Abstract
This study reports four cases of transmission of the rare hepatitis C virus genotype 4 which occurred in a hemodialysis unit and originated from a single source of infection. Phylogenetic analyses performed with the NS5b domain showed that all four patients with secondary infections were infected with virus strains very similar to that of the index patient. Cross infections probably resulted from breaches in safety precautions and lack of dialysis machine sterilization.
In France, the overall prevalence of anti-hepatitis C virus (HCV) antibody in hemodialysis units is 10 to 15% but may be as high as 30% in some units (11). Blood transfusions, particularly those carried out before the screening of blood donors, have certainly been involved in the initial spread of HCV in this population (8). Molecular analyses of viral strains have shown that nosocomial cross infections of nontransfused patients were possible (1, 5). Percutaneous procedures, sharing of dialysis equipment, and failure to implement elementary hygienic measures may constitute routes of transmission from patient to patient. Our study reports four cases of transmission of the rare HCV genotype 4 which probably originated from brief contacts with a single carrier patient.
Patients went to an autodialysis unit (unit A) for 4-h dialysis sessions three times per week on five dialysis machines (Fresenius, Bad Hombourg, Germany). There were two dialysis sessions (morning and afternoon) every day. No dedicated areas or dedicated machines were used for HCV-infected patients. The dialyzers, filters, and tubes were never reused. All dialysis machines were disinfected by chemical sterilization (Dialox; CFPO, Paris, France) at the end of the day, after the two sessions.
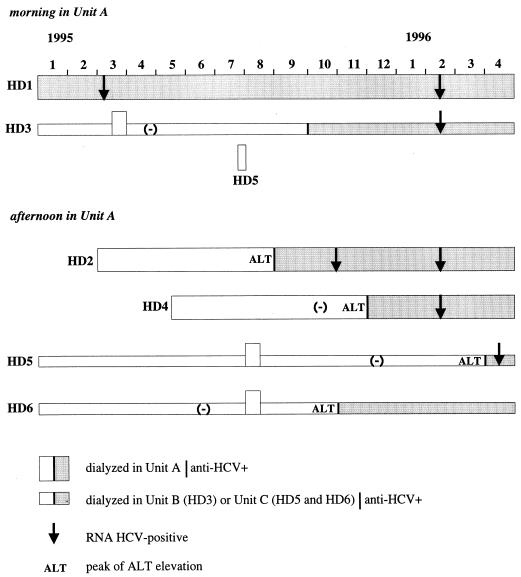
One patient (HD2), last determined to be anti-HCV antibody negative in October 1994, had an increase in alanine aminotransferase (ALT) levels at the end of July 1995 (Fig. 1). He was found to have anti-HCV antibody (Ortho HCV 3.0 and Chiron RIBA HCV 3.0 SIA, both from Ortho Diagnostic Systems, Raritan, N.J.) in September 1995 and was infected by a genotype 4 virus (Inno-LiPA HCV II; Innogenetics, Zwijnaarde, Belgium). A second patient (HD3), originally treated at another dialysis center (unit B), spent only 2 weeks in unit A in March 1995. He last tested negative in April 1995, was found to be positive in October 1995, and was infected with a genotype 4 virus. A third patient (HD4) tested HCV negative on 3 October 1995, had an increase in ALT levels on 6 November, and was found to be anti-HCV antibody positive 10 days later. She was also infected with a genotype 4 virus. None of these three patients had been given a blood transfusion or a kidney transplant. Dialysis records showed that they had all been in contact with a fourth patient, HD1, who was infected with a genotype 4 virus. A fifth patient (HD5) had tested HCV positive in April 1996 after an increase in ALT levels in March. This patient was infected with a genotype 4 virus and had been briefly present in autodialysis unit A during the summer of 1995 (a single morning session in July with patient HD1 and afternoon sessions with patients HD2 and HD4 during 2 weeks in August). The last negative test for this patient was in December 1995.
FIG. 1.
Chronology of HCV transmission events in dialysis unit A.
