Highlights
-
•
EV-71 infection induces intracellular HNRNPA2B1 expression, suggesting that HNRNPA2B1 is involved in EV-71 infection in SK-N-SH cells.
-
•
Redistribution of HNRNPA2B1 from the nucleus to the cytoplasm in SK-N-SH cells during EV-71 infection.
-
•
Down-regulation of HNRNPA2B1 inhibited EV-71 replication in SK-N-SH cells, indicating that HNRNPA2B1 is a key factor for EV-71 infection.
Keywords: HFMD, SK-N-SH cells, EV-71, HNRNPA2B1
Abstract
Objective
To investigate the effect of heterogeneous nuclear ribonucleoprotein A2B1 (HNRNPA2B1) on the replication of enterovirus 71 (EV-71) in SK-N-SH cells.
Methods
The mRNA and protein expression of HNRNPA2B1 in SK-N-SH cells were detected by real-time quantitative PCR (qRT-PCR) and western blotting (WB), respectively. WB was used to detect HNRNPA2B1 protein expression in the nucleus and cytosol. The localization of HNRNPA2B1 protein in the nucleus and cytosol was detected by immunofluorescence (IF). The expression of HNRNPA2B1 was inhibited by small interfering RNA (si-HNRNPA2B1). Viral RNA, viral structural protein VP1, and viral titer were detected by qRT-PCR, WB, and viral dilution counting, respectively.
Results
EV-71 infection significantly upregulates the expression of HNRNPA2B1 in SK-N-SH cells. EV-71 infection promotes HNRNPA2B1 nucleus-cytoplasm redistribution. Down-regulation of HNRNPA2B1 expression significantly inhibited EV-71 replication.
Conclusion
HNRNPA2B1 protein redistributed from nucleus to cytoplasm and is highly expressed in the cytoplasm during EV-71 infection. Inhibition of HNRNPA2B1 levels effectively inhibits EV-71 replication in SK-N-SH cells.
1. Introduction
Enterovirus 71 (EV-71) is one of the most important pathogens causing severe hand, foot and mouth disease (HFMD) in infants and children. Severe HFMD is considered a persistent global public health threat because of its aggressive illnesses and the difficulty of prevention and treatment (Bajaber and Ramanathan, 2021). There are no effective drugs to prevent and treat severe infections caused by EV-71, and drugs such as ribavirin and interferon are only used for reducing the symptoms of infection (Wang and Li, 2019). Therefore, it is particularly essential to search for more effective antiviral drugs to prevent and treat severe HFMD.
EV-71 is an envelope-free, single-positive-stranded RNA virus with a genome consisting of 7400 kb encoding a single polyprotein that is cleaved into four structural proteins (VP1, VP2, VP3, and VP4) and seven nonstructural proteins (2A, 2B, 2C, 3A, 3B, 3C and 3D) (Yi et al., 2011). The lifecycle of EV-71 consists of five steps: virus adsorption to host cells, entry into host cells, viral RNA replication, protein translation, and assembly and release of intact viral particles (Baggen et al., 2018). Studies have shown that host cell molecules can participate in the entire lifecycle of the virus to regulate viral infection (Jin et al., 2018).
Heterogeneous nuclear ribonucleoproteins (HNRNPs) are a class of RNA binding proteins (RBPs) with a highly conserved structure. They are not only involved in gene transcription, post-transcriptional modification, and maturation of precursor mRNAs but are also associated with mRNA stability and nucleus-to-cytoplasm transport (Kim et al., 2000). The HNRNPs protein family consists of 20 major HNRNPs proteins and some minor HNRNPs proteins (Geuens et al., 2016; Will and Lührmann, 2011). HNRNPs control viral infection by interacting directly with viral RNA or indirectly with host molecules and appear to be a potential class of antiviral targets (Wang et al., 2014; Fan et al., 2015; Pingale et al., 2020). Recently, it has been reported that HNRNPs are involved in enterovirus infection, including HNRNPK, HNRNPC, and HNRNPA1 (Shih et al., 2011). HNRNPA2B1, a core member of the HNRNPs family, is usually located in the nucleus and can undergo a nucleoplasmic shuttling to the cytoplasm under cellular stress. Despite the importance of HNRNPA2B1 during DNA and RNA virus infections (Humphries and Fitzgerald, 2019; Wang et al., 2019; Li and Nagy, 2011), the role of HNRNPA2B1 in enterovirus EV-71 infection is still unknown. In order to understand the role of HNRNPA2B1 in EV-71 infection, we focus on the expression of HNRNPA2B1 in viral infection and the effect of inhibition of the HNRNPA2B1 on viral replication and infection. This paper will provide new perspectives for searching pathogenesis of EV-71 infection.
