Abstract
Sensitive detection of circulating tumor cells (CTCs) from patients’ peripheral blood facilitates on-demand monitoring of tumor progression. However, clinically significant capture of renal cell carcinoma CTCs (RCC-CTCs) remains elusive due to their heterogenous surface receptor expression. Herein, a novel capture platform is developed to detect RCC-CTCs through integration of dendrimer-mediated multivalent binding, a mixture of antibodies, and biomimetic cell rolling. The nanoscale binding kinetics measured using atomic force microscopy reveal that dendrimer-coated surfaces exhibit an order of magnitude enhancement in off-rate kinetics compared to surface without dendrimers, which translated into cell capture improvements by ~60%. Selectin-induced cell rolling facilitates surface recruitment of cancer cells, further improving cancer cell capture by up to 1.7-fold. Lastly, an antibody cocktail targeting four RCC-CTC surface receptors, which included epithelial cell adhesion molecule (EpCAM), carbonic anhydrase IX (CA9), epidermal growth factor receptor (EGFR), and hepatocyte growth factor receptor (c-Met), improves the capture of RCC cells by up to 80%. The optimal surface configuration outperforms the conventional assay solely relying on EpCAM, as demonstrated by detecting significantly more CTCs in patients’ samples (9.8±5.1 vs. 1.8±2.0 CTCs mL−1). These results demonstrate that the newly engineered capture platform effectively detects RCC-CTCs for their potential use as tumor biomarkers.
Keywords: circulating tumor cells, renal cell carcinoma, dendrimer-mediated multivalent binding effect, biomimetic cell rolling, liquid biopsy
1. Introduction
Circulating tumor cells (CTCs) are known to play a key role in cancer spreading, or metastasis (Baccelli et al. 2013). The cells enter the bloodstream from a primary tumor burden, aggregate at distant organs, and migrate into metastatic lesions (Hou et al. 2011; Murlidhar et al. 2016; Nguyen et al. 2009). Given that the presence of CTCs in the bloodstream is likely associated with disease status, a myriad of CTC enrichment, quantification, and characterization strategies have been developed to test their potential as a tumor biomarker (Bu et al. 2017a; Bu et al. 2016). Growing evidence suggests that CTC counts and their phenotypic features are correlated with prognosis and disease progress for multiple types of malignancy, including breast, colon, and prostate cancers (Bidard et al. 2014; Bu et al. 2017b; Bu et al. 2017c; Cohen et al. 2008; de Bono et al. 2008).
Renal cell carcinoma (RCC) is one of the 10 most common cancers in the US, and yet the disease lacks a reliable biomarker. This clinical gap could be potentially addressed by CTCs (Shingarev and Jaimes 2017). Several approaches have attempted to utilize CTCs as biomarkers for estimating therapeutic response and metastatic potential of RCCs (Liu et al. 2016; Maertens et al. 2017; Nel et al. 2016). Despite these efforts, significant challenges remain in capturing RCC-derived CTCs with existing cancer cell isolation techniques, primarily due to their highly heterogeneous phenotypes. Specifically, the main obstacle is that the commonly used CTC-targeting molecule, epithelial cell adhesion molecule (EpCAM), is down-regulated in clear cell RCC (ccRCC), which accounts for approximately 75% of RCC cases (Brodaczewska et al. 2016). Moreover, EpCAM is expressed in less than 40% of RCC primary tumors, indicating that conventional EpCAM-targeting CTC isolation methods would not achieve effective detection of RCC-CTCs (Liu et al. 2016; Rossi et al. 2012). Other CTC isolation strategies, such as CD45-based leukocyte depletion (Bluemke et al. 2009) and size-based filtration methods (El-Heliebi et al. 2013), have also been found ineffective, detecting only a few CTCs per 7.5 mL of blood from only 53% (81/154 patients) and 54% (15/28) of RCC patients, respectively (Bluemke et al. 2009; El-Heliebi et al. 2013).
Our platform incorporates three independently effective CTC isolation strategies that overcome previous barriers and enable highly sensitive RCC-CTC capture. First, capture antibodies were conjugated to generation 7 (G7) poly(amidoamine) (PAMAM) dendrimers that provide a high density of coverage and facilitate multivalent binding at the nanoscale (Jeong et al. 2020; Jeong et al. 2018; Myung et al. 2011; Myung et al. 2015; Poellmann et al. 2018). This confers a strong adhesion force between the target cells and the capture surface by forming concurrent, multiple binding pairs in a small area, improving CTC arrest and retention (Myung et al. 2011). Second, a naturally occurring cell rolling mechanism was mimicked by employing E-selectin within a flow chamber. E-selectin exhibits fast association and dissociation kinetics with tumor cells, which allows highly efficient surface recruitment of tumor cells from the bulk flow (Myung et al. 2010). Third, a mixture of capture antibodies would recognize a more diverse set of RCC-CTCs than the anti-EpCAM antibody (aEpCAM) alone. A new antibody cocktail consisting of antibodies against carbonic anhydrase IX (CA9), epidermal growth factor receptor (EGFR), and hepatocyte growth factor receptor (HGFR, or also known as tyrosine-protein kinase Met; c-Met) was thus employed. These receptors were chosen, since they are expressed by ccRCC tumors (Takacova et al. 2013), overexpressed in cases of RCC progression (Hynes and Lane 2005), and found on RCCs with high metastatic potential (Lalani et al.).
Through integration of the three strategies, we prepared a novel surface configuration that is capable of capturing RCC-CTCs at significantly improved sensitivity, compared to other methods that have been reported elsewhere. In this paper, our capture surfaces were engineered, characterized, and optimized through a systematically designed study, ranging from nanoscale, quantitative analysis to clinical validation. The multivalent binding effects of the dendrimer surfaces functionalized with the RCC-specific antibodies were quantitatively compared using atomic force microscopy (AFM) in terms of nano Newton (nN)-scale binding kinetics. The nanoengineered capture surfaces were then tested using in vitro cell lines to assess any synergistic effect in RCC cell capture efficiency when combined with biomimetic cell rolling. Lastly, our capture surfaces were clinically tested using blood samples from RCC patients. Here we present a series of results indicating that our newly engineered capture system enables clinically significant detection of highly heterogeneous RCC-CTCs, presenting a potential liquid biopsy method for RCC diagnosis, prognosis, and surveillance.