To show that these five patients were infected with the same virus, part of the NS5b domain of HCV RNA was analyzed (12). Viral RNA was extracted by a modified version of the acid guanidinium thiocyanate-phenol-chloroform method, and then reverse transcription and the first run of PCR were performed in a single tube (7) with primers 242 (position −8304) and 243 (position +7904) (6). Amplified products were analyzed by electrophoresis in a 1.5% agarose gel. Nested PCR was carried out with primers 122 (position +7935) and 123 (position −8250) (15) or primers 554 (position +7935) and 555 (position −8227) (3) when no amplicons were detected after the first run.
Amplicons were purified from a 1% low-melting-point agarose gel, cloned (Original TA cloning kit; Invitrogen, San Diego, Calif.), and sequenced by the dideoxynucleotide chain termination method (14). Two clones per sample were analyzed. NS5b sequences (nucleotides 7975 to 8196) from HD1, HD2, HD3, HD4, and HD5 were analyzed in comparison with four local but unrelated HCV genotype 4 controls (T1 to T4) and 21 genotype 4 sequences retrieved from data banks.
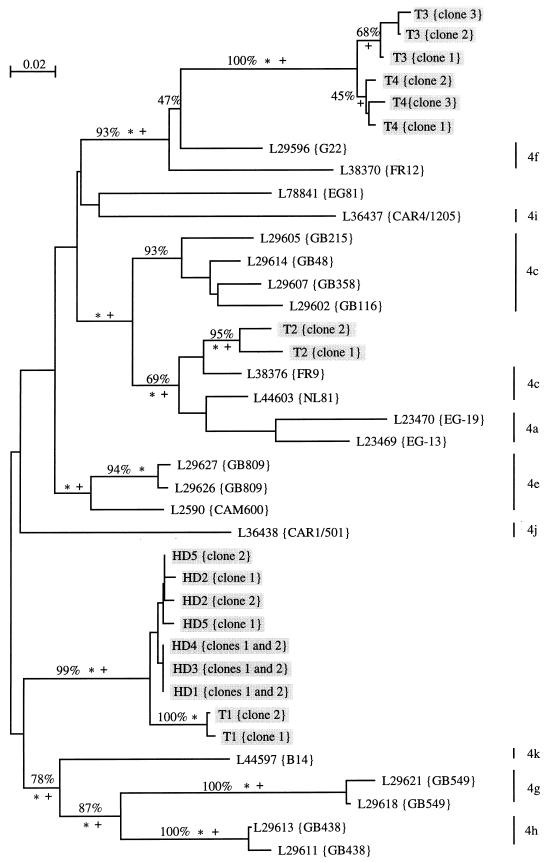
A tree including the interpretations derived from three phylogenetics methods (9) is shown in Fig. 2. Most of the HCV genotype 4 sequences deposited in the data bank were from African patients. Our type 4 samples (patients and controls) could not be assigned to any specific subtype. The viral sequences of the five hemodialyzed patients were included in the same distinct branch of the tree.
FIG. 2.
Unrooted phylogenetic tree of 38 genotype 4 hepatitis C virus isolates derived from analysis of a partial NS5b sequence (nucleotides 7975 to 8196) retrieved from the EBI databank or by using the Entrez software from the National Center for Biotechnology Information (about 600 sequences). Sequences were aligned manually, and preliminary phylogenetic analyses were used to identify a subset of 120 sequences representative of each monophyletic taxon. More detailed analyses were restricted thereafter to type 4 sequences. The topology was obtained by a maximum-likelihood method (asterisks indicate branches at a P value of <0.01]. Monophyletic taxons also retrieved in the most parsimonious tree are indicated by plus signs. Percentages show branches identified by bootstrap resampling (500 replicates) by using neighbor-joining analysis (13). Trees were drawn with the njplot program for the Macintosh computer (M. Gouy, CNRS URA 243, Université Claude Bernard, Lyon, France). Subtypes of sequences retrieved from data banks were obtained from the persons who deposited the sequences (16, 17). Sequences of viruses from our patients (HD) and from four unrelated controls from the same administrative region (T) are shaded.