2. Materials and methods
2.1. Cells and virus
Human neuroblastoma cells (SK-N-SH cells) were purchased from the Shanghai Academy of Sciences cell bank. SK-N-SH cells were cultured in OPTI-DMEM complete medium (Gibco, USA) containing 10 % fetal bovine serum (FBS, Gibco, USA) at 37°C in a 5 % CO2 incubator. EV-71 virus (GenBank accession number
OP191657) was isolated from pharyngeal swab specimens of patients with hand, foot, and mouth disease, and human rhabdomyosarcoma cells (RD cells) were cultured in OPTI-DMEM medium containing 2 % FBS for viral amplification. The viral solution was stored at -80°C.
2.2. Main reagents
Protease inhibitor (PMSF, Beyotime, China); Cell RIPA lysate (Beyotime, China); Cell nuclear staining solution dihydrochloride (DAPI, Beyotime, China). Antibodies: anti-VP1 (gift from Zhenjiang First People's Hospital), anti-HNRNPA2B1 (Abcam, USA), anti-GAPDH (Abclonal, China), anti-histone (Abcam, USA), anti-tubulin (Abcam, USA); anti-HRP-IgG (Abcam, USA), anti-IgG H&L (Alexa Fluor® 594) fluorescent secondary antibody (Abcam, USA), Nuclear Plasma Isolation Kit (Beyotime, China), LipofectamineTM 2000 Transfection Reagent (Thermo Fisher Scientific, USA), si-HNRNPA2B1 (Ribobio, China).
Primer: EV-71: forward, 5’-GCTCTATAGGAGATAGTGTGAGTAGGG-3’, and reverse, 5’-ATGACTGCTCACCTGCGTGTT-3’. HNRNPA2B1: forward, 5’-GCTTAAGCTTTGAAACCACAGA-3’; and reverse, 5’-CTTGATCTTTTGCTTGCAGGAT-3’. The GAPDH primers were purchased from Shanghai Bioengineering Company.
2.3. qRT-PCR analysis
Trizol reagent was used to extract total RNA, and 1000 ng RNA was used for reverse transcription after determining the total RNA concentration. The EV-71 and HNRNPA2B1 RNA expression were detected by SYBR ® Green qPCR SuperMix Universal Kit (Thermo Fisher Scientific, USA), and the results were calculated using 2−ΔΔCt. GAPDH mRNA was used as an internal control for measuring cellular RNA expression.
2.4. WB analysis
Enhanced cell RIPA lysate containing PMSF (Beyotime, China) was applied for total protein extraction according the manufacture's protocol. Briefly, 100 μL lysate was added into cells per 24-well plate. After mixing for 10 s, the mixture was centrifuged at 12000 × g for 5 min. Protein samples were kept on ice throughout the process. Extracted proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene fluoride membranes (BioRad, USA) as previously described (Wu et al., 2019). The membranes were then blocked in tris-buffered saline containing Tween (TBST) containing 5 % non-fat dried milk for 1 h. Then, the membranes were incubated with primary antibodies overnight at 4°C. After 3 washes in TBST, membranes were incubated with secondary antibodies for 1 h at room temperature and then were washed for 3 times. Finally, the proteins signal was visualized by enhanced chemiluminescence (ECL) substrate.
2.5. IF assay
24-well culture plates were incubated overnight until cell density was approximately 50–60 %. Cells were washed 3 times with 1 × PBS for 15 min. 200 μL of 4 % paraformaldehyde was added to cells, fixed for 15 min and then washed 5 times with 1 × PBS for 15 min. 200 μL of 0.2 % Triton X-100 was added to each well for 20 min at room temperature. Anti-HNRNP2AB1 antibody was added to cells and incubated overnight in a wet box at 4°C. Cells were washed 5 times with 1 × PBS/T for 15 min. Fluorescent secondary antibody was added and incubated for 1 h at room temperature. 1 × PBS/T was used to wash 5 times for 15 min. Cytosolic dye DAPI was added into cells for 10 min away from light. 1 × PBS/T was used to wash 5 times for 15 min again. Neutral resin was used to block the slices. Fluorescent slices were visualized with a confocal laser scanning microscope.