2. Materials and Methods
2.1. Materials
Anti-human EpCAM/TROP1 polyclonal antibody (aEpCAM), anti-human CA9 antibody (aCA9), anti-human EGFR antibody (aEGFR), anti-human anti-human HGF R/c-Met polyclonal antibody (ac-Met), recombinant human EpCAM/TROP1 Fc Chimera, and recombinant human E-selectin/Fc Chimera (E-selectin) were all purchased from R&D systems (Minneapolis, MN). All polyethylene glycol (PEG) linkers, including 5000 MW carboxy-PEG-amine (denoted as PEG) and 5000 MW methoxy-PEG-amine (denoted as mPEG) were acquired from Nektar Therapeutics (Huntsville, AL). 1-(3-Dimethylaminopropyl)-3-EthylcarbodiimideHydrochloride (EDC) and N-Hydroxy-Succinimide (NHS) were obtained from Sigma–Aldrich (St. Louis, MO). Goat anti-Human IgG (H+L) antibody was obtained from Pierce Biotechnology, Inc. (Rockford, IL). Epoxy-coated glass slides were provided by Tekdon Inc. (Myakka City, FL). G7 PAMAM dendrimers were purchased from Dendritech Inc. (Midland, MI). Live/dead viability kit and CellTracker Green were obtained from Thermo Fisher Scientific (Waltham, MA). All other chemicals were obtained from Sigma-Aldrich (St. Louis, MO).
2.2. Capture Surface Preparation
The RCC-CTC isolation device is composed of two distinct regions: (a) a capture region, which consists of dendrimers functionalized with RCC-targeting capture agents and (b) a rolling region, which has an E-selectin coat on its surface for efficient CTC recruitment. Schematic diagrams of the fabrication process are provided in Figure 1 and can also be found in our previous literatures (Myung et al. 2010; Myung et al. 2015). The capture region was prepared by sequential conjugation of PEG, G7 PAMAM dendrimers, and different RCC-CTC capture agents on an epoxide glass slide. The epoxy-coated glass slides were covered with three-well polydimethylsiloxane (PDMS) gaskets (Figure S2). The confined area was then PEGylated at a concentration of 1 mg/mL in deionized water (DW) for 12 h at room temperature (RT). Partially carboxylated G7 PAMAM dendrimers (1 mg/mL in 0.1 M phosphate buffer; pH 9) were prepared (Figure S3) and immobilized on the PEGylated surface for 12 h at RT, following EDC (3.8 mg/mL in DW)/NHS (4.4 mg/mL in DW) activation. Four different antibodies, including aEpCAM, aCA9, aEGFR, and ac-Met, were then conjugated to the dendrimers via EDC/NHS chemistry. The dendrimer-conjugated surface was incubated with antibodies at a concentration of 5 μg/mL for each (total 20 μg/mL in PBS), at 4 °C for 12 h. In case of functionalizing the surface with a single type of antibody, the surface was incubated at an antibody concentration of 20 μg/mL. The three-well PDMS gaskets were replaced with one-well gaskets and filled with E-selectin at a concentration of 5 μg/mL in PBS for 4 h at RT. The slide surfaces were then blocked with mPEG (1 mg/mL in PBS; RT) to prevent non-specific binding. The functionalized RCC-CTC capture slides were used within 72 h.
Figure 1.
Summary of our capture surfaces specifically engineered to detect RCC-CTCs: (A) A schematic diagram illustrating the capture mechanism using dendrimer-mediated multivalent binding effect, cell rolling, and RCC-specific antibody cocktail; (B) An overview of surface preparation of our RCC-CTC capture device, consisting of rolling and capture domains, followed by assembly into a flow system. The multipatterned capture regions were prepared by sequential surface immobilization of PEG (1 mg/mL in DW; overnight; RT), partially carboxylated G7 PAMAM dendrimers (1 mg/mL in 0.1 M phosphate buffer at pH 9; overnight; RT), and RCC-specific capture agents (5 μg/mL for each antibody in PBS; overnight; 4°C), followed by addition of E-selectin (5 μg/mL in PBS; 4 h; RT) to fabricate the rolling regions; Capture efficiencies of the surface measured under various flow rates (12.5, 25, and 50 μL/min) using (C) ACHN (a papillary RCC cell line) and (D) HL-60 (a promyeloblast leukemia cell line) cells. Note that the optimal capture flow rate was found to be 25 μL/min where the highest capture of RCC cells with minimal non-specific capture of HL-60 cells was observed. Error bars: standard deviations.
2.3. Cell Preparation
The human RCC cell lines ACHN and 786-O cells were used in this study as representatives for papillary RCC and ccRCC models, respectively. Each cell line was grown in Eagle’s Minimum Essential Media (EMEM) and Roswell Park Memorial Institute (RPMI) 1640 media, respectively, supplemented with 10% FBS (v/v) and 1% penicillin/streptomycin (v/v) under humidified conditions with 5% CO2 levels at 37 °C. The human promyeloblast cell line HL-60 was cultured in Icsove’s Modified Dulbecco’s Medium (IMDM) with 20% FBS (v/v) and 1% penicillin/streptomycin (v/v). Cells were then fluorescently labelled with CellTracker Green as described previously (Bu et al. 2017c). The cells were treated with trypsin and spiked into either cell culture media or peripheral blood mononuclear cell (PBMC) layer at a concentration of ~2,500 cells/mL.
2.4. Surface Characterization and Cellular Analysis
Partially modified G7 PAMAM dendrimers were characterized with 1H NMR, as described elsewhere (Jeong et al. 2020). The amount of antibodies functionalized on the PEGylated and dendrimer-conjugated surfaces were compared using a bicinchoninic acid (BCA) assay (Micro BCA Protein kit, Thermo Fisher Scientific). AFM was used to measure the surface roughness and determine the dissociation rate of the aEpCAM-PEGylated or aEpCAM-dendrimer conjugated capture surfaces. Western blot and bead binding assays were performed to quantify the expression of the RCC-targeting molecules used in this study. The viability of the captured cancer cells was validated using a two-colored live/dead cell viability assay (Thermo Fisher Scientific). See Supporting Information for additional details.
