Abstract
Launch of in-house sensitive cell-free deoxyribonucleic acid (cfDNA) mould polymerase chain reaction (PCR) assays increased detection of moulds meeting suspected healthcare-associated infection (HAI) criteria. Definition was based on time from admission and mould detection in culture or via molecular methods. We created a modified mould HAI algorithm incorporating clinical context into the case definition, which allowed for better capture of possible mould HAIs, decreased number of investigations, and improved utilization of Infection Prevention and Control (IPC) resources.
Keywords: healthcare-associated, moulds, outbreak, infection prevention
Background
The Centers for Disease Control and Prevention (CDC) defines a suspected mould outbreak in a hospital setting as any cluster of invasive mould infections that appears to be above the baseline rate or is otherwise concerning to healthcare staff, especially if occurs >1 week after admission (The Centers for Disease Control and Prevention, 2022). A definitive diagnosis of a proven mould infection is challenging and is usually established through identification of a mould in culture or on histopathology (De Pauw et al., 2008).
In November 2020, our institution’s Microbiology Laboratory launched newly validated cell-free deoxyribonucleic acid (cfDNA) fungal polymerase chain reaction (PCR) assays for the detection of various pathogenic fungi in plasma, fluid, or tissue specimens. Order options include a full mould PCR panel (Aspergillus, Mucorales agents, Scedosporium, and Fusarium), as well as separate Aspergillus spp. Polymerase chain reaction and Mucorales agents (Mucor, Rhizopus, and Rhizomucor spp.) PCR. The specificity of these assays is 99.5%, while sensitivity in plasma specimens is 56.3% for Aspergillus spp. and 91.7% for Mucorales agents. Overall positive predictive value (PPV) and negative predictive value (NPV) of the test are 86% and 99%, respectively (Senchyna et al., 2021).
Subsequently, there was an increase in detection of these moulds in 2021 relative to prior years leading to an increased number of investigations done by the Infection Prevention and Control (IPC) department to evaluate for possible healthcare-associated infection (HAI).
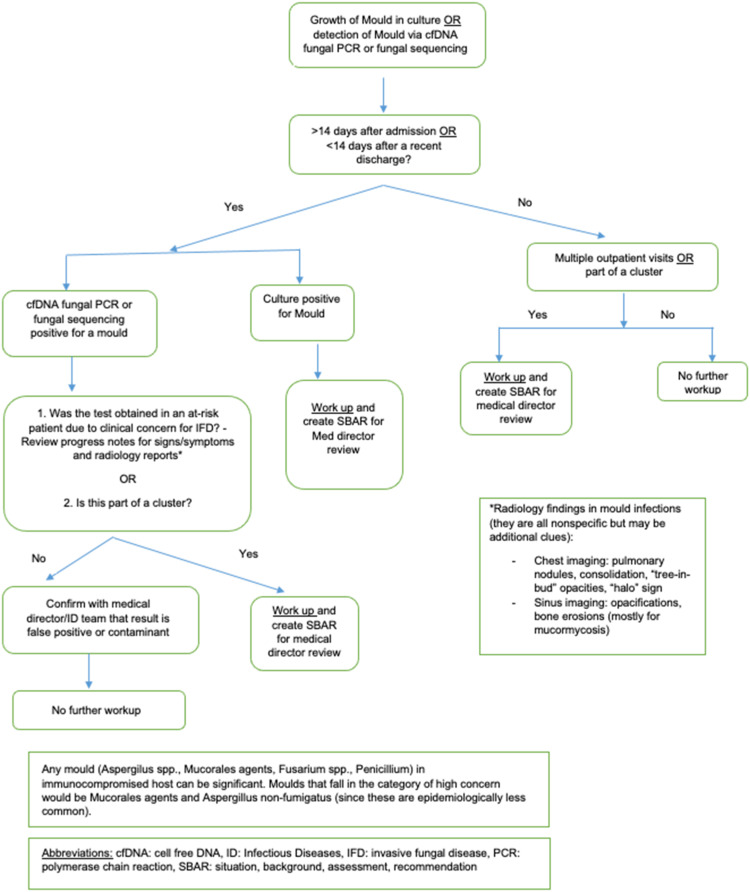
In March 2021, the IPC team developed and implemented a modified HAI algorithm for moulds that incorporated the established time-based surveillance as well as a review of the medical record. This algorithm incorporated clinical concern for invasive mould infection based on host risk factors, symptoms, and radiology reports. Cases that failed to meet this additional element and that were not part of a cluster were not considered to be HAI and did not undergo further investigation.
The purpose of this retrospective analysis was to describe the effect of the new diagnostic cfDNA mould PCR panels on the rate of positive tests, the need for HAI investigation based on previous definitions, and the impact of the modified mould HAI algorithm on the number of mould HAI investigations.