2.6. Preparation of cytoplasmic and nuclear extracts
The cytoplasm and nucleus of SK-N-SH cells were extracted with a Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, China) following the manufacturer's instructions. Before using extraction reagent A and nucleoprotein extraction reagent, PMSF was added at a final concentration of 1 mM. Cells were detached with a cell scraper. After adding 200 μL of extraction reagent A, cells were shaked vigorously for 5 s, and then moved to ice for 10 min. After adding extraction reagent B, cells were shaked vigorously for 5 s, and then moved to ice for 1 min; Cell supernatant (cytoplasmic protein) was obtained after centrifuging at 12,000 g for 5 min at 4°C. The remaining samples at the bottom were added 50 μL cellular nucleoprotein extraction reagent. After being shaked vigorously for 20 s, the samples were placed on ice for 2 min. Supernatant (nucleoprotein) was obtained after centrifuging at 4°C 12,000 g for 10 min.
2.7. Virus titration
EV-71 virus was serially diluted 10 times in OPTI-DMEM medium to obtain 8 concentrations of virus solution (100-10−7). 104 of RD cells/well were inoculated in 96-well culture plates. When the cells were at a density around 60 %, cells was inoculated with 1 concentration of virus solution in turn according to the virus concentration gradient. Un-infected cells were used as a negative control. All cells were incubated for 1 h at 37°C with 5 % CO2. The virus solution was removed and the cells were washed 3 times with 1 × PBS. OPTI-DMEM with 2 % FBS medium was added to incubate continuously for 7 days. Well numbers for cytopathic lesions were observed and recorded daily. The viral titer was determined by measuring TCID50 in serial dilutions according to the Karber method.
2.8. RNA interference
Si-HNRNPA2B1 and control siRNA with scrambled sequence (si-NC) were purchased from Ribo Biotech (Guangzhou, China). LipofectamineTM 2000 (Thermo Fisher Scientific, USA) was used for si-RNAs transfection. Briefly, si-RNAs and LipofectamineTM 2000 were diluted in OPTI-MEM without FBS, and then mixed at room temperature for 20 min. Cells were incubated with the mixture in DMEM without FBS at 37°C with 5 % CO2. After 6 h of transfection, the culture media was replaced with fresh DMEM with 10 % FBS. Cells were cultured continuously for next experiments.
2.9. Statistical analysis
Each independent experiment was repeated at least three times. Statistical analysis was performed using GraphPad Prism 6.0 software. p < 0.05 was statistically significant.
3. Results
3.1. EV-71 infection may induce intracellular HNRNPA2B1 expression
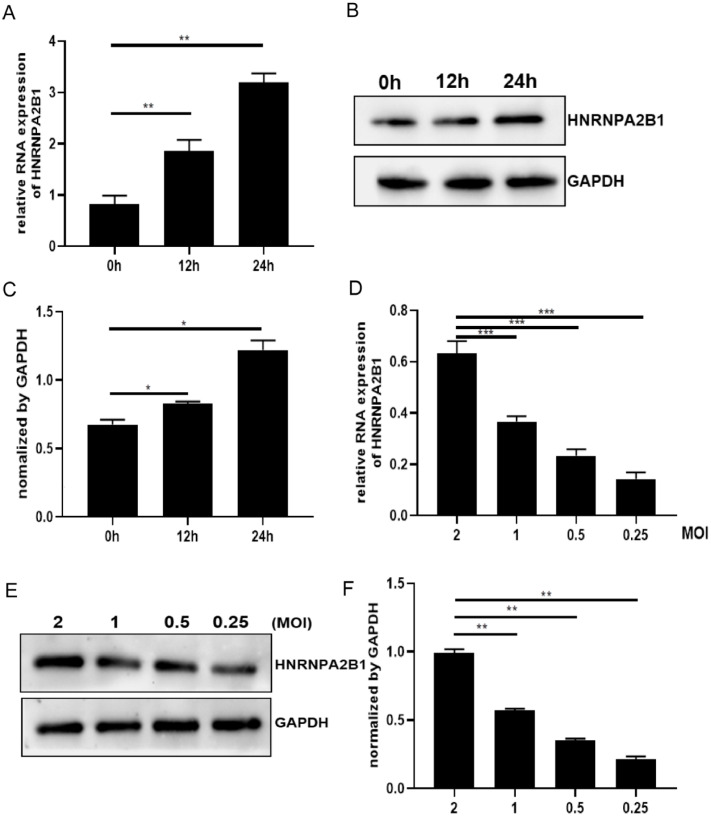
Cellular molecules are abnormally expressed in response to viral infection. To demonstrate the effect of EV-71 infection on HNRNPA2B1, we infected SK-N-SH cells with EV-71 virus and collected the cells at 0 h, 12 h and 24 h post-infection to assay the mRNA and protein of intracellular HNRNPA2B1. As shown in Fig. 1A–C, both HNRNPA2B1 mRNA levels and protein expression increased depend on virus-infected time (Fig. 1A–C). We also utilized different dilutions of the virus (2, 1, 0.5 and 0.25) to infect SK-N-SH cells, and then detected the effect of viral load on intracellular HNRNPA2B1. As shown in Fig. 1D–F, both HNRNPA2B1 mRNA levels and protein expression decreased with decreasing viral dilutions (Fig. 1D–F). The above results indicated that EV-71 infection may induce HNRNPA2B1 expression in SK-N-SH cells.
