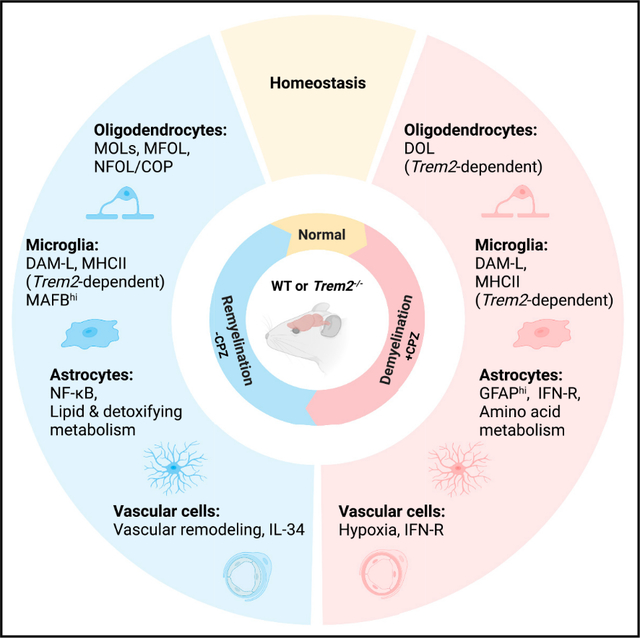
SUMMARY
Demyelination is a hallmark of multiple sclerosis, leukoencephalopathies, cerebral vasculopathies, and several neurodegenerative diseases. The cuprizone mouse model is widely used to simulate demyelination and remyelination occurring in these diseases. Here, we present a high-resolution single-nucleus RNA sequencing (snRNA-seq) analysis of gene expression changes across all brain cells in this model. We define demyelination-associated oligodendrocytes (DOLs) and remyelination-associated MAFBhi microglia, as well as astrocytes and vascular cells with signatures of altered metabolism, oxidative stress, and interferon response. Furthermore, snRNA-seq provides insights into how brain cell types connect and interact, defining complex circuitries that impact demyelination and remyelination. As an explicative example, perturbation of microglia caused by TREM2 deficiency indirectly impairs the induction of DOLs. Altogether, this study provides a rich resource for future studies investigating mechanisms underlying demyelinating diseases.
In brief
Hou et al. identify key transcriptional signatures of brain glial and vascular cells during demyelination and remyelination induced by cuprizone using single-nucleus RNA sequencing. They reveal cell-cell interactions that impact these processes and demonstrate that a population of demyelination-associated oligodendrocytes is dependent on a TREM2-mediated microglia response.
Graphical Abstract
INTRODUCTION
Myelin is a multilayered lipid sheath produced by oligodendrocytes (OLs) and Schwann cells that wrap around neuronal axons to facilitate the propagation of electrical impulses.1 In the central nervous system (CNS), white matter is enriched for myelinated axons, whereas neuronal somas are mostly located in the gray matter. Loss of myelin sheaths, a hallmark of multiple sclerosis (MS) as well as leukoencephalopathies and cerebral vasculopathies, results in axon degeneration and impairment of neuronal connectivity and functions.2–4 Damage of myelin integrity has also been reported in several neurodegenerative diseases, including Alzheimer’s disease.5 Recent single-cell transcriptional profiling studies have highlighted the heterogeneity of OLs and microglia responses in human MS and the mouse model of experimental autoimmune encephalomyelitis (EAE) caused by an autoimmune attack against myelin components.6–10 However, the transcriptomes of all brain cell types in response to myelin damage caused by non-autoimmune conditions is incompletely understood. Therefore, we analyzed global transcriptomic changes at the single-cell level in a commonly used mouse model of CNS demyelination that is induced by the toxic agent cuprizone (CPZ).11,12
CPZ is a copper chelator that disrupts mitochondrial functions.13 Oral administration of 0.2% CPZ in mice causes selective OL cell death and massive demyelination in the corpus callosum (CC) and the cerebral cortex; removal of CPZ from the diet is followed by progressive remyelination.14,15 OL death initiates within the first 2 weeks of CPZ treatment, inducing demyelination along with subsequent microgliosis and astrogliosis. Peak demyelination occurs around 4–5 weeks of CPZ treatment. Meanwhile, oligodendrocyte