2.5. Peripheral Blood Mononuclear Cell Isolation
Patients’ blood samples were obtained from Duke University Hospital with institutional review boards (IRB) approval. The samples were collected in heparin-treated tubes. PBMC layers were then isolated from 2 mL of human whole blood using Ficoll density gradient separation, as described in previous literatures (Myung et al. 2019). They were maintained in 100 μL DMEM supplemented with 10% FBS (v/v) and 1% penicillin/streptomycin (v/v).
2.6. Cancer Cell Capture
The prepared cell suspensions or PBMC layers were infused through the flow chamber. Cells were withdrawn into the flow chamber using a syringe pump (New Era pump 505 Systems Inc., Farmingdale, NY) at a flow rate of 25 μL/min for 20 min. Cells were then kept on the capture slides for 5 min before being washed backwards (pushing) for another 20 min at a washing flow rate of 50 μL/min. For the cell line experiments, images were taken with an inverted microscope (Zeiss Axiocam 503 mon, Carl Zeiss, Germany) and the number of captured cells was counted using Image J software (NIH). For the clinical samples, the capture slides were removed from the flow chamber and processed through a sequence of immunofluorescence staining assays (Bu et al. 2016). See Supporting Information for additional details.
3. Results and Discussion
3.1. RCC-CTC Capture Concept and Flow Channel Fabrication
Fig. 1A and S1 depict the three strategies that were incorporated into the RCC-CTC isolation device. The chip surface consisted of a rolling region followed by a capture region consisting of G7 PAMAM dendrimers conjugated with multiple antibodies which worked together to arrest CTCs from a high throughput flow. The cell rolling region utilized a rapid on/off binding by E-selectin for CTC recruitment from flow (Myung et al. 2018a; Myung et al. 2014; Myung et al. 2010). The CTCs recruited to the surface were then secured in the capture region, which was comprised of four different RCC-targeting antibodies conjugated to dendrimers.
The surface preparation process is illustrated in Fig. 1B, along with additional details provided in Supporting Information (Fig. S2–S4). The functionalized glass slide was assembled with a flow chamber, generating two flow channels of 8 mm wide, 42 mm long, and 110 μm high for each. Cells were infused through the chamber, as described in our previous publications (Myung et al. 2010; Myung et al. 2015).
3.2. Optimization of Flow Condition
Flow conditions were optimized in vitro using surfaces with the same configuration described above but functionalized with aEpCAM instead of the multiple antibodies. Two cell lines were employed: ACHN (EpCAM-positive) and HL-60 cells as models for papillary RCC-CTCs (Ueda et al. 2013) and leukocytes, (Myung et al. 2010) respectively. To optimize the flow condition, we tested multiple flow rates at 12.5, 25.0, and 50.0 μL/min, corresponding to shear stresses at 0.09, 0.18, and 0.36 dyne/cm2, respectively, with infusion time of 20 min. Cells were then incubated for 5 min and washed in the reverse flow direction for another 20 min at twice the capture flow rate to remove the cells that are non-specifically bind on the surface. As shown in Fig. 1C, the capture efficiency of ACHN cells decreased drastically at a flow rate from 25.0 to 50.0 μL/min(54.7 ± 7.9% vs. 31.9 ± 4.9%; p = 0.013). This experimental result is well supported by our numerical simulation (Fig. S5) where a majority of the cells failed to interact with the capture surface at the higher flow rate.
Although a similar number of ACHN cells was captured at the both flow rates of 12.5 and 25.0 μL/min, the domains where the cells were captured on the surfaces was notably different (Fig. 1C). The ratios of the cells within the capture region functionalized with dendrimer-aEpCAM to total cell capture were 76.2 ± 4.5% and 90.2 ± 6.6% at 12.5 μL/min and 25.0 μL/min, respectively (p = 0.038). The capture efficiency of HL-60 cells (Fig. 1D) was also significantly different at 12.5 μL/min vs. 25.0 μL/min (10.3 ± 5.3% vs. 4.7 ± 1.2%; p = 0.050), indicating that the rolling-based capture, originally designed to enrich the CTC population, may negatively affect the purity of tumor cells captured on the surface at a lower flow rate. Considering the objective of this optimization was to maximize the capture efficiency and purity of tumor cells, we chose the flow rate of 25.0 μL/min for subsequent experiments in this study.
3.3. Enhanced Binding with Dendrimer-coated Surface
The effect of adding dendrimers to the capture surface configuration for RCC-CTC capture was first assessed by quantifying and comparing amounts of antibodies immobilized on the surfaces, with and without dendrimers. We anticipated that hyperbranched nanostructure of dendrimers would accommodate more antibodies on the capture surface and increase the local antibody density, thereby enhancing overall CTC capture, as we previously reported (Myung et al. 2011). The BCA assay results confirmed an 1.65-fold increase (p = 0.006) in antibody density on the dendrimer surfaces compared to the surfaces without dendrimers (Fig. 2A). Height measurements using AFM (Fig. 2B and C) also revealed clear differences in nanoscale topography. The root mean square (rms) representing quantitative roughness (Rq) of the dendrimer surfaces was 7.6 ± 0.4 nm, more than 2-fold higher than the roughness of the PEG-antibody surfaces (p = 0.005).
Figure 2.
Improved binding kinetics and cell capture using dendrimer-mediated multivalent binding effect: (A) An increased amount of antibodies immobilized onto the dendrimer-coated surface, compared to the epoxy surfaces with and without PEG. AFM analysis on the surfaces revealing (B) an increase in surface roughness and (C) an increased surface texture as observed in the AFM images, upon surface modification with PEG (1 mg/mL in DW; overnight; RT) and subsequently with partially modified G7 PAMAM dendrimers (1 mg/mL in 0.1 M phosphate buffer pH 9; overnight; RT). (D) Enhanced adhesion (rupture forces) from the dendrimer surface, compared to the PEG surface, indicating that the dendrimers mediate multivalent binding effect, measured using AFM. The Bell-Evans models plotted for the aEpCAM-PEGylated and the aEpCAM-dendrimer conjugated capture surfaces demonstrate that the dendrimer surface lowers the dissociation rate by one order of magnitude. (E) Enhanced retention of the surface-bound RCC cells (ACHN) on the dendrimer surfaces (bottom images), compared to the PEG surfaces (top images), upon washing at 50 μL/min for 20 min. (F) Enhanced capture efficiency of the dendrimer surface with the antibody cocktail for both ACHN and 786-O cells at a flow cycle consisting of 20 min infusion at 25 μL/min, 5 min incubation, and 20 min reverse washing at 50 μL/min. Error bars: standard deviations.