At the beginning of the episode reported in our study, three of 17 regular patients (17.6%) were anti-HCV positive (two with genotype 1b and one [HD1] with genotype 4). The medical staff, which consisted of two nurses (one for each session), were anti-HCV negative. In the last 3 years, no seroconversion to HCV genotype 1b has been observed. In contrast, our study clearly demonstrated that nosocomial transmission of the rare HCV genotype 4 has occurred in this hemodialysis unit.
Patients HD2, HD3, HD4, and HD5 had not been given blood transfusion or transplantation. No major incident such as profuse bleeding during dialysis or after the sessions had been reported. Patients HD1 and HD3 were dialyzed during the morning session, whereas patients HD2 and HD4 attended the afternoon sessions. We cannot formally exclude the possibility that some overlapping might have occurred. Since in dialysis unit A there was no sterilization procedure between the morning and afternoon sessions, sharing of the same dialysis equipment was considered to have been the most probable route of HCV transmission from patient HD1 to patients HD2 and HD4. The implication of dialysis machines in HCV transmission has not been formally demonstrated (2, 4), but in our cases, dialysis equipment was the only regular means of contact between the infected patients.
Patient HD3 underwent dialysis during the same session as patient HD1 for 2 weeks and then never returned to unit A. In his case, HCV transmission presumably resulted from a breach in the safety precautions, although no major mishap was reported during the study period. A prospective multicenter study carried out in Belgium on the incidence and risk factors for HCV in hemodialysis units demonstrated the importance of universal precautions to alleviate the risk of nosocomial transmission. Risk of seroconversion for patients dialyzed next to anti-HCV-positive patients was higher in dialysis units where such precautions were not implemented (8). In our dialysis unit, safety measures may have been overlooked in emergency situations; nurses may not have changed gloves between patients, for example.
For patient HD5, there was a very long delay (7 months) between the presumed contamination and seroconversion. It has been reported that seroconversion after HCV infection may be greatly delayed (up to 10 months) in dialysis patients, perhaps due to the compromised immune response known to occur in chronic renal failure (10). After the completion of this work, we undertook a survey of the dialysis unit where patient HD5 had been transferred (unit C). To our surprise, we found a sixth patient (HD6) (Fig. 1) infected by HCV serotype 4 (Murex HCV Serotyping 1-6 assay; Murex Diagnostics, Chatillon, France). Reexamination of the records revealed that patient HD6 had been briefly dialyzed in unit A during the summer of 1995 along with patient HD5. She had seroconverted to HCV 3 months later—that is, 5 months before HD5 (Fig. 1). Thus, HD5 may have been infected in unit C via contacts with patient HD6. We cannot further explore this hypothesis since patient HD6 is no longer viremic.
Molecular analysis of viral strains made it possible to demonstrate that an outbreak of HCV infections originating from a common source occurred in dialysis unit A. The lack of sterilization between the two daily sessions and the nonimplementation of elementary safety precautions were probably the causes of the cross infections. After these episodes, a sterilization procedure was introduced after each dialysis run and universal prevention measures were tightened. No new HCV seroconversions have been reported since.
Acknowledgments
This work was supported by grants from the Association pour la Recherche sur le Cancer, Villejuif, France (ARC 415/94), and the Association Recherche et Partage des Caisses d’Epargne Ecureuil, Paris, France.
