Abstract
A novel multiplex single-cell genomic and immunophenotypic strategy leverages the sensitivity of MRD detection and distinguishes leukemic and preleukemic subpopulations.
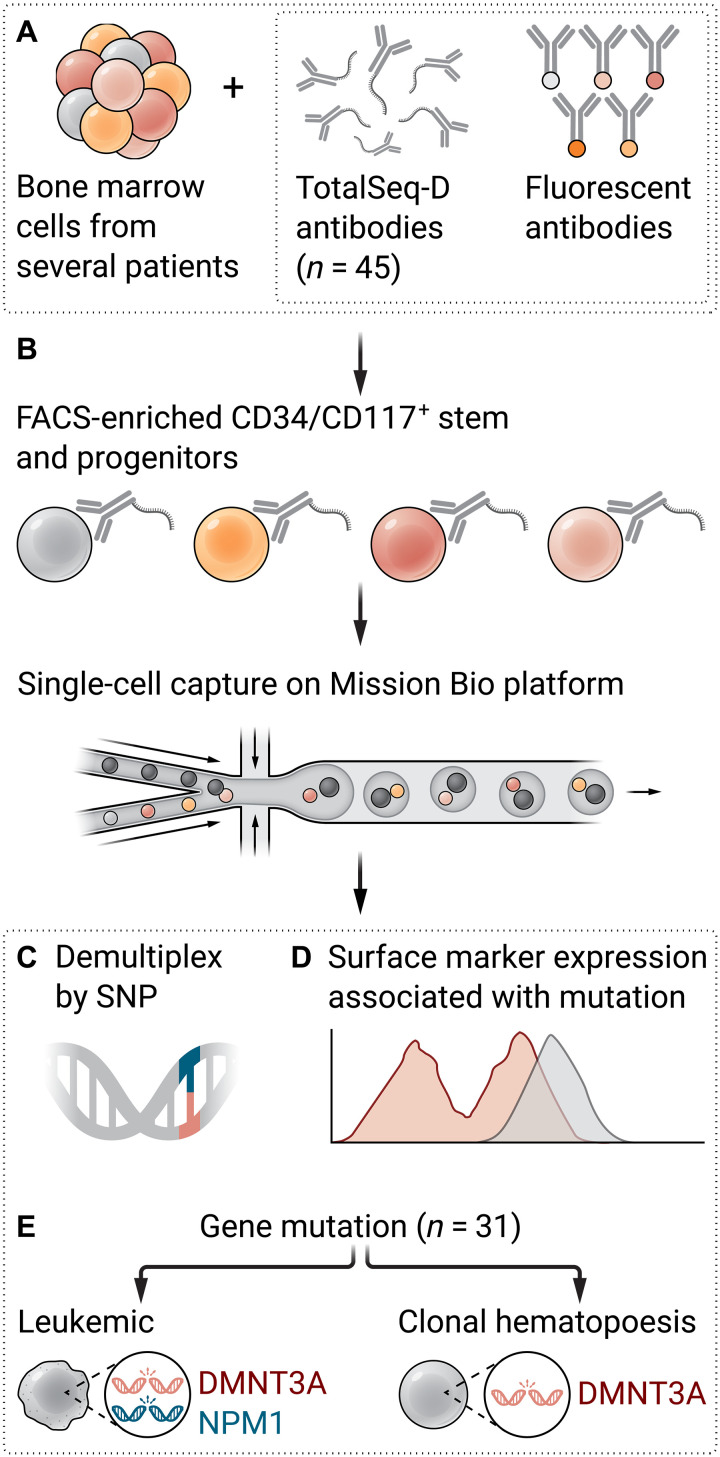
Fig. 1. A novel multiplex single-cell genomic and immunophenotypic strategy leverages the sensitivity of MRD detection and MRD clonal architecture to identify leukemic and preleukemic subpopulations.
(A) AML samples from different patients are pooled and stained with TotalSeq-D antibody cocktail and fluorescent antibodies. (B) Cells are sorted based on CD34/CD117 expression for stem and progenitors and captured on Mission Bio scDNA-seq platform. (C) Samples are demultiplexed by SNP after sequencing. (D) Unique surface marker expression pattern correlates with mutation or combinations of mutations. (E) Leukemic clones are distinguished from clonal hematopoiesis clones based on the acquisition of additional mutations. Illustration: Ashley Mastin/Science Advances.