Next, we assessed the multivalent binding effect mediated by dendrimers, which would contribute to the overall increase in capture efficiency of the surfaces. AFM force spectroscopy was employed to obtain quantitative evidence of such binding effect by comparing various surface configurations. The interaction forces between an AFM probe coated with recombinant EpCAM and capture surfaces functionalized with antibodies were recorded. Representative retraction curves are shown in Fig. S6 where the characteristic unbinding patterns of multivalent binding were observed.[24] The surfaces with dendrimers required greater force before separation in a range of pulling rates (40–400 nN/s) than the surfaces without (rupture force of 689 vs. 430 nN at a pulling rate of 40 nN/s). The results were fitted to the Bell-Evans model to obtain the kinetic off-rate (dissociation rate, koff) (Evans and Ritchie 1997), as described in Fig. 2D. The dissociation rate from the dendrimer surfaces was an order of magnitude lower than the surface without dendrimers (4.00 × 10−1 s−1 vs. 1.34 × 10 s−1). All of these results support that the dendrimers immobilized on the capture surfaces successfully mediated the strong multivalent binding effect.
In vitro binding affinity was further evaluated by measuring the retention of captured cells under persistent flow. The suspended ACHN cells were allowed to settle down in a flow chamber for 30 min, then subjected to flow for 20 min at 50 μL/min. Dendrimer surfaces functionalized with aEpCAM retained 96.8 ± 1.2% of the cells (Fig. 2E), compared to 90.1 ± 0.7% of cells remaining on the same surfaces without dendrimers (p = 0.001). We then evaluated the dendrimer-mediated capture with a mixture of antibodies (the final capture surface configuration) using ACHN cells, along with a ccRCC cell line, 786-O (Brodaczewska et al. 2016). Our final capture surface configuration resulted in an increased in capture efficiency, compared to the control surfaces without dendrimers by 1.61-fold and 1.21-fold for ACHN (p = 0.007) and 786-O cells (p < 0.001), respectively (Fig. 2F). These in vitro results further confirmed that the dendrimer-mediated multivalent binding effect substantially increased adhesion between RCC cells and the capture surface, enhancing the overall capture efficiency of the RCC cells.
3.4. Enhanced Capture with E-selectin-induced cell rolling
Our group has used E-selectin in previous studies to enhance the capture of lung and breast cancer cells (Myung et al. 2010; Myung et al. 2018b). Fast flowing cancer cells form transient adhesions with E-selectin, resulting in physical rolling that subsequently increases the contact time between cells and the capture region, and enhances capture overall (Myung et al. 2010). We hypothesized that the E-selectin-induced cell rolling would similarly enhance RCC-CTC capture, since published literature suggests that the overexpression of carbohydrate antigens including Sialyl Lewis X (sLex) correlate with poor prognosis of RCCs (Kajiwara et al. 2005; Kobayashi et al. 2007).
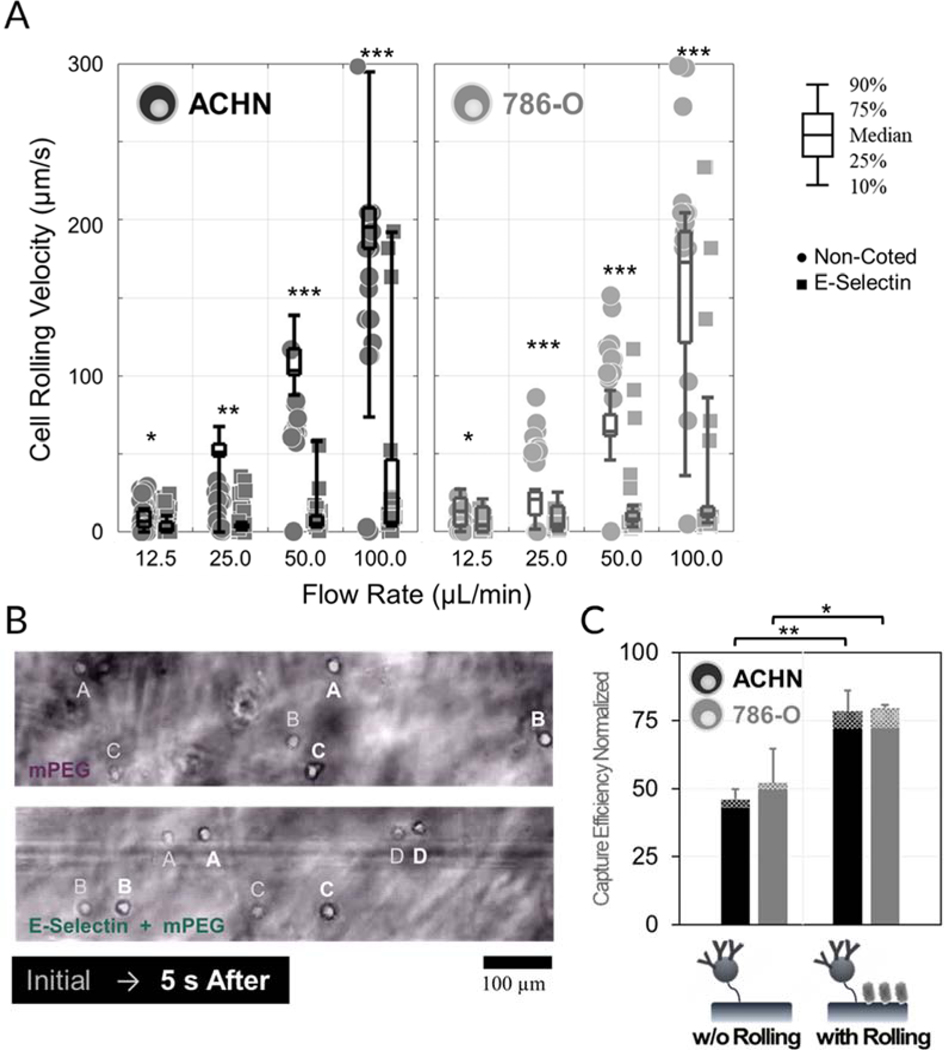
The rolling velocities of the RCC cell lines on surfaces coated with E-selectin and methoxy PEG (mPEG, 5 kDa) were compared after 10 min incubation inside the chamber (Fig. 3A and B). Cells were rinsed from the surface at flow rates ranging from 12.5 to 100 μL/min. Interaction with E-selectin significantly reduced the flow velocity for both RCC cell lines at all flow rates tested (p ˂ 0.050). Notably, both RCC cell lines exhibited stable rolling under the flow rate below 50 μL/min, whereas over 10% of cells failed to resist the shear stress at 100 μL/min and traveled toward outlet at the speed of stream velocity. It should be also noted that the low affinity interaction with selectins induces stationary adhesion of RCC cells at low shear rate conditions (flow rates of 12.5 and 25.0 μL/min), arresting them at a rolling region instead of recruiting them at a capture region. These data suggest that flow channels be washed at a flow rate of 50 μL/min following the optimum capture flow rate of 25 μL/min in order to completely remove cells in the rolling regions. The capture efficiency of the device was then tested with and without an E-selectin rolling region. Fig. 3C shows that the capture efficiencies were significantly enhanced when rolling regions were included: 1.70-fold and 1.53-fold for ACHN (p = 0.003) and 786-O cells (p = 0.019), respectively. These results indicate that cell rolling greatly enhanced CTC capture via their recruitment to the capture region of the device, thus increasing their chance of interacting with antibodies.