Fig. 1.
EV-71 infection may induce intracellular HNRNPA2B1 expression. (A-C) SK-N-SH cells were infected with EV-71 with MOI=2. Un-infected (0h) and EV-71-infected cells (12 and 24 h) were collected. (A) HNRNPA2B1 mRNA levels in SK-N-SH cells were detected by qRT-PCR analysis. (B) HNRNPA2B1 protein expression was detected by WB analysis. Due to HNRNPA2B1 is a nuclear protein, enhanced RIPA lysate was applied to achieve a more complete recovery of HNRNPA2B1 protein. (D–F) Cells were infected with different MOI (2, 1, 0.5, and 0.25) and were collected at 24 h post-infection time. (D) HNRNPA2B1 mRNA levels in SK-N-SH cells were detected by qRT-PCR analysis. (E) HNRNPA2B1 protein expression was detected by WB analysis. (C, F) Graphic representation of HNRNPA2B1 abundance after Image J analysis. Expression was normalized using the abundance of the endogenous protein GAPDH. Data are from three independent experiments with each experiment performed in triplicate. *p < 0.05, **p < 0.01, ***p < 0.001.
3.2. Redistribution of HNRNPA2B1 during EV-71 infection
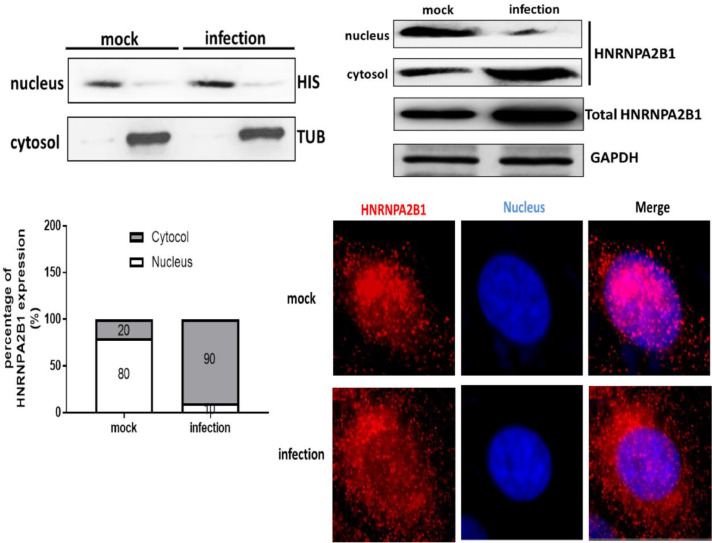
HNRNPA2B1 protein is mainly localized in the nucleus while the cells are not subjected to external stimuli. However in viral infection, the localization of HNRNPA2B1 can be altered from the nucleus to the cytoplasm. Several cell lines (RD cells and Hela cells) have been shown to alter the localization of HNRNPs in EV-71 infection. SK-N-SH cell line is a nerve cell line that is commonly used to study the pathogenesis of severe HFMD. We firstly isolated the nucleus and cytoplasm of un-infected and EV-71-infected cells, and detected the expression of cytoplasmic marker protein tubulin and nucleus marker protein histone to identify the isolation efficiency (Fig. 2A). Next, to demonstrate the effect of EV-71 infection on the redistribution of HNRNPA2B1, we examined the protein expression of HNRNPA2B1 in the cytoplasm and nucleus, respectively. As shown in Fig. 2B, the protein content of HNRNPA2B1 in the nucleus of un-infected cells was significantly higher than that in the EV-71-infected cells, while the protein content of HNRNPA2B1 in the cytoplasm of infected cells was significantly higher than that in the un-infected cells (Fig. 2B). Analysis using Image J software yielded that the protein content of HNRNPA2B1 in the cytoplasm of un-infected cells accounted for only 20 % of the total protein content of HNRNPA2B1, while in the cytoplasm of infected cells accounted for up to 90 % of the total protein content (Fig. 2C). Meanwhile, the IF assay results showed that HNRNPA2B1 was mainly accumulated in the nucleus in un-infected cells, and HNRNPA2B1 was significantly redistributed in the cytoplasm in EV-71-infected cells (Fig. 2D). The results in Fig. 2 demonstrated that HNRNPA2B1 is able to relocate in EV-71-infected nerve cells.