Acute myeloid leukemia (AML) is a relentless blood cancer. Achieving long-term remission remains a daunting task, even with initial treatment success. The looming threat of relapse casts a shadow on patient outcomes, underscoring the crucial role of measurable residual disease (MRD) in driving disease recurrence and therapy resistance. However, current methods for MRD detection are limited in their ability to simultaneously achieve high sensitivity and high specificity. Multicolor flow cytometry (MFC) is limited by 0.1% sensitivity (1); real-time quantitative polymerase chain reaction (RT-qPCR) often is only applicable to known mutations and fusion genes; bulk-tumor next-generation sequencing (NGS) cannot distinguish between driver and bystander mutations. These limitations necessitate the urgent development of sensitive and specific MRD detection techniques.
Single-cell DNA sequencing (scDNA-seq) technology has emerged as a powerful tool to study clonal architecture in AML and overcome these limitations by distinguishing between subpopulations sharing the same “founder” mutations (acquired early in leukemic development). However, without the transcriptome or surface markers, the state or types of these single cells within clonal architecture cannot be defined, making it difficult to translate them to clinical disease monitoring. In this issue of Science Advances, Robinson et al. (2) present a novel multiplex single-cell genomic and immunophenotypic strategy, aiming to leverage the sensitivity of MRD detection and MRD clonal architecture.
INCREASING MRD DETECTION SENSITIVITY BY ENRICHMENT
MRD clones responsible for relapse primarily reside in immature compartments that are usually positive for stem and progenitor markers, including CD34, CD33, and CD117. With the increasing availability of fluorescence-assisted cell sorting (FACS) in clinical settings, more and more physicians are considering flow enrichment based on these surface markers as an upstream step in common MRD detection methods. This approach is expected to enable the identification and profiling of a greater number of MRD cells and their associated mutations, thereby enhancing the accuracy and sensitivity of MRD assessment. For instance, CD34+ enrichment of peripheral blood from patients with AML increased the sensitivity in detecting MRD by deep DNA sequencing (3).
Robinson et al. went a step further by first examining the marker expression on their AML blast samples from the bone marrow, confirming that they are either CD34- or CD117-positive before sorting. Next, they used the Mission Bio Tapestri single-cell sequencing platform to sequence 31 genes recurrently involved in myeloid malignancies of patients with European ancestry using CD34+/CD117+ sorted AML cells.
To overcome the low throughput of this scDNA-seq platform and limited patient material, the authors multiplexed their samples via pooling and developed a computational approach to deconvoluting different individuals by germline single-nucleotide polymorphisms (SNPs) covered by the custom scDNA panel. Through a rigorous limiting dilution experiment in which AML blasts from three genetically distinct AML samples were mixed with 10 million normal bone marrow mononuclear cells at different ratios and in replicates, scDNA-seq with enrichment had an estimated sensitivity of 0.0077% when using a threshold of detecting ≥1 mutated cell and 0.0578% for the detection of ≥3 mutated cells.
While the sensitivity of scMRD is on par with deep NGS (0.01 to 0.001%), single-cell data are by nature sparse, and the results and interpretations need to be benchmarked with bulk NGS. The authors demonstrated that of 30 samples, 23 can be consistently interpreted by both methods. scMRD better identifies relapse-related mutations than bulk NGS (unsorted), confirming that the relapse-related clones are within the CD34+/CD117+ populations.
SAME MUTATION, DIFFERENT CLONES
A pivotal drawback of using bulk NGS data to interpret MRD and outcome is that it cannot distinguish between MRD clones and mutated clonal hematopoiesis (CH)/preleukemic cells. This distinction is vital as not all preleukemic clones lead to relapse, and accurate identification of MRD clones can profoundly affect therapeutic decision-making. The utility of scMRD extends beyond MRD detection, as it enables the study of clonal architecture and the characterization of leukemic and preleukemic subpopulations. The authors surveyed 30 AML MRD samples and showcased three distinct clones within one patient: preleukemic DNMT3A, bystander JAK2, and relapse-related DNMT3A/NPM1. This level of precision offers critical rationales for targeting specific mutations over others, leading to more effective therapeutic strategies.
