Abstract
To overcome the instability of viral RNA, we carried out hepatitis C virus (HCV) RNA detection in dried serum spotted onto filter paper. The spotted serum samples were stored at room temperature and then processed for PCR assay at intervals of 1, 2, 3, and 4 weeks. The results showed that serum HCV RNA is stable in a dried condition, as it was detectable in spotted serum samples stored for 4 weeks at room temperature. Furthermore, although the HCV RNA titer showed an ∼10-fold reduction in virus yield in dried serum stored at room temperature for 4 weeks, the PCR results of frozen serum samples and dried serum samples matched completely. This storage method facilitates transport and analysis by nucleic acid amplification techniques even when freezing conditions are not available.
Hepatitis C virus (HCV) is a major etiological agent of posttransfusion and community-acquired hepatitis. Detection of circulating HCV antibody (anti-HCV) and PCR of HCV RNA are the two major techniques used to diagnose HCV infection. Since HCV RNA detection with reverse transcription (RT)-PCR is more direct and sensitive than anti-HCV testing, it has become one of the diagnostic standards for HCV infection. Presently, RT-PCR is the only reliable method that allows early detection of HCV infection. To assess a dried serum spot on filter paper as a source of material for HCV RNA detection, we investigated the stability of dried serum spots. This study describes the method of HCV RNA detection from a dried serum spot on filter paper.
Serum samples from eight patients who were seropositive for anti-HCV were used in this study. In addition, four serum samples from anti-HCV-negative patients were used for negative controls. A 50-μl aliquot of each serum sample was spotted directly onto filter paper (GF/C glass microfiber filter, 2.1 cm in diameter; Whatman Ltd., Kent, United Kingdom), which was put into a well of a flat-bottomed 24-well tissue culture plate (Nunc, Roskilde, Denmark). The serum spots were allowed to air dry and were then stored at room temperature (20 to 25°C). For recovery of RNA from these serum spots, the filters were suspended in 300 μl of an in-house lysis buffer (guanidinium thiocyanate-phenol) containing guanidinium thiocyanate buffer (4.2 M guanidinium thiocyanate, 0.5% Sarkosyl, and 25 mM Tris-HCl [pH 8.0]) and an equal volume of phenol, 150 mM sodium acetate (pH 5.2), and 0.35% 2-mercaptoethanol. The filters were incubated at 37°C for 30 min and then transferred to 1.5-ml microtubes, followed by precipitation with isopropanol. The resulting pellet was resuspended in 50 μl of RNase-free water, and then 10 μl of this RNA solution (= 10 μl of serum) was used as a template to detect HCV RNA. RT-PCR was performed by the one-step method combined with a cDNA synthesis reaction, followed by PCR in a single tube as described previously (1). That is, the first PCR was combined with the RT step in a single tube containing 40 μl of a reaction buffer made up as follows: 10 units of RNase inhibitor (Promega, Madison, Wis.), 100 units of Moloney murine leukemia virus reverse transcriptase (Gibco BRL, Tokyo, Japan), 100 ng of each outer primer, a 200 μM concentration of each of the four deoxynucleosides, 2 unit of AmpliTaq Gold DNA polymerase (Perkin-Elmer, Norwalk, Conn.), and 1× reaction buffer containing 1.5 mM MgCl2. To obtain an automatic hot-start reaction, we used AmpliTaq Gold DNA polymerase instead of regular thermostable DNA polymerase. The sequence of primers used for HCV PCR was designed from the 5′ untranslated region of the HCV genome (isolate HC-J1; GenBank