Fig. 2.
Redistribution of HNRNPA2B1 during EV-71 infection (A) Cytoplasmic protein (tubulin, TUB) and nucleoprotein (histone, HIS) were verified by WB analysis. (B) Un-infected and EV-71-infected cells were collected at 24 h post-infection time. HNRNPA2B1 protein expression was detected by WB analysis. (C) Image J analysis was performed to scan the protein abundance. The percentage of HNRNPA2B1 protein in the nucleus and cytoplasm in SK-N-SH cells was statistically analyzed. GAPDH was used as a control for qRT-PCR and WB analysis. (D) HNRNPA2B1 protein localization in SK-N-SH cells was detected by IF assay. Red signals, HNRNPA2B1. DAPI signals, nuclei. Scale bar = 5 μm. Data are from three independent experiments with each experiment performed in triplicate.
3.3. Down-regulation of HNRNPA2B1 inhibited EV-71 replication
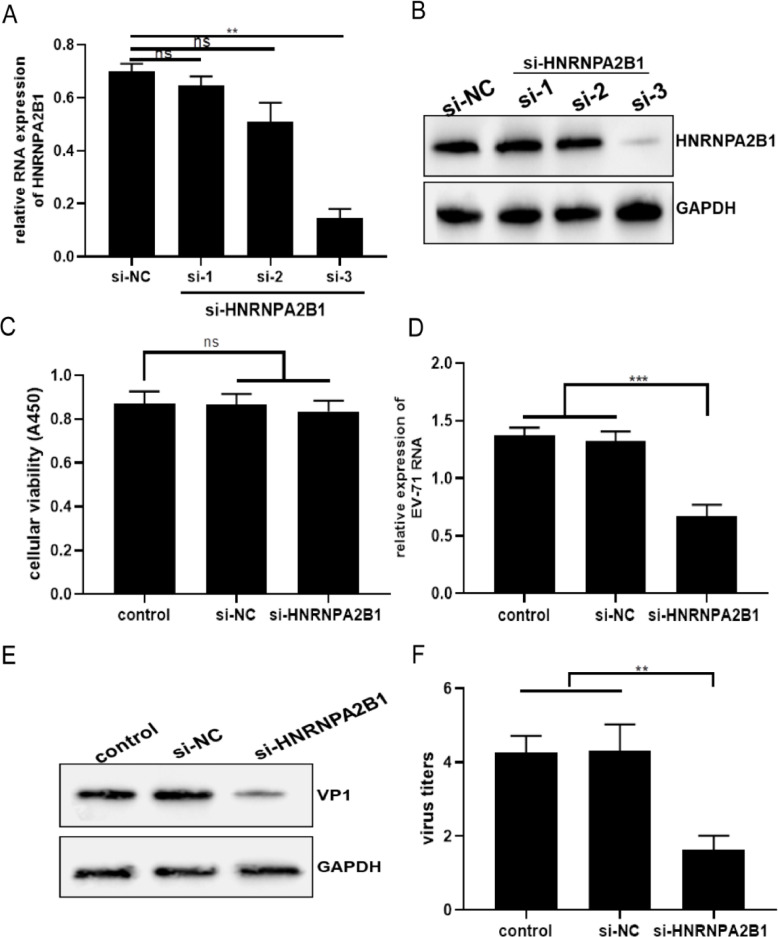
Three candidates si-HNRNPA2B1 (si-1, 2, 3) were designed and selected to examine the transfection effect of si-HNRNPA2B1 on the expression of endogenous HNRNPA2B1 in SK-N-SH cells. Cells were transfected with si-NC and si-HNRNPA2B1s by liposome for 48 h, and the intracellular HNRNPA2B1 expression were detected. As shown in Fig. 3A and B, compared with the control si-NC group, the HNRNPA2B1 mRNA levels and protein expression in the cells of the No. 3 candidate si-HNRNPA2B1 showed a significant decrease, indicating that No. 3 si-HNRNPA2B1 can successfully reduce the expression of endogenous HNRNPA2B1 (Fig. 3A and B). In addition, we performed the cellular toxicity assay to eliminate non-specific inhibition of viral replication due to the toxicity of HNRNPA2B1 knockdown (Fig. 3C). Next, we demonstrated the effect of down-regulation of HNRNPA2B1 expression on viral replication using No. 3 si-HNRNPA2B1. Viruses were added to cells transfected with si-NC and si-HNRNPA2B1, and cells were collected for detecting intracellular viral RNA and VP1 protein expression. Both viral RNA levels and VP1 protein expression were significantly reduced in cells of the si-HNRNPA2B1 group compared to the si-NC group (Fig. 3D and E). Subsequently, we verified the effect of down-regulation of HNRNPA2B1 on viral titers by detecting viral titers in cell supernatants. The viral titer in the supernatant of cells in the si-HNRNPA2B1 group was significantly lower than that in the si-NC group (Fig. 3E). These results suggested that down-regulation of HNRNPA2B1 expression in SK-N-SH cells can significantly reduce the replication of EV-71.