progenitor cells (OPCs) proliferate and differentiate, giving rise to newly formed OLs that generate new myelin sheaths.16 Both microglia and astrocytes participate in demyelination and remyelination processes: reactive microglia promote myelin debris clearance17 and subsequent remyelination,18 whereas astrocytes facilitate microglia recruitment to damaged myelin19 and secrete growth factors that promote OPC proliferation and differentiation.16 Analysis of microglia transcriptomes at the single-cell level in the CPZ model divulged a stereotypic response to demyelination characterized by proliferation and expression of genes indicative of activation, phagocytosis, and lipid metabolism that is dependent on the microglial receptors TREM2 and MERTK.20–22 Transcriptomic analysis also suggested predominant upregulation of the cholesterol-synthesis pathway in OLs during remyelination.23,24
Here, we deciphered the dynamic molecular responses of virtually all brain cell types to demyelination and remyelination utilizing single-nucleus RNA sequencing (snRNA-seq) and constructed a network of cell-cell interactions that may mediate these transcriptional changes. Moreover, we experimentally validated the effects of CPZ-induced demyelination and cellular responses in mice deficient in interleukin-33 (IL-33) signaling and TREM2, two key molecules mediating microglia functions.
RESULTS
CPZ-mediated myelin alterations induce global transcriptional changes
We fed 8-week-old male wild-type (WT) mice with 0.2% CPZ or regular chow and set up three experimental groups: (1) demyelination (5-week CPZ treatment), (2) remyelination (5-week CPZ treatment followed by 2-week regular chow), and (3) control (regular chow throughout the time course). Tissues from cerebral cortex and CC were collected after each treatment for snRNA-seq analysis (Figure 1A). Examination of CC by electron microscopy (EM) corroborated de- and remyelination (Figures S1A and S1B). In total, 58,079 nuclei from all three groups were analyzed after quality control (Figure S1C). Unsupervised clustering yielded 19 populations, annotated as various neuronal subsets, OLs, astrocytes, microglia, OPCs, and vascular cells, based on cluster marker genes and known cell-type markers25 (Figures 1B, 1C, S1D, and S1E; Table S1). CPZ treatment mostly affected the frequency of OL and microglia nuclei (Figures 1D and S1F). OL nuclei were drastically purged by CPZ treatment but fully recovered after CPZ withdrawal; microglia nuclei increased ~3-fold after demyelination and remained elevated throughout remyelination (Figures 1D and S1F). Microglia and OLs also bore the most differentially expressed genes (DEGs) during both de- and remyelination (Figure 1E and Table S2). The frequency of astrocyte nuclei slightly dipped during demyelination but significantly rose during remyelination (Figure 1D), whereas DEGs were substantial during both de- and remyelination (Figure 1E). A marked number of DEGs was also noted in vascular cells and OPCs during de- and remyelination, although their nuclei frequencies did not obviously change (Figures 1D and 1E). These results suggest that CPZ predominantly affects brain glial and vascular cells.
Figure 1. CPZ-mediated myelin alterations induce global transcriptional changes.