Methods
This retrospective descriptive study was conducted at a 361-bed, freestanding pediatric medical center in Northern California. Using the institutional IPC surveillance database, we identified mould cases in the period from January 2017 to November 2022. Patients with a mould detected >2 weeks after admission or <2 weeks after discharge, and who had an oncologic diagnosis or history of hematopoietic stem cell transplantation (HSCT) were included. Diagnosis was via molecular testing (cfDNA fungal PCR panel or fungal sequencing) or conventional diagnostics with culture and/or pathology. The intervention of interest was the application of the modified mould algorithm (Figure 1) by the IPC team, which was applied to all potential cases after implementation in March 2021. The primary outcome was a mould case that resulted in an HAI investigation. When applicable, Fisher’s exact test was used for the analysis of the primary outcome. A two-sided p value < .05 was considered statistically significant.
Figure 1.
Modified healthcare-associated infection algorithm for moulds.
Results
There were 19 cases with moulds identified in clinical specimens obtained from oncology and HSCT patients during the 6-year study period from 2017 to 2022 that met the inclusion criteria (Table 1). There were 9 (47.4%) Aspergillus spp., 2 (10.5%) Fusarium spp., and 8 (42.1%) Mucorales agents cases. Twelve (63.1%) mould cases occurred in the post-cfDNA fungal PCR era (November 2020 or later), and 6/8 (75%) of the Mucorales agents cases occurred in that time period alone.
Table 1.
Mould cases and investigations for potential healthcare-associated infection (HAI) by year between 2017 and 2022.
| Year | Positive cases | Diagnosis | Patient location(s) | HAI organism(s) identified | Diagnostic method | HAI investigations |
|---|---|---|---|---|---|---|
| 2017 | 3 | AML | 1. PICU: PPR, NPA HemOnc: PPR, NPA | Rhizomucor spp. | Culture | 0 |
| AML | 1. PICU, PPR, NPA HemOnc: PPR, NPA | Rhizomucor pusillus | Sequencing | 0 | ||
| ALL | 1. PICU: PPR HemOnc: PPR, NPA | Aspergillus fumigatus | Culture | 0 | ||
| 2018 | 2 | ALL | 1. PICU: PPR | Aspergillus fumigatus | Culture | 0 |
| ALL | 1. PICU: PPR | Aspergillus spp. | Sequencing | 0 | ||
| 2019 | 1 | AML | 1. HemOnc: PPR | Fusarium solani | Culture | 1 |
| 2020 | 1 | AML | 1. SCT: PPR | Fusarium fukikuroi | Culture | 0 |
| 2021 ( a pre) | 4 | ALL | 1. HemOnc: PPR | Aspergillus niger | Culture | 1 |
| ALL, HSCT | 1. PICU: PPR 2. SCT: PPR | Aspergillus terreus (n = 2) | cfDNA | |||
| HSCT | 1. SCT: PPR | Mucorales agents | cfDNA | 1 | ||
| 2021 ( a post) | 5 | HSCT | 1. PICU: PPR, NPA 2. SCT: PPR | Mucorales agents (n = 2) | cfDNA | 2 |
| HSCT | 1. SCT: PPR 2. PICU: PPR, NPA PICU: PPR | Aspergillus fumigatus (n = 2) | Culture and cfDNA | 0 | ||
| ALL | 1. HemOnc: PPR | Rhizopus oryzae (n = 1) | Culture and cfDNA | 1 | ||
| 2022 | 3 | AML | 1. HemOnc: PPR | Mucor spp. | Culture | 1 |
| ALL | 1. HemOnc: PPR PICU: PPR | Rhizopus microspores | cfDNA and sequencing | 0 | ||
| ALL | 1. PICU: NPR, PPA PICU: PPR | Aspergills spp. | cfDNA | 0 |
cfDNA, cell-free deoxyribonucleic acid; PCR, polymerase chain reaction; HAI, healthcare-associated infection; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; SCT, stem cell transplantation; PICU, pediatric intensive care unit; HemOnc, hematology oncology unit; PPR, positive pressure room; NPA, negative pressure anteroom.
aRefers to pre- and post implementation of the modified mould HAI algorithm.
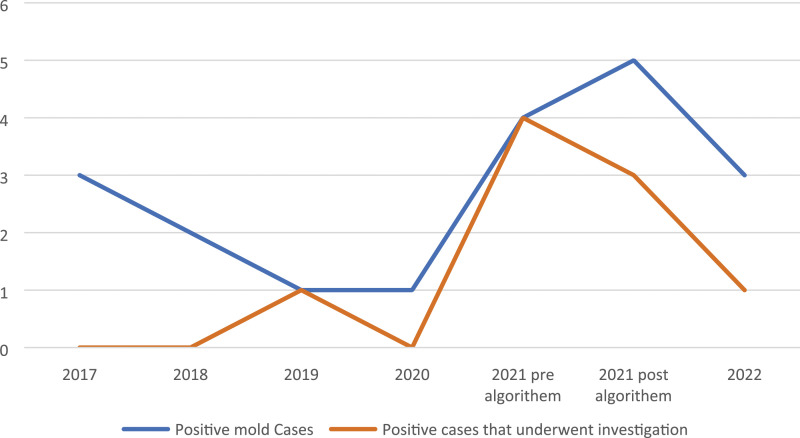
The average positivity rate per month was 0.15 between 2017 and 2020 compared to 0.5 per month in 2021–2022 after the introduction of the cfDNA mould PCR panels (p < .05). We observed that mould HAI investigations increased in parallel with increased number of positive mould results following the launch of the cfDNA fungal PCR assays (Figure 2). All four positive mould results meeting the HAI time definition underwent investigation in the first 3 months of 2021 alone (4/4, 100%). After implementation of the modified mould HAI protocol, four out of eight positive mould cases required investigation (4/8, 50%; Table 1).