accession no. D10749). The sequences of the HCV-specific primers were 5′-GCGACACTCCACCATAGAT-3′ (primer 19, sense) and 5′-GCTCATGGTGCACGGTCTA-3′ (primer 20, antisense) for the outer primer pairs (329 bases) and 5′-CTGTGAGGAACTACTGTCT-3′ (primer 21, sense) and 5′-ACTCGCAAGCACCCTATCA-3′ (primer 22, antisense) for the inner primer pairs (268 bases). The thermocycler was programmed first to incubate the samples for 50 min at 37°C for the initial RT step and then to preheat the samples at 95°C for 10 min to activate AmpliTaq Gold DNA polymerase. Subsequently, there followed 50 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 30 s with a Perkin-Elmer 2400 or 9700 thermal cycler. For the second reaction, 2 μl (1/20 volume) of the first PCR product was added to a tube containing the second set of each inner primer, deoxynucleosides, AmpliTaq Gold DNA polymerase, and PCR buffer, as in the first reaction, but without reverse transcriptase and the initial 50-min incubation at 37°C. Thermocycling for 50 cycles was done as above, but the annealing temperature was set at 60°C instead of 55°C for the second-round PCR. By this nested RT-PCR method, it was possible to detect one copy of HCV RNA (data not shown). The PCR products were detected by electrophoresis on 2% agarose gels stained with ethidium bromide and evaluated under UV light. The sizes of the PCR products were estimated according to the migration pattern of a 50-bp DNA ladder (Pharmacia Biotech, Piscataway, N.J.). All PCR assays were performed in duplicate to confirm specificity. The specificities of the amplification products from three serum samples, estimated to be 268 bases, were confirmed by purification with a QIAquick gel extraction kit (Qiagen Inc., Chatsworth, Calif.). The recovered PCR products were subjected to direct sequencing from both directions with the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer). Sequences of amplified cDNA were determined by a sequencer (ABI model 373A; Applied Biosystems, Foster City, Calif.).
Spotted serum samples were stored at room temperature and then processed for the PCR assay at intervals of 1, 2, 3, and 4 weeks. The results revealed that HCV RNA is stable in a dried condition, as it was detectable in spotted serum samples stored for 4 weeks at room temperature (Fig. 1). That is, HCV RNA was detected in all tested serum samples obtained from HCV-infected patients but not from uninfected patients. Furthermore, we determined the PCR titers of HCV RNA by 10-fold serial dilution of a frozen serum sample stored at −80°C until use; the same serum sample was then stored at room temperature for 4 weeks to investigate any decrease in sensitivity. The results revealed that there was a 10-fold reduction in virus yield in six of eight tested serum samples from anti-HCV-positive patients but no reduction in the remaining two samples (Fig. 2; Table 1). However, although the HCV RNA titer decreased in the serum spots, the PCR results of frozen serum samples and dried, spotted serum samples stored for 4 weeks matched completely in 12 tested samples (Fig. 3). The specificity of the PCR products of 268 bases from three patients in serum spots was confirmed to be the HCV genome by sequence analysis.
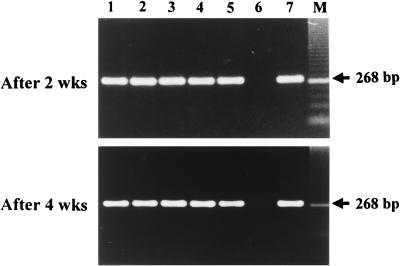
FIG. 1.
Detection of HCV RNA in dried serum spots on filter paper by nested RT-PCR. HCV RNA was detected in dried serum spots obtained at intervals of 2 and 4 weeks, respectively. Lanes 1 through 5 and 7 are samples from HCV-infected patients, and lane 6 is a sample from an uninfected patient.
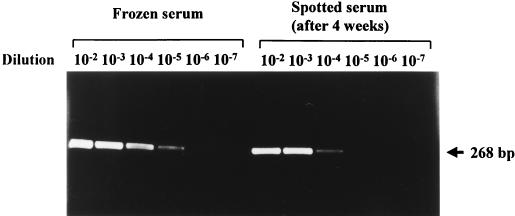
FIG. 2.