Figure 3.
Selectin-induced cell rolling enhances surface recruitment of both ACHN and 786-O RCC cell lines to the capture surface: (A, B) Interaction with E-selectin significantly reduces the flow velocity of RCC cell lines under various washing flow rates (12.5, 25, 50, and 100 μL/min). (C) Cell rolling greatly enhances CTC capture by recruiting cancer cells to the capture region of the device at a flow cycle consisting of 20 min infusion at 25 μL/min, 5 min incubation, and 20 min reverse washing at 50 μL/min. Error bars: standard deviations.
Biomimetic cell rolling has been reported to be beneficial for enhancing the capture of various CTC subtypes, particularly the type of cells that express less epithelial cell adhesion molecules, such as EpCAM and E-cadherin (Myung et al. 2019). These markers are often downregulated in ccRCC.(Brodaczewska et al. 2016; Maru et al. 2013) As a result, 786-O cells had significantly faster rolling speeds than ACHN cells did, although we still observed enhanced capture of 786-O cells when E-selectin was included. Notably, interactions with selectins resulted in a significant reduction in rolling velocity of 786-O cells, reaching down to 6% of that on the same surfaces without E-selectin at a flow rate of 50 μL/min. All of our results indicate that reduced rolling velocity through interactions between cells and E-selectin contributes to the enhanced capture efficiency, which justifies the addition of E-selectin to the capture surface configuration despite its added complexity.
3.5. Enhanced Capture with Multiple Antibodies
The expression of four, targetable RCC surface markers was quantitatively measured using a western blot assay (Fig. 4A and B). EpCAM expression was not detected in the ccRCC 786-O cell, indicating the inherent limitation of the CTC techniques solely relying on aEpCAM for RCC cells. The three additional targets were thus selected to minimize missing RCC-CTCs that do not express EpCAM. CA9 helps tumor cells maintain a neutral pH and is commonly expressed in RCC tissues (Tostain et al. 2010). More importantly, it is the only transmembrane member of carbonic anhydrase (CA) family that is present on an extracellular catalytic site (Shin et al. 2011). The second target, EGFR, has been proven to be a successful target in lung and prostate CTC isolation(Myung et al. 2015; Okegawa et al. 2016). The growth factor is also present on up to 80% of clear cell RCC tumors (Hynes and Lane 2005). Finally, c-Met was chosen as a target based on the presence of the protein across various RCC subtypes, particularly on the metastatic RCC sites (Lalani et al. 2017). In addition, we also performed a fluorescence bead assay to quantitatively confirm the relative surface expression of the 4 targets from ACHN and 786-O cell lines (Fig. 4C and D). The results of this bead binding assay were consistent with the western blot results.
Figure 4.
Application of multiple capture agents enables effective RCC-CTC capture: (A, B) The expression of RCC-targeting molecules, EpCAM, EGFR, c-Met, and CA9, on RCC cancer cell lines was quantified using western blot assay (C, D) RCC cells were incubated with antibody-functionalized beads for 30 min, followed by PBS washing (1 min; three times), to investigate whether the target molecules are present on the cell surface. The gray circle on Fig. 4C denotes a single well of 96 well plate. (E, F) Cooperation of multiple RCC-targeting capture agents significantly improved RCC cell capture. None of the single antibody showed capture efficiency >60%, whereas the antibody mixture exhibiting ~80% for both ACHN and 786-O cells (p < .05). Flow chamber experiments were conducted at a flow cycle consisting of 20 min infusion at 25 μL/min, 5 min incubation, and 20 min reverse washing at 50 μL/min. Error bars: standard deviations.
Next, we tested the ability of each antibody to capture CTCs in the flow channel. Capture efficiency of the both cell lines was strongly associated with the amount of target molecules expressed on each cell line (Fig. 4E and F) and was highest for the mixture of 4. None of the antibodies alone exhibited a capture efficiency over 60% for either cell line. Notably, aEpCAM-functionalized surfaces demonstrated a capture efficiency of only 14.1 ± 4.0% for the ccRCC model (786-O), highlighting the difficulty of RCC-CTC screening with conventional aEpCAM-based approaches. The cooperation of multiple RCC-targeting capture agents may likely be attributed to the significantly enhanced RCC cell capture (Fig. S7). Both ACHN and 786-O cell lines achieved capture efficiencies of approximately 80%, significantly outperforming all capture surfaces functionalized with a single antibody only (p<0.031).
