81–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcikiewicz EP, Abdulreda MH, Zhang X, Moy VT. Force spectroscopy of LFA‐1 and its ligands, ICAM‐1 and ICAM‐2. Biomacromolecules. 2006;7(11):3188–3195. 10.1021/bm060559c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcikiewicz EP, Zhang X, Moy VT. Force and compliance measurements on living cells using atomic force microscopy (AFM). Biol Proced Online. 2004;6:1–9. 10.1251/bpo67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C‐L, Abiona O, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel AG, Benton DJ, Xu P, Roustan C, Martin SR, Rosenthal PB, et al. SARS‐CoV‐2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin‐cleavage effects. Nat Struct Mol Biol. 2020;27(8):763–767. 10.1038/s41594-020-0468-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaohui Zhang FR, Xu AJ, Moy VT. Atomic force microscopy of protein‐protein interactions. New York, NY: Springer US; 2009. [Google Scholar]
- Xu J, Zhang Y. How significant is a protein structure similarity with TM‐score = 0.5? Bioinformatics. 2010;26(7):889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS‐CoV‐2 by full‐length human ACE2. Science. 2020;367(6485):1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Petitjean SJL, Koehler M, Zhang Q, Dumitru AC, Chen W, et al. Molecular interaction and inhibition of SARS‐CoV‐2 binding to the ACE2 receptor. Nat Commun. 2020;11(1):4541. 10.1038/s41467-020-18319-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalpoor H, Shapourian H, Akbari A, Shahveh S, Haghshenas L. Increased neuropilin‐1 expression by COVID‐19: a possible cause of long‐term neurological complications and progression of primary brain tumors. Hum Cell. 2022;35(4):1301–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazhytska M, Kodra A, Hoagland DA, Frere J, Fullard JF, Shayya H, et al. Non‐cell‐autonomous disruption of nuclear architecture as a potential cause of COVID‐19‐induced anosmia. Cell. 2022;185(6):1052–1064. e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu R, Canena D, Sikora M, Klausberger M, Seferovic H, Mehdipour AR, et al. Force‐tuned avidity of spike variant‐ACE2 interactions viewed on the single‐molecule level. Nat Commun. 2022;13(1):7926. [DOI] [PMC free article] [PubMed] [Google Scholar]