REFERENCES
- 1.Allander T, Medin C, Jacobson S H, Grillner L, Persson M A. Hepatitis C transmission in a hemodialysis unit: molecular evidence for spread of virus among patients not sharing equipment. J Med Virol. 1994;43:415–419. doi: 10.1002/jmv.1890430417. [DOI] [PubMed] [Google Scholar]
- 2.Caramelo C, Navas S, Alberola M L, Bermejillo T, Reyero A, Carreño V. Evidence against transmission of hepatitis C virus through hemodialysis ultrafiltrate and peritoneal fluid. Nephron. 1994;66:470–473. doi: 10.1159/000187867. [DOI] [PubMed] [Google Scholar]
- 3.Chan S W, McOmish F, Holmes E C, Dow B, Peutherer J F, Follet E, Yap P L, Simmonds P. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J Gen Virol. 1992;73:1131–1141. doi: 10.1099/0022-1317-73-5-1131. [DOI] [PubMed] [Google Scholar]
- 4.Chiaramonte S, Tagger A, Ribero M L, Grossi A, Milan M, La Greca G. Prevention of viral hepatitis in dialysis units: isolation and technical management of dialysis. Nephron. 1992;61:287–289. doi: 10.1159/000186907. [DOI] [PubMed] [Google Scholar]
- 5.De Lamballerie X, Olmer M, Bouchouareb D, Zandotti C, De Micco P. Nosocomial transmission of hepatitis C virus in hemodialysis patients. J Med Virol. 1996;49:296–302. doi: 10.1002/(SICI)1096-9071(199608)49:4<296::AID-JMV7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 6.Enomoto N, Takada A, Nakao T, Date T. There are two major types of hepatitis C virus in Japan. Biochem Biophys Res Commun. 1990;170:1021–1025. doi: 10.1016/0006-291x(90)90494-8. [DOI] [PubMed] [Google Scholar]
- 7.François M, Dubois F, Brand D, Bacq Y, Guerois C, Mouchet C, Tichet J, Goudeau A, Barin F. Prevalence and significance of hepatitis C virus (HCV) viremia in HCV antibody-positive subjects from various populations. J Clin Microbiol. 1993;31:1189–1193. doi: 10.1128/jcm.31.5.1189-1193.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jadoul M, Cornu C, Van Ypersele De Strihou C the UCL Collaborative Group. Incidence and risk factors for hepatitis C seroconversion in hemodialysis: a prospective study. Kidney Int. 1993;44:1322–1326. doi: 10.1038/ki.1993.385. [DOI] [PubMed] [Google Scholar]
- 9.Le Pogam S, Dubois F, Christen R, Raby C, Cavicchini A, Goudeau A. Comparison of DNA enzyme immunoassay and line probe assays (Inno-LiPA HCV I and II) for hepatitis C virus genotyping. J Clin Microbiol. 1998;36:1461–1463. doi: 10.1128/jcm.36.5.1461-1463.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okuda K, Hayashi H, Kobayashi S, Irie Y. Hepatitis C infection in dialysis units. In: Rizzetto M, Purcell R H, Gerin J L, Verme G, editors. Viral hepatitis and liver disease. Turin, Italy: Minerva Medica; 1997. pp. 590–597. [Google Scholar]
- 11.Olmer M, Bouchouareb D, Zandotti C, De Micco P, De Lamballerie X. Transmission of the hepatitis C in a hemodialysis unit: evidence for a nosocomial infection. Clin Nephrol. 1997;47:263–270. [PubMed] [Google Scholar]
- 12.Power J P, Lawlor E, Davidson F, Holmes E C, Yap P L, Simmonds P. Molecular epidemiology of an outbreak of infection with hepatitis C virus in recipients of anti-D immunoglobulin. Lancet. 1995;345:1211–1213. doi: 10.1016/s0140-6736(95)91993-7. [DOI] [PubMed] [Google Scholar]
- 13.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 14.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simmonds P, Holmes E C, Cha T A, Chan S W, McOmish F, Irvine B, Beall E, Yap P L, Kolberg J, Urdea M S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 16.Stuyver L, Wyseur A, Van Arnhem W, Lunel F, Laurent-Puig P, Pawlotsky J M, Kleter B, Bassit L, Nkengasong J, van Doorn L J, Maertens G. Hepatitis C virus genotyping by means of 5′UR/core line probe assays and molecular analysis of untypeable samples. Virus Res. 1995;38:137–157. doi: 10.1016/0168-1702(95)00052-r. [DOI] [PubMed] [Google Scholar]
- 17.Stuyver L, Wyseur A, van Arnhem W, Hernandez F, Maertens G. Second-generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol. 1996;34:2259–2266. doi: 10.1128/jcm.34.9.2259-2266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]