3.6. CTC Detection from Human Blood
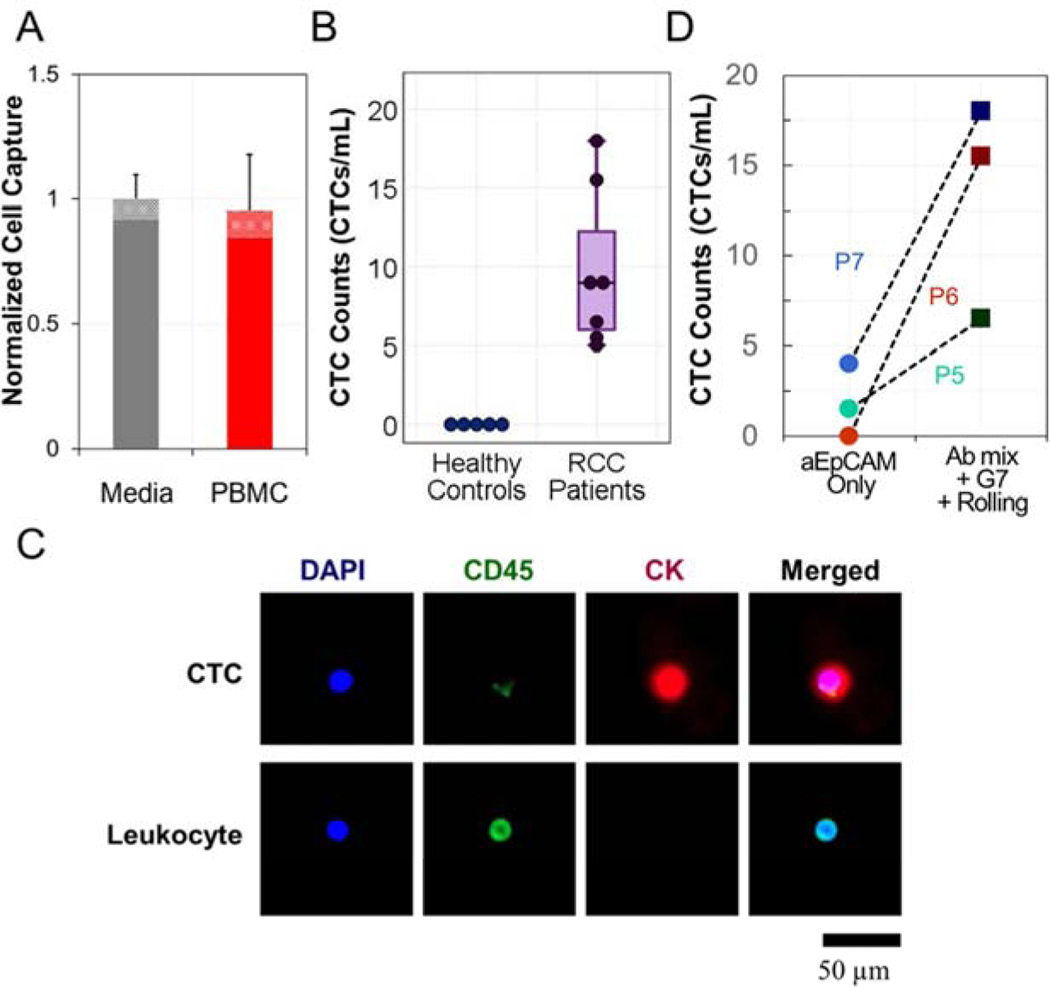
As the last step prior to the clinical pilot study, the ACHN and 786-O cell lines were labeled with a fluorescent marker and spiked into human peripheral blood mononuclear cells (PBMC) isolated from 2 mL whole blood from healthy donors and resuspended to a volume of 100 μL media. The suspensions were used to determine the sensitivity and specificity of our final capture surface configuration, consisting of E-selectin, G7 PAMAM dendrimers, and a mixture of antibodies against EpCAM, EGFR, CA9, and c-Met (Fig. 5A). Cell viability following capture was over 90% (Fig. S8), and the surfaces resisted non-specific adhesion of over 99.96% of PBMCs ((2.9 ± 1.5) × 103 PBMCs/1mL whole blood; n = 5). The results strongly suggest that cells isolated on the capture surface have sufficient quality and purity for downstream analyses such as genomic sequencing in future work.
Figure 5.
The synergistic combination of the three isolation strategies enhanced RCC-CTC capture: (A) Comparison of capture efficiency for ACHN cells when spiked into different cell suspensions. No significant differences were found between cells spiked in culture media and in PBMC layer. (B) CTCs were detected from all seven RCC patients’ samples, while no cancer-like cells were found from the five healthy controls. PBMC layers obtained from 2 mL human whole blood was subjected to RCC-CTC capture device. (C) Representative IHC images of a captured CTC and leukocyte, co-stained with CK (red), CD45 (green), and DAPI (blue). Cell images were obtained from P7 sample. (D) Our device significantly outperformed the conventional assay relaying on EpCAM only, showing a significantly higher number of CTCs from all three RCC patients’ samples. Error bars: standard deviations.
Finally, a clinical pilot study was conducted with 12 human blood samples that were comprised of 7 RCC patients and 5 healthy controls (Table S1). Unlike the other CTC capture platforms, our capture surface does not contain any visible patterns; thus, the slides can be directly applied for IHC analysis. Cells exhibiting CK+/CD45−/DAPI+ were defined as CTCs as in previous studies (Bu et al. 2017b). The detailed criteria regarding CTC validation is supported in the Supplementary Materials (Fig. S9). As demonstrated in Fig. 5B and C, CTCs were detected in all patient samples and ranged from 4 to 18 CTCs/mL. No CK+/CD45−/DAPI+ cells were observed in healthy controls using our counting algorithm. A limited number of samples was also tested on surfaces targeting only EpCAM, without dendrimers or E-selectin coatings, resulting in an average 7.28-fold (p = 0.073) lower capture than our optimized capture surfaces (Fig. 5D).