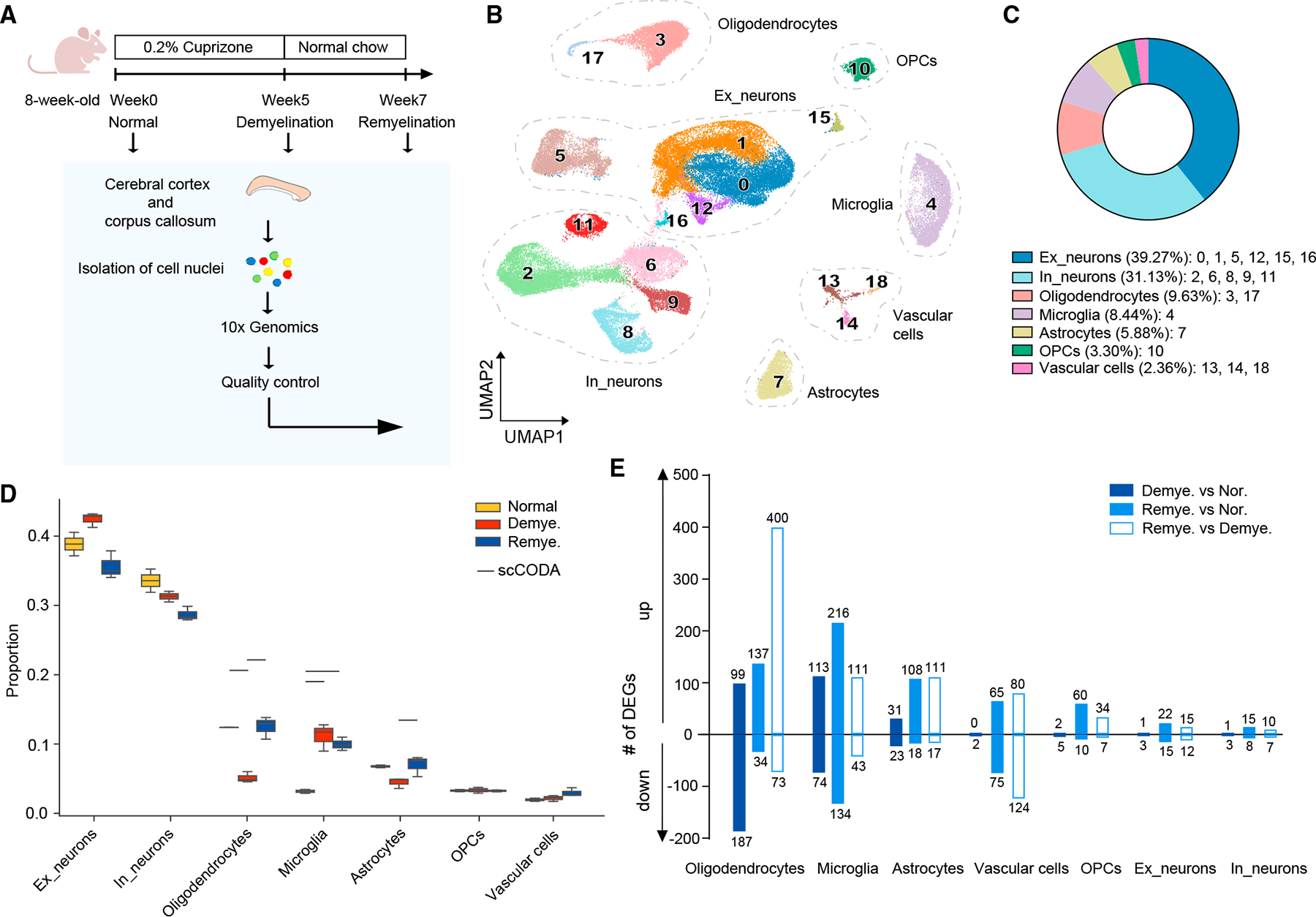
(A) Schematic diagram of the experimental strategy.
(B) UMAP plot of 58,079 nuclei showing 19 distinguished clusters with cell-type identities determined by expression of specific markers. n = 2 mice in normal, n = 3 in demyelination, and n = 3 in remyelination condition.
(C) Pie chart showing the frequency of each cell type across all conditions.
(D) Cell-type compositional analysis across different conditions. Statistically credible changes, as tested by scCODA, are denoted with bars on top. n = 2 mice innormal, n = 3 in demyelination, and n = 3 in remyelination condition.
(E) Number of differentially expressed genes (DEGs) in each cell type by pairwise comparisons between demyelination vs. normal (Demye. vs. Nor.), remyelinationvs. normal (Remye. vs. Nor.), or remyelination vs. demyelination (Remye. vs. Demye.). Log2(fold change) > 0.5, adjusted p < 0.05, non-parametric Wilcoxon rank-sum test, Bonferroni correction.