Figure 2.
Positive mould cases between 2017 and 2022 and number of cases investigated for possible HAI plotted over time.
Investigations included environmental air sampling, high efficiency particulate air (HEPA) evaluation by the hospital engineering department, and unit entrance and exit evaluations. Of note, the hospital conducts regular maintenance on HEPA filters every 6 months, which includes visual inspections and cleanouts but routine air sampling is not conducted except in the event of water incursion or leaks or dust producing construction activity in the area or as indicated during HAI investigations. Targeted additional evaluations to identify potential sources of infection were conducted based on previously published reports. For example, Rhizopus oryzae investigation included additional analysis of appropriate sterile nasal packing product use, and Aspergillus spp. investigation included evaluation of linen supply and nursing and wound care practices (Repetto et al., 2012). None of the investigations found a confirmed hospital source of infection.
Discussion and conclusions
The CDC and our institution’s definitions for possible mould HAIs are based on the time from admission and diagnosis of a mould using fungal cultures, histopathology, or molecular diagnostics (About Healthcare-Associated Mould Outbreaks | Fungal Diseases | CDC, 2022). A definitive diagnosis is rare and difficult to establish (De Pauw et al., 2008). Upon implementation of highly sensitive cfDNA fungal PCR assays in our institution, detection of moulds, especially Mucorales agents, increased. This led to institutional concern for a possible hospital-acquired source resulting in multiple extensive HAI investigations with no specific hospital source identified. Thus, there was a need to modify the definition and add clinical context to determine which cases should trigger a full HAI investigation.
A modified algorithm was created to include clinical aspects including host immune status, symptoms, and imaging findings suggestive of a true infection. Since sole detection of a mould via molecular testing such as cfDNA or fungal sequencing may represent colonization rather than an infection, the clinical aspects in the algorithm guide the IPC specialists to determine if this represents a true infection and if it warrants an HAI investigation and specific actions. Cases and recommendations are then presented in the form of an SBAR (situation, background, assessment, and recommendation) communication tool to the medical directors of IPC, who are ID specialists, for final review and confirmation. As a result, HAI investigations became more targeted and 50% fewer investigations needed to be performed. Nevertheless, the total number of investigations was still higher than in previous years.
The potential considerations for increased mould infections in addition to the possibility of an in-hospital exposure and use of a sensitive molecular test are as follows: changes to anti-fungal prophylaxis, types of immunosuppressive regimens, patients’ baseline colonization with moulds, and environmental exposures from changes to fungal ecology due to climate and geographic factors. While we have not been able to identify a singular cause for these infections, extensive investigations for a hospital source have not been revealing. Our study has limitations. It is a retrospective study in a single center with a small sample size. Additionally, mould infections are likely multifactorial making it challenging to identify a unifying cause.
This retrospective analysis shows the impact of advanced diagnostic techniques on IPC work around mould HAI investigations. Our experience highlights the gap between surveillance definition based mainly on cultures and histopathology versus the inclusion of novel sensitive molecular diagnostics. We propose that incorporating clinical criteria into case definitions is critical to better assess potential mould HAIs and utilize IPC departmental resources optimally, especially at a time when the sensitivity and specificity of these newer testing technologies are evolving.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD
Ayelet Rosenthal https://orcid.org/0000-0002-1143-829X
References
- De Pauw B, Walsh TJ, Donnelly JP, et al. (2008) Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) consensus group. Clinical Infectious Diseases : An Official Publication of the Infectious Diseases Society of America 46(12): 1813–1821. DOI: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto EC, Giacomazzi CG, Castelli F. (2012) Hospital-related outbreaks due to rare fungal pathogens: a review of the literature from 1990 to June 2011. European Journal of Clinical Microbiology & Infectious Diseases 31(11): 2897–2904. DOI: 10.1007/s10096-012-1661-3. [DOI] [PubMed] [Google Scholar]
- Senchyna F, Hogan CA, Murugesan K, et al. (2021) Clinical accuracy and impact of plasma cell-free dna fungal polymerase chain reaction panel for noninvasive diagnosis of fungal infection. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America 73(9): 1677–1684. DOI: 10.1093/cid/ciab158. [DOI] [PubMed] [Google Scholar]
- The Centers for Disease Control and Prevention (2022) About Healthcare-Associated Mold Outbreaks. Available at: https://www.cdc.gov/fungal/outbreaks/healthcare-associated-mold-outbreaks.html [Google Scholar]