Previous publications have demonstrated RCC-CTC isolation in human blood samples through various techniques.(Bluemke et al. 2009; El-Heliebi et al. 2013; Gradilone et al. 2011; Liu et al. 2016) Although in vitro results using such techniques were comparable to our results presented here, the CTC numbers isolated from patient blood were all lower than our counts (Fig. S10). More specifically, EpCAM-based detection (Gradilone et al. 2011), size-based isolation (El-Heliebi et al. 2013), leukocyte depletion (Bluemke et al. 2009), and other techniques (Liu et al. 2016) all showed similar sensitivity to our capture system against cultured RCC cell lines. However, a significant difference was noted in enumerating clinical CTCs. Although CTC counts could be significantly affected by differences in patient cohort, sample size, sample volume, and CTC counting algorithm among different CTC capture platforms, our RCC-optimized capture surface achieved remarkably greater counts (1.5 to 9.1-fold) from patient-derived blood samples than all reported values to our knowledge. This suggests the high in vitro sensitivity of our device could be successfully translated to clinical CTC analysis.
Densely functionalized, quadruple antibody cocktails conjugated to a dendrimer-coated surface, along with E-selectin inducing biomimetic cell rolling, enabled efficient capture of different RCC-CTC subtypes, demonstrating promising potential for our capture system to utilize CTCs as a biomarker for RCC.
4. Conclusion
The impact of clinically significant CTC detection and isolation cannot be overstated, given the potential role that CTCs play as a biomarker in cancer metastasis. The capture surface tailored to RCC-CTCs herein achieved a significantly improved CTC enumeration for both RCC in vitro cells and clinical CTCs from RCC patients, indicating the potential clinical significance of our results. The success of our device is attributed to the synergistic effect from three distinct capture components, i.e., dendrimer-mediated strong multivalent binding, E-selectin-based cell rolling, and a combination of the 4 RCC-specific capture antibodies. The successful development of this CTC capture technology may potentially lead to the development of a reliable biomarker of RCC, which may ultimately realize personalized care by providing actionable information to physicians for optimal treatment decisions. Considering that this technology provides a modular platform for CTC detection, the impact of our results could be further expanded to other malignancies.
Supplementary Material
Highlights.
Three effective CTC isolation strategies were integrated for RCC-CTC capture
A new platform has been tested from nanoscale analysis to clinical application
Dendrimer nanoparticles exploited the strong multivalent binding to the tumor cells
Recruitment of the flowing CTCs via cell rolling enhanced capture of RCC cells
The combination of the 4 antibodies allowed sensitive detection of RCC cells
Acknowledgements
This study was partially supported by DRP program of the Wisconsin Head & Neck Cancer SPORE Grant (P50-DE026787) and by Sponsored Research Program (SRP) from Capio Biosciences, Inc.
Hong and A.Z. Wang are co-founders of Capio Biosciences, Inc., a biotech startup that is commercializing CapioCyte™ CTC technology. M.J. Poellmann is an employee of Capio Biosciences.
Zhang has received research funding (to Duke) from Abbvie, Acerta, Janssen, Merck,
Merrimack, Novartis, OmniSeq, Pfizer, and PGDx; speaking fees from Exelixis,
Genentech/Roche, Sanofi Aventis, and Genomic Health; and has consulted for AstraZeneca, Bristol-Myers Squibb, Foundation Medicine, Janssen, Pfizer, Pharmacyclics, Amgen, and Sanofi Aventis. Stock ownership/employment (spouse) from Capio Biosciences.
Footnotes
CRediT author statement
Jiyoon Bu: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – Original Draft, Writing – Review & Editing, Visualization. Ashita Nair: Methodology, Investigation. Luke Kubiatowicz: Investigation, Writing – Original Draft. Michael Poellmann: Methodology, Investigation, Writing – Original Draft. Woo-jin Jeong: Investigation. Marco Reyes-Martinez: Investigation. Andrew J. Armstrong: Resources, Supervision. Daniel George: Resources, Supervision. Andrew Wang: Conceptualization, Resources, Supervision. Tian Zhang: Conceptualization, Resources, Supervision, Project administration, Funding acquisition. Seungpyo Hong: Conceptualization, Methodology, Writing – Original Draft, Writing – Review & Editing, Supervision, Project administration, Funding acquisition.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, Klein C, Saini M, Bäuerle T, Wallwiener M, Holland-Letz T, Höfner T, Sprick M, Scharpff M, Marmé F, Sinn HP, Pantel K, Weichert W, Trumpp A, 2013. Nat. Biotechnol 31(6), 539–544. [DOI] [PubMed] [Google Scholar]
- Bidard FC, Peeters DJ, Fehm T, Nolé F, Gisbert-Criado R, Mavroudis D, Grisanti S, Generali D, Garcia-Saenz JA, Stebbing J, Caldas C, Gazzaniga P, Manso L, Zamarchi R, de Lascoiti AF, De Mattos-Arruda L, Ignatiadis M, Lebofsky R, van Laere SJ, Meier-Stiegen F, Sandri MT, Vidal-Martinez J, Politaki E, Consoli F, Bottini A, Diaz-Rubio E, Krell J, Dawson SJ, Raimondi C, Rutten A, Janni W, Munzone E, Carañana V, Agelaki S, Almici C, Dirix L, Solomayer EF, Zorzino L, Johannes H, Reis-Filho JS, Pantel K, Pierga JY, Michiels S, 2014. Lancet Oncol. 15(4), 406–414. [DOI] [PubMed] [Google Scholar]
- Bluemke K, Bilkenroth U, Meye A, Fuessel S, Lautenschlaeger C, Goebel S, Melchior A, Heynemann H, Fornara P, Taubert H, 2009. Cancer Epidemiol. Biomarkers Prev. 18(8), 2190–2194. [DOI] [PubMed] [Google Scholar]