Demyelination induces a subset of SERPINA3N+ oligodendrocytes
To further delineate the impact of CPZ on oligodendrocyte lineage transcriptomes, we reclustered oligodendrocyte lineage cells into eight subclusters that were defined based on published markers as mature oligodendrocytes (MOLs, clusters 0, 2, 4), myelin-forming oligodendrocytes (MFOLs, cluster 5), newly formed oligodendrocytes/differentiation-committed oligodendrocytes (NFOL/COPs, cluster 6), OPCs (cluster 1), proliferating OPCs (POPCs, cluster 7), and demyelination-associated oligodendrocytes (DOLs, cluster 3)26 (Figures 2A and S2A). Demyelination was associated with a marked reduction of MOLs and MFOLs (Figures 2B and 2C); these cells expressed genes encoding components of actin filaments (Anln, Gsn, and Stmn4), cholesterol-synthesis enzymes (Hmgcs1), prostaglandin D2 synthase (Ptgds), Il33, and myelin components, such as Plp1, Mal, Cldn11, and Mog (Figures 2D and 2E). PTGDS has been shown to promote myelination in the peripheral nervous system in vivo,27 whereas the role of IL-33 in CNS remyelination remains controversial.28,29 Oligodendrocytes remaining after demyelination formed the DOL cluster (Figures 2B and 2C); these cells expressed genes indicative of proteolytic inhibition (Serpina3n),30 senescence and death (Cdkn1a), and anti-oxidative response (Moxd1) (Figures 2D and 2E). DOLs also expressed growth differentiation factor 15 (Gdf15), a neurotrophic factor of the transforming growth factor β superfamily elevated in the serum of patients with MS,31,32 Apoe and Abca1 that mediate cholesterol transport, and Col5a3 that drives the deposition of fibrillar collagen and extracellular matrix (ECM)33 (Figures 2D and 2E). In parallel with decline of MOLs/MFOLs and expansion of DOLs during demyelination, we observed a slightly compensatory increase of OPCs and NFOL/COPs (Figure 2C). We validated the appearance of SERPINA3N+ DOLs during both demyelination and remyelination by co-staining the medial CC for SERPINA3N and the mature oligodendrocyte marker CC1 (Figures 2F and 2G); we corroborated the expansion of OPCs during demyelination by immunostaining the lateral CC for NG2 (Figures S2B and S2C).