Comparison of HCV RNA titers in frozen serum and in the same serum spotted onto filter paper and stored at room temperature for 4 weeks. A 10-fold reduction in virus yield in spotted serum samples was noted.
TABLE 1.
Comparison of HCV RNA titers by nested RT-PCR of frozen serum samples and of the same serum samples spotted onto filter paper
| Sample | Relative titer in 100 μl of serum
|
|
|---|---|---|
| Frozen serum | Dried seruma | |
| 1 | 105 | 104 |
| 2 | 103 | 102 |
| 3 | 104 | 104 |
| 4 | 103 | 102 |
| 5 | 103 | 102 |
| 6 | 104 | 103 |
| 7 | 103 | 103 |
| 8 | 104 | 103 |
Serum spots were stored at room temperature for 4 weeks.
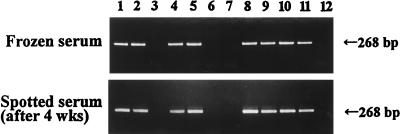
FIG. 3.
Comparison of detection rates of HCV RNA in frozen serum and dried serum spots. The PCR results between the two groups matched completely. Lanes 1, 2, 4, 5, 8, 9, 10 and 11 are samples from HCV-infected patients, and the other lanes are samples from uninfected patients.
Dried blood spots have been used for many years to screen for metabolic disorders such as phenylketonuria and sickle cell disease (3, 4). It has also been used for anonymous screening of newborns to assess the seroprevalence of human immunodeficiency virus (HIV) among childbearing women and for the diagnosis of perinatal HIV infections with HIV DNA (2, 5). From our preliminary results reported here, dried serum spotted on filter paper may be useful for HCV RNA detection by the RT-PCR technique, although potential limitations might cause false-negative results for samples with extremely low virus titers. This technique has the potential to facilitate large field studies by simplifying all aspects of sample collection, storage, and shipment. Furthermore, this method will be particularly valuable for small-volume samples and large, population-based studies in which cold storage and transportation present special problems, as is often the case in developing countries. In addition, the use of serum spots on filter paper as specimens is especially suitable for large-scale efforts, since the specimens can be mailed after collection at room temperature. However, further studies should be performed with a large number of clinical specimens under conditions that more closely simulate those of actual specimen transport before a final recommendation can be made concerning the optimal conditions for transport of blood samples used for HCV PCR assays.
Acknowledgments
We thank Takeshi Kurata, National Institute of Infectious Diseases, for continuous encouragement during this study.
This study was supported in part by a Grant-in-Aid for Science Research from the Ministry of Education, Science and Culture of Japan and the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Abe K, Edamoto Y, Park Y N, Nomura A M Y, Taltavull T C, Tani M, Thung S N. In situ detection of hepatitis B, C and G virus nucleic acids in human hepatocellular carcinoma tissues from different geographic regions. Hepatology. 1998;28:568–572. doi: 10.1002/hep.510280239. [DOI] [PubMed] [Google Scholar]
- 2.Fiscus S A, Brambilla D, Grosso L, Schock J, Cronin M. Quantitation of human immunodeficiency virus type 1 RNA in plasma by using blood dried on filter paper. J Clin Microbiol. 1998;36:258–260. doi: 10.1128/jcm.36.1.258-260.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrick M D, Dembure P, Guthrie R. Sickle-cell anemia and other hemoglobinopathies. N Engl J Med. 1973;288:1265–1268. doi: 10.1056/NEJM197306142882403. [DOI] [PubMed] [Google Scholar]
- 4.Guthrie R, Susi A. A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics. 1963;32:338–343. [PubMed] [Google Scholar]
- 5.Hoff R, Berardi V P, Weiblen B J, Mahoney-Trout L, Mitchell M L, Grady G F. Seroprevalence of human immunodeficiency virus among childbearing women. N Engl J Med. 1988;318:525–530. doi: 10.1056/NEJM198803033180901. [DOI] [PubMed] [Google Scholar]