- Brodaczewska KK, Szczylik C, Fiedorowicz M, Porta C, Czarnecka AM, 2016. Mol. Cancer 15(1), 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu J, Cho Y-H, Han S-W, 2017a. RSC Advances 7(78), 49684–49693. [Google Scholar]
- Bu J, Kang YT, Kim YJ, Cho YH, Chang HJ, Kim H, Moon BI, Kim HG, 2016. Lab Chip 16(24), 4759–4769. [DOI] [PubMed] [Google Scholar]
- Bu J, Kang YT, Lee YS, Kim J, Cho YH, Moon BI, 2017b. Biosens. Bioelectron 91, 747–755. [DOI] [PubMed] [Google Scholar]
- Bu J, Kim YJ, Kang YT, Lee TH, Kim J, Cho YH, Han SW, 2017c. Biomaterials 125, 1–11. [DOI] [PubMed] [Google Scholar]
- Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ, 2008. J. Clin. Oncol 26(19), 3213–3221. [DOI] [PubMed] [Google Scholar]
- de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D, 2008. Clin. Cancer Res. 14(19), 6302–6309. [DOI] [PubMed] [Google Scholar]
- El-Heliebi A, Kroneis T, Zöhrer E, Haybaeck J, Fischereder K, Kampel-Kettner K, Zigeuner R, Pock H, Riedl R, Stauber R, Geigl JB, Huppertz B, Sedlmayr P, Lackner C, 2013. J. Transl. Med 11(1), 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Ritchie K, 1997. Biophys. J 72(4), 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradilone A, Iacovelli R, Cortesi E, Raimondi C, Gianni W, Nicolazzo C, Petracca A, Palazzo A, Longo F, Frati L, Gazzaniga P, 2011. Anticancer Res. 31(12), 4219–4221. [PubMed] [Google Scholar]
- Hou JM, Krebs M, Ward T, Sloane R, Priest L, Hughes A, Clack G, Ranson M, Blackhall F, Dive C, 2011. Am. J. Pathol 178(3), 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes NE, Lane HA, 2005. Nat. Rev. Cancer 5(5), 341–354. [DOI] [PubMed] [Google Scholar]
- Jeong W. j., Bu J, Han Y, Drelich AJ, Nair A, Král P, Hong S, 2020. J. Am. Chem. Soc 142(4), 1832–1837. [DOI] [PubMed] [Google Scholar]
- Jeong W-J, Bu J, Kubiatowicz LJ, Chen SS, Kim Y, Hong S, 2018. Nano Converg. 5(1), 38–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara H, Yasuda M, Kumaki N, Shibayama T, Osamura Y, 2005. Tokai J. Exp. Clin. Med 30(3), 177–182. [PubMed] [Google Scholar]
- Kobayashi H, Boelte KC, Lin PC, 2007. Curr. Med. Chem 14(4), 377–386. [DOI] [PubMed] [Google Scholar]
- Lalani AA, Gray KP, Albiges L, Callea M, Pignon JC, Pal S, Gupta M, Bhatt RS, McDermott DF, Atkins MB, Woude GFV, Harshman LC, Choueiri TK, Signoretti S, 2017. Oncotarget 8(61), 103428–103436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Tian Z, Zhang L, Hou S, Hu S, Wu J, Jing Y, Sun H, Yu F, Zhao L, Wang R, Tseng HR, Zhau HE, Chung LW, Wu K, Wang H, Wu JB, Nie Y, Shao C, 2016. Oncotarget 7(37), 59877–59891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens Y, Humberg V, Erlmeier F, Steffens S, Steinestel J, Bögemann M, Schrader AJ, Bernemann C, 2017. Oncotarget 8(50), 87710–87717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maru S, Ishigaki Y, Shinohara N, Takata T, Tomosugi N, Nonomura K, 2013. J. Urol 189(5), 1921–1929. [DOI] [PubMed] [Google Scholar]
- Murlidhar V, Rivera-Báez L, Nagrath S, 2016. Small 12(33), 4450–4463. [DOI] [PubMed] [Google Scholar]
- Myung JH, Cha A, Tam KA, Poellmann M, Borgeat A, Sharifi R, Molokie RE, Votta-Velis G, Hong S, 2019. Anal. Chem 91(13), 8374–8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung JH, Eblan MJ, Caster JM, Park SJ, Poellmann MJ, Wang K, Tam KA, Miller SM, Shen C, Chen RC, Zhang T, Tepper JE, Chera BS, Wang AZ, Hong S, 2018a. Clin. Cancer Res. 24(11), 2539–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung JH, Gajjar KA, Chen J, Molokie RE, Hong S, 2014. Anal. Chem 86(12), 6088–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung JH, Gajjar KA, Saric J, Eddington DT, Hong S, 2011. Angew. Chem. Int. Ed 50(49), 11769–11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung JH, Launiere CA, Eddington DT, Hong S, 2010. Langmuir 26(11), 8589–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung JH, Park SJ, Wang AZ, Hong S, 2018b. Adv. Drug Deliv. Rev 125, 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung JH, Roengvoraphoj M, Tam KA, Ma T, Memoli VA, Dmitrovsky E, Freemantle SJ, Hong S, 2015. Anal. Chem 87(19), 10096–10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel I, Gauler TC, Bublitz K, Lazaridis L, Goergens A, Giebel B, Schuler M, Hoffmann AC, 2016. PLoS One 11(4), e0153018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DX, Bos PD, Massagué J, 2009. Nat. Rev. Cancer 9(4), 274–284. [DOI] [PubMed] [Google Scholar]
- Okegawa T, Itaya N, Hara H, Tambo M, Nutahara K, 2016. Int. J. Mol. Sci 17(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poellmann MJ, Bu J, Hong S, 2018. Nanomedicine (Lond) 13(18), 2327–2340. [DOI] [PubMed] [Google Scholar]
- Rossi E, Fassan M, Aieta M, Zilio F, Celadin R, Borin M, Grassi A, Troiani L, Basso U, Barile C, Sava T, Lanza C, Miatello L, Jirillo A, Rugge M, Indraccolo S, Cristofanilli M, Amadori A, Zamarchi R, 2012. Br. J. Cancer 107(8), 1286–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H-J, Rho SB, Jung DC, Han I-O, Oh E-S, Kim J-Y, 2011. J. Cell Sci 124(7), 1077. [DOI] [PubMed] [Google Scholar]
- Shingarev R, Jaimes EA, 2017. Am. J. Physiol. Renal Physiol 313(2), F145–F154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacova M, Bartosova M, Skvarkova L, Zatovicova M, Vidlickova I, Csaderova L, Barathova M, Breza J, Bujdak P, Pastorek J, Pastorekova S, 2013. Oncol. Lett 5(1), 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostain J, Li G, Gentil-Perret A, Gigante M, 2010. Eur. J. Cancer 46(18), 3141–3148. [DOI] [PubMed] [Google Scholar]
- Ueda K, Ogasawara S, Akiba J, Nakayama M, Todoroki K, Sanada S, Suekane S, Noguchi M, Matsuoka K, Yano H, 2013. PLoS One 8(10), e75463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.