Fig. 3.
Down-regulation of HNRNPA2B1 inhibited EV-71 replication (A, B) SK-N-SH cells were transfected with si-NC and si-HNRNPA2B1 for 48 h. (A) HNRNPA2B1 mRNA levels in SK-N-SH cells were detected by qRT-PCR analysis. (B) HNRNPA2B1 protein expression in SK-N-SH cells was detected by WB analysis. (C) Cellular toxicity assay was performed using CCK8 kit. The cells of control group were treated with OPTI-DMEM. (D-F) After being transfected with si-NC and si-HNRNPA2B1, un-infected and EV-71-infected cell lysates were prepared for detecting HNRNPA2B1 mRNA levels and protein expression. Un-infected and EV-71-infected cell supernatant was prepared for detecting viral titers. (D) EV-71 RNA levels in SK-N-SH cells were detected by qRT-PCR analysis. (E) EV-71 VP1 protein expression in SK-N-SH cells was detected by WB analysis. (F) Measurement of viral titers. GAPDH was used as a control for qRT-PCR and WB analysis. Data are from three independent experiments with each experiment performed in triplicate. **p < 0.01, ***p < 0.001, ns=no significance.
4. Discussion
Understanding virus-cell interactions in virus multiplication is crucial for comprehending viral pathogenesis. Various RBPs are implicated in the replication of cytoplasmic picornaviruses (Flather and Semler, 2015, Fitzgerald et al., 2013). HNRNPI was the first host protein shown to interact with, and promote translation from, the IRES regions of picornaviruses (Hunt and Jackson, 1999). HNRNPK knockout inhibits dengue virus type 2 (DENV-2) multiplications, evidencing that HNRNPK is a host factor required for DENV-2 (Geuens et al., 2016). It has been reported that RBPs are involved in EV-71 infection. Poly (rC)-Binding Protein 2 (PCBP2) was been showed to be required for EV-71 translation (Sweeney et al., 2014). Both HNRNPA1 and far upstream element-binding protein 1 (FBP1) can interact with EV-71 IRES. Depletion of them resulted in reducing EV-71 gene translation (Lin et al., 2009, Huang et al., 2011). It is also evidenced that HNRNPA2B1 participated in viral replication and pathogenesis. For example, HNRNPA2B1 plays an inhibitory role in the replication of influenza A virus (IAV) in host cells (Wang et al., 2014). In this paper, we initially showed the high expression of the host protein HNRNPA2B1 in EV-71-infected SK-N-SH cells. Down-regulation of HNRNPA2B1 expression could inhibit EV-71 replication and infection in SK-N-SH cells, indicating that HNRNPA2B1 is involved in EV-71 infection in nerve cells.
High neurotropism makes EV-71 susceptible to neuronal progenitor cells and nerve cells, resulting in cell damage and the development of severe HFMD (Ong and Wong, 2015). HNRNPs reside usually in the nucleus and can undergo the redistribution from nucleus to cytoplasm under enterovirus infection. It has been reported that the HNRNPs can regulate the EV-71 transcription and translation from nucleus to cytoplasm (Levengood et al., 2013; Cammas et al., 2007). However, the redistribution phenomenon of HNRNPA2B1 in EV-71 infected nerve cells has not been clearly reported. In our study, we investigated and confirmed that EV-71 infection induced re-localization of HNRNPA2B1 protein from the nucleus to the cytoplasm in SK-N-SH cells. The redistribution in EV-71-infected nerve cells may be a novel mechanism of severe HFMD caused by EV-71.