Figure 2. Demyelination induces SERPINA3N+ oligodendrocytes.
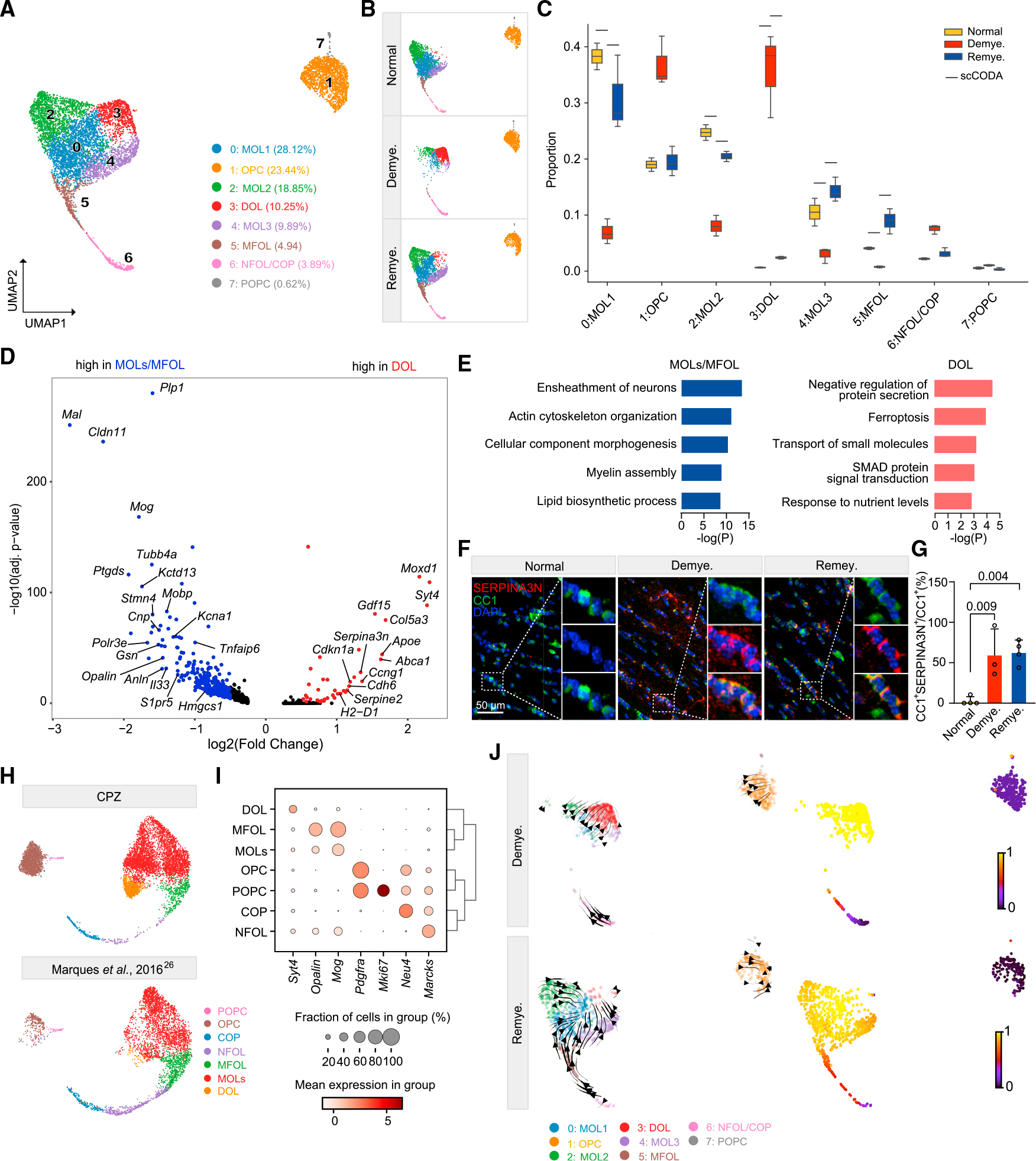
(A) UMAP plot of reclustered oligodendrocyte lineage nuclei identifying eight subclusters. n = 7,251 oligodendrocyte lineage nuclei.
(B) UMAP plot of oligodendrocyte subclusters split by condition. n = 2,572 nuclei in normal, n = 1,839 nuclei in demyelination, and n = 2,840 nuclei in remyelination.
(C) Subcluster compositional analysis across different conditions. Statistically credible changes, as tested by scCODA, are denoted with bars on top. n = 2 mice in normal, n = 3 in demyelination, and n = 3 in remyelination condition.
(D) Volcano plot showing significant DEGs in DOLs (subcluster 3) vs. MOLs/MFOLs (subclusters 0, 2, 4, 5). Log2(fold change) > 0.5, adjusted p < 0.05, non-parametric Wilcoxon rank-sum test, Bonferroni correction.
(E) Pathways enriched in MOLs/MFOLs (left panel) and DOLs (right panel). p value by hypergeometric test.
(F) Representative immunofluorescence (IF) images of SERPINA3N and CC1 staining in the medial corpus callosum (MCC) in each condition. CC1, green;SERPINA3N, red; DAPI, blue. Scale bar, 50 μm.
(G) Quantification of SERPINA3N+CC1+/CC1+ ratio across different conditions. p value by one-way ANOVA with Tukey’s multiple comparisons test. Data are presented as mean ± SD. n = 3 mice/condition.
(H) Integration of oligodendrocyte lineage cells from Marques et al.26 and the current CPZ dataset. UMAP plot split by datasets.
(I) Dot plot of marker genes enriched in each subpopulation of the integrated data in (H).
(J) UMAP plot of oligodendrocyte lineage cells overlaid with the RNA velocity streamlines derived from the basic model, colored by annotated subcluster (left).
Corresponding velocity pseudo-time shown on the right. One sample per condition is shown.
See also Figures S2 and S3; Table S2.