Moreover, the integration of mutation and immunophenotypic information (42 surface markers by TotalSeq-D, BioLegend) enhances MRD detection and may hold valuable insights into targeted therapy strategies by identifying genotype-specific protein expression patterns. For example, TP53 clones have higher CD71 expression. More importantly, clones harboring co-mutations indicate unique immunophenotypes within the same sample. For example, DNMT3A/NPM1 co-mutated cells highly express granulocytic/monocytic markers such as CD11b and CD16. These findings could serve as the “ground-state truth” for any studies that investigate co-mutation phenotypes and therapeutic vulnerabilities.
FUTURE MRD ASSESSMENT
While the scMRD assay shows promising results in enhancing MRD detection, multiple limitations exist. Notably, cell recovery during the capture process is generally low (14%) and variable, which may affect sensitivity. Future work should focus on optimizing cell recovery for a more accurate representation of the bulk tumor. In addition, computational demultiplexing (36% recovery rate) and allele dropout pose challenges. New version 3 chemistry from Mission Bio might improve these numbers, but this has yet to be confirmed by users. While cell-surface hashtag antibodies can be used for better demultiplexing accuracy than SNP, allele dropout with current scDNA-seq cannot be corrected, necessitating further refinements to enhance the accuracy of the approach.
The study’s emphasis on comparative analyses with existing MRD assays, such as bulk NGS, is crucial for gauging the clinical utility of the scMRD approach and validating its superiority over current methods. Large-scale scMRD profiling of longitudinal AML samples with diverse outcomes and genetic background is needed to make solid claims, such as which marker on which co-mutation clone indicates a high risk of relapse or the duration of remission. Further expanding the repertoire of antibodies available is also needed to detail the differentiation stages of AML clones or immune interactions. In addition, the boutique workflow (sorting, scDNA-seq with antibody staining), which involves high specialization and long turnaround, would require streamlining to make the approach accessible for clinical diagnosis.
The proposed multiplex scMRD assay represents a promising peek into the future of MRD detection in AML. By combining single-cell genomic and immunophenotypic profiling, this approach offers unprecedented insights into disease persistence, clonal architecture, and therapeutic vulnerabilities. Notably, the approach can also delineate donor versus recipient cells after allogeneic transplant; potentially transplant failure causes such as relapse versus de novo AML from donor (which would affect treatment). The single-cell multimodal technologies may bring us closer to realizing the goal of durable remissions and improved patient outcomes. Collaborative efforts, further optimization, and large-scale clinical validation will undoubtedly propel us toward a future where the mysteries of MRD in AML are unraveled.
References
- 1.Heuser M., Freeman S. D., Ossenkoppele G. J., Buccisano F., Hourigan C. S., Ngai L. L., Tettero J. M., Bachas C., Baer C., Béné M. C., Bücklein V., Czyz A., Denys B., Dillon R., Feuring-Buske M., Guzman M. L., Haferlach T., Han L., Herzig J. K., Jorgensen J. L., Kern W., Konopleva M. Y., Lacombe F., Libura M., Majchrzak A., Maurillo L., Ofran Y., Philippe J., Plesa A., Preudhomme C., Ravandi F., Roumier C., Subklewe M., Thol F., van de Loosdrecht A. A., van der Reijden B. A., Venditti A., Wierzbowska A., Valk P. J. M., Wood B. L., Walter R. B., Thiede C., Döhner K., Roboz G. J., Cloos J., 2021 update on MRD in acute myeloid leukemia: A consensus document from the European LeukemiaNet MRD working party. Blood 138, 2753–2767 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson T., Bowman R. L., Persaud S., Liu Y., Gao Q., Zhang J.-P., Sun X., Sciambi A., Llanso A., Famulare C., Goldberg A. D., Dogan A., Roshal M., Levine R. L., Xiao W., Single cell genotypic and phenotypic analysis of measurable residual disease in acute myeloid leukemia. Sci. Adv. 9, eadg0488 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stasik S., Burkhard-Meier C., Kramer M., Middeke J. M., Oelschlaegel U., Sockel K., Ehninger G., Serve H., Müller-Tidow C., Baldus C. D., Röllig C., Bornhäuser M., Platzbecker U., Thiede C., Deep sequencing in CD34+ cells from peripheral blood enables sensitive detection of measurable residual disease in AML. Blood Adv. 6, 3294–3303 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]