Apart from affecting viral RNA replication, the redistribution of nucleoprotein influence cellular homeostasis in viral infection. HNRNPA2B1 protein is produced by alternative splicing of a single gene and has been shown to regulate RNA alternative splicing, RNA trafficking, and telomere maintenance. Down-regulation of HNRNPA2B1 inhibits EV-71 replication, possibly by directly blocking the binding of HNRNPA21B1 to viral genes, just as HNRNPA can directly bind to EV-71 5′-terminal noncoding region (5′UTR). Moreover, HNRNPA2B1 may indirectly regulate the other host protein, altering the accessibility of HNRNPA2B1 for host transcripts. Several recent articles have shown that HNRNPA2B1 could modulate N6-methyladenosine-dependent miRNA maturation processing and function in the trafficking of mRNA and miRNA (Liu and al., 2022; Zhao et al., 2020; Alarcon et al., 2015). What's interesting is that miRNAs have long been shown to play an important role in EV-71 infection (Wu et al., 2015). The above studies show an important and meaningful relationship between HNRNPA2B1, miRNAs and EV-71, which could be one of the deep mechanisms why inhibition of HNRNPA2B1 can reduce EV-71 replication.
The dynamic balance of the intracellular environment and the host-virus interaction must influence the entire process of EV-71 infection. Our study only provides preliminary evidence that HNRNPA2B1 is associated with EV-71 replication and infection, but the questions remain to be addressed. As a core member of HNRNPs, does HNRNPA2B1 also possess the ability to bind to the EV-71 viral 5′UTR or other components of the virus? From our research, we could hypothesis HNRNPA2B1 play a positive role in EV-71 infection. However, EV-71 3C protease with RNA-binding activity also can cleave host factors (Weng et al., 2009; Lei et al., 2013). Research showed HNRNPA1 is a novel target of EV-71 3C protease (Li et al., 2019). So, how can we prove and explain the complex relationship between HNRNPA2B1 and EV-71 in the future studies? It is worth further exploring the mechanism of HNRNPA2B1 in EV-71 infection.
In summary, our study is the first to report the involvement of HNRNPA2B1 in EV-71 replication in nerve cells. In EV-71-infected SK-N-SH cells, redistribution of HNRNPA2B1 significantly affected EV-71 replication. Inhibition intracellular HNRNPA2B1 levels markedly attenuated the replication ability of EV-71. The findings of this study lay the foundation for further exploration of the role of HNRNPA2B1 in EV-71 infection and offer more hints for studying the mechanism of severe HFMD.
Headings
-
•
EV-71 infection promoted HNRNPA2B1 redistribution from the nucleus to the cytoplasm in SK-N-SH cells.
-
•
Inhibition of HNRNPA2B1 reduced EV-71 replication in SK-N-SH cells.
CRediT authorship contribution statement
Jing Wu: Conceptualization, Visualization, Writing – original draft. Jian Lu: Conceptualization, Visualization. Lingxiang Mao: Formal analysis. Meiqin Xu: Formal analysis. Lu Dai: Writing – original draft. Yun Wang: Formal analysis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Data will be made available on request.
References
- Bajaber N., Ramanathan B. Methods in screening antiviral drugs against enterovirus 71. Methods Mol. Biol. 2021;2296:167–184. doi: 10.1007/978-1-0716-1358-0_9. [DOI] [PubMed] [Google Scholar]
- Wang H., Li Y. Recent progress on functional genomics research of enterovirus 71. Virol. Sin. 2019;34(1):9–21. doi: 10.1007/s12250-018-0071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L., et al. The virology and developments toward control of human enterovirus 71. Crit. Rev. Microbiol. 2011;37(4):313–327. doi: 10.3109/1040841X.2011.580723. [DOI] [PubMed] [Google Scholar]
- Baggen J., et al. The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Micro. 2018;16(6):368–381. doi: 10.1038/s41579-018-0005-4. [DOI] [PubMed] [Google Scholar]
- Jin Y., et al. Antiviral and inflammatory cellular signaling associated with enterovirus 71 infection. Viruses. 2018;10(4) doi: 10.3390/v10040155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., et al. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 2000;298(3):395–405. doi: 10.1006/jmbi.2000.3687. [DOI] [PubMed] [Google Scholar]
- Geuens T., Bouhy D., Timmerman V. The hnRNP family: insights into their role in health and disease. Hum. Genet. 2016;135(8):851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3(7) doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhou J., Du Y. hnRNP A2/B1 interacts with influenza a viral protein NS1 and inhibits virus replication potentially through suppressing NS1 RNA/protein levels and NS1 mRNA nuclear export. Virology. 2014;449:53–61. doi: 10.1016/j.virol.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B., et al. Heterogeneous ribonucleoprotein K (hnRNP K) binds miR-122, a mature liver-specific MicroRNA required for hepatitis C virus replication. Mol. Cell. Proteom. 2015;14(11):2878–2886. doi: 10.1074/mcp.M115.050344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingale K.D., Kanade G.D., Karpe Y.A. Heterogeneous nuclear ribonucleoproteins participate in hepatitis E virus replication. J. Mol. Biol. 2020;432(7):2369–2387. doi: 10.1016/j.jmb.2020.02.025. [DOI] [PubMed] [Google Scholar]
- Shih S.R., Stollar V., Li M.L. Host factors in enterovirus 71 replication. J. Virol. 2011;85(19):9658–9666. doi: 10.1128/JVI.05063-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries F., Fitzgerald K.A. hnRNPA2B1: fueling antiviral immunity from the nucleus. Mol. Cell. 2019;76(1):8–10. doi: 10.1016/j.molcel.2019.09.021. [DOI] [PubMed] [Google Scholar]
- Wang L., Wen M., Cao X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science. 2019;(6454):365. doi: 10.1126/science.aav0758. [DOI] [PubMed] [Google Scholar]
- Li Z., Nagy P.D. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol. 2011;8(2):305–315. doi: 10.4161/rna.8.2.15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., et al. Exosomal MicroRNA-155 inhibits enterovirus A71 infection by targeting PICALM. Int. J. Biol. Sci. 2019;15(13):2925–2935. doi: 10.7150/ijbs.36388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flather D., Semler B.L. Picornaviruses and nuclear functions: targeting a cellular compartment distinct from the replication site of a positive-strand RNA virus. Front. Microbiol. 2015;6:594. doi: 10.3389/fmicb.2015.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K.D., et al. Viral proteinase requirements for the nucleocytoplasmic relocalization of cellular splicing factor SRp20 during picornavirus infections. J. Virol. 2013;87(5):2390–2400. doi: 10.1128/JVI.02396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt S.L., Jackson R.J. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999;5(3):344–359. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney T.R., et al. The mechanism of translation initiation on type 1 picornavirus IRESs. EMBO J. 2014;33(1):76–92. doi: 10.1002/embj.201386124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.Y., et al. hnRNP A1 interacts with the 5′ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J. Virol. 2009;83(12):6106–6114. doi: 10.1128/JVI.02476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.N., et al. Far upstream element binding protein 1 binds the internal ribosomal entry site of enterovirus 71 and enhances viral translation and viral growth. Nucleic. Acids. Res. 2011;39(22):9633–9648. doi: 10.1093/nar/gkr682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong K.C., Wong K.T. Understanding enterovirus 71 neuropathogenesis and its impact on other neurotropic enteroviruses. Brain Pathol. 2015;25(5):614–624. doi: 10.1111/bpa.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levengood J.D., et al. High-affinity interaction of hnRNP A1 with conserved RNA structural elements is required for translation and replication of enterovirus 71. RNA Biol. 2013;10(7):1136–1145. doi: 10.4161/rna.25107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas A., et al. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol. Biol. Cell. 2007;18(12):5048–5059. doi: 10.1091/mbc.E07-06-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., et al. m(6)A reader hnRNPA2B1 drives multiple myeloma osteolytic bone disease. Theranostics. 2022;12(18):7760–7774. doi: 10.7150/thno.76852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., et al. m(6)A-binding proteins: the emerging crucial performers in epigenetics. J. Hematol. Oncol. 2020;13(1):35. doi: 10.1186/s13045-020-00872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon C.R., et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162(6):1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., et al. The role of microRNAs in enteroviral infections. Braz. J. Infect. Dis. 2015;19(5):510–516. doi: 10.1016/j.bjid.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng K.F., et al. Enterovirus 71 3C protease cleaves a novel target CstF-64 and inhibits cellular polyadenylation. PLoS Pathog. 2009;5(9) doi: 10.1371/journal.ppat.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., et al. Cleavage of interferon regulatory factor 7 by enterovirus 71 3C suppresses cellular responses. J. Virol. 2013;87(3):1690–1698. doi: 10.1128/JVI.01855-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.L., et al. EV71 3C protease induces apoptosis by cleavage of hnRNP A1 to promote apaf-1 translation. PLoS One. 2019;14(9) doi: 10.1371/journal.pone.0221048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.