Withdrawal of CPZ and remyelination led to reconstitution of MOLs and MFOLs paralleled by reduction of DOLs (Figure 2C). Correspondingly, most of the genes down- and upregulated during demyelination returned to homeostatic expression levels (Figure S2D). A few genes rose throughout de- and remyelination, including pleiotrophin (Ptn), a secreted growth factor that functions as a neuromodulatory peptide in neurodevelopment34 and promotes remyelination35 (Figure S2D). During remyelination, OPCs expressed elevated levels of transcription factors such as Olig1 and Sox10 that drive OL development and maturation,36,37 suggesting that remyelination and developmental myelination follow a similar trajectory (Figure S2E).
To corroborate this conclusion, we integrated oligodendrocyte lineage cells in our CPZ model with those from mouse juvenile and adult CNS.26 Results showed that OLs in the CPZ model followed a trajectory from OPCs to MOLs similar to that occurring in development (Figures 2H and 2I). In addition, DOLs were predominantly enriched in the CPZ model (Figure 2H). To further determine the trajectory toward DOLs, we calculated RNA velocity in demyelination and remyelination, which was projected onto the existing uniform manifold approximation and projection (UMAP).38 The results showed movement of MOLs toward DOLs during demyelination as well as movement from NFOL/COPs toward MOLs during remyelination (Figure 2J). The trajectories were further corroborated by velocity pseudo-time calculation (Figure 2J).
To compare oligodendrocytes in the CPZ model with other neurodegenerative disease models, we integrated our CPZ oligodendrocytes with those in the 5XFAD model of Alzheimer’s disease (AD),39 which revealed two distinct DOL subpopulations (DOL1 and DOL2) (Figures S2F and S2G). Expressing Serpina3n and C4b, DOL1 appeared during both de- and remyelination as well as in the 5XFAD model (Figures S2H–S2J). DOL2 was more demyelination specific and highly expressed stress-response genes such as Cdkn1a, Gdf15, and Moxd1 (Figures S2H–S2J). Moreover, a subset of interferon-responsive (IFN-R) oligodendrocytes was present in both models (Figures S2H and S2I).
Finally, we compared CPZ oligodendrocyte signatures with those in human MS. We selected genes upregulated in pairwise comparisons between demyelination vs. normal or remyelination vs. demyelination in CPZ; these gene sets were compared with genes upregulated in various pathological regions of progressive patients with MS vs. controls from two public datasets,6,8 including normal-appearing white matter, demyelinated lesion core, active, chronic active (CA), chronic inactive, remyelinated lesions, and periplaque regions. We found that limited, yet crucial, genes were shared between human MS oligodendrocyte and mouse CPZ signatures (Figures S2K–S2N; Tables S2 and S3). The cholesterol-synthesis gene HMGCS1 was upregulated in MS tissues in both human datasets (Figures S2K, S2M, S2O, and S2P) as well as during remyelination following CPZ in mice (Figures S2K and S2M). Co-staining of the medial CC for HMGCS1 and the mature oligodendrocyte marker CA2 validated the significant reduction of HMGCS1+ oligodendrocytes during demyelination, followed by a recovery during remyelination (Figures S2Q–S2S). Genes encoding components involved in formation of and myelin itself, such as PLP1 and CNP, were also enriched during remyelination in the CPZ model and in several pathological MS states, including active and CA lesions in one dataset (Figure S2P). This implies that demyelination in MS is paralleled by a compensatory process of remyelination.
IL-33-ST2 pathway attenuates demyelination
Patients with MS reportedly have increased levels of IL-33 in the serum and cerebrospinal fluid.40,41 IL-33 was thought to be predominantly released by astrocytes,42,43 especially in humans.40,41 However, our snRNA-seq dataset revealed predominant expression of Il33 mRNA in oligodendrocytes, which dipped to the lowest level during demyelination (Figures S3A and S3B). We validated nuclear IL-33 protein in oligodendrocytes at steady state and following remyelination, whereas it was absent during demyelination (Figures S3C and S3D). These results suggest that IL-33 is stored in oligodendrocyte nuclei in steady state, released into the extracellular space upon their death, and re-constituted de novo in the nuclei of new oligodendrocytes. We also detected scarce Il33 expression in astrocytes, which did not change across various conditions either in our snRNA-seq data (Figures S3E and S3F) or at the protein level (Figures S3G and S3H).
Since IL-33 is known to elicit repair mechanisms in spinal cord injury by acting on microglia and astrocytes,42 we further examined the impact of IL-33 signaling on CPZ-induced demyelination in mice lacking the IL-33 cognate receptor ST2 encoded by Il1rl1 (Figure S3I). Microglia expressed ST2 in steady state, but this waned after 2 weeks of CPZ treatment (Figurse S3J and S3K). Luxol fast blue staining of the CC revealed more pronounced demyelination in Il1rl1−/− than in WT mice after 2 weeks of CPZ treatment, but comparable demyelination after 5 weeks of CPZ treatment (Figures S3L and S3M). EM confirmed a lower percentage of myelinated axons in Il1rl1−/− mice after 2 weeks of CPZ treatment (Figures S3N and S3O). Exacerbated demyelination in Il1rl1−/− mice was associated with heightened microglia activation, measured by expression of apolipoprotein E (APOE) in microglia on a per-cell basis (Figures S3P–S3R), whereas GFAP+ activated astrocytes were similar in Il1rl1−/− and WT mice (Figures S3S and S3T). These data suggest that IL-33-ST2 interaction attenuates CPZ-induced demyelination at an early stage.
Astrocytes adopt distinct metabolic and stress-response signatures in demyelination and remyelination
Although astrocyte abundance was minimally affected by CPZ (Figures 1D and 3A), marked transcriptional changes were evident during de- and remyelination (Figure 1E). Demyelination induced upregulation of the classical activation genes Gfap and vimentin (Vim) together with Serpina3n and C4b (Figures 3B and S4A). Upregulated genes also encoded the heterodimeric amino acid transporters SLC7A5/SLC3A2 and SLC7A11/SLC3A2. The SLC7A5/SLC3A2 complex imports essential amino acids that promote mammalian target of rapamycin (mTOR) activation.44 The SLC7A11/SLC3A2 complex mediates the exchange of extracellular L-cystine and intracellular L-glutamate across the plasma membrane.45 Cystine intake facilitates synthesis and release of glutathione that attenuates oxidative stress and ferroptosis.46,47 Epoxide hydrolase (EphX1), which has been associated with anti-oxidant activity and senescence,48 was also upregulated (Figures 3B and S4A). Upregulation of Stat3 suggested potential involvement of this signaling pathway in the astrocyte response. Additionally, astrocytes upregulated IFN-R genes such as H2-d1 and Ifi27 during demyelination (Figures 3B and S4A). Pathway analysis of DEGs corroborated the involvement of mTOR, anti-oxidative, anti-apoptotic, STAT3, and IFN pathways (Figure S4B). During remyelination, astrocytes adopted a distinct signature marked by detoxifying metabolic pathways and activation of nuclear factor κB (NF-κB) (Irak2, Nfkbia) (Figures 3C and S4C–S4E); the former included Slc39a14, a transporter of divalent metals that reduces their overload during inflammation,49 and glutamate dehydrogenase (Glud1), a major enzyme for glutamate detoxification.50 In addition, pathway analysis highlighted the synthesis of cholesterol and metabolism of fatty acids in astrocytes during remyelination (Figure S4F), which may be supplied to neurons and other cells.51
Figure 3. Astrocytes adopt distinct stress-response signatures in demyelination and remyelination.
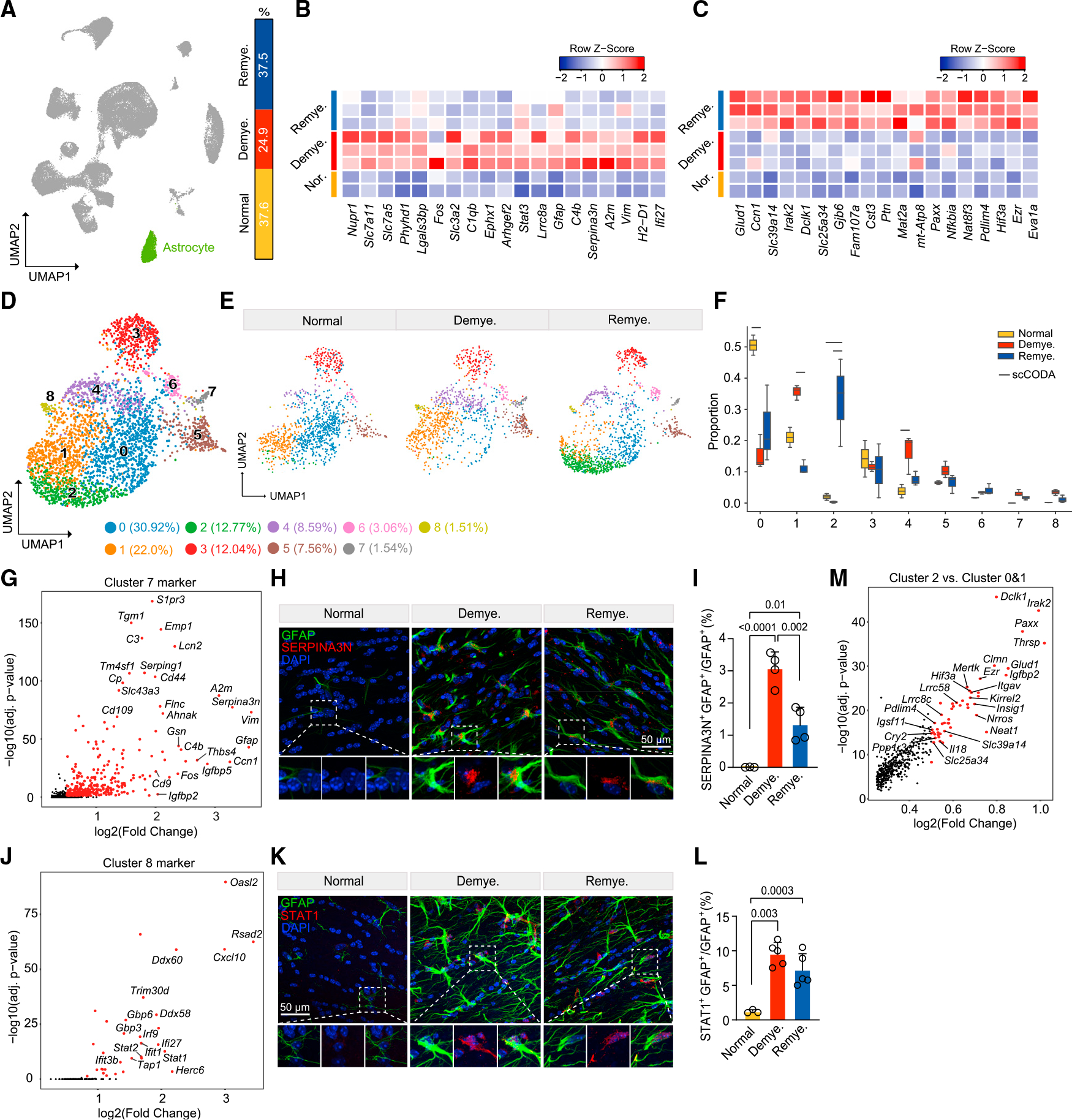
(A) UMAP plot showing the astrocyte cluster (cluster 7 in Figure 1B), and relative frequency across different conditions.
(B and C) Heatmap showing average expression of top astrocyte DEGs upregulated in demyelination vs. normal (B) and in remyelination vs. demyelination (C). Log2(fold change) > 0.5, adjusted p < 0.05, non-parametric Wilcoxon rank-sum test, Bonferroni correction. Each column represents one sample. Color scheme represents row Z score.
(D) UMAP plot of reclustered astrocytes identifying nine subclusters. n = 3,305 astrocyte nuclei.
(E) UMAP plot of astrocyte subclusters split by condition. n = 1,114 astrocyte nuclei in normal, n = 986 in demyelination, and n = 1,205 in remyelination.
(F) Subcluster compositional analysis across different conditions. Statistically credible changes, as tested by scCODA, are denoted with bars on top. n = 2 mice in normal, n = 3 in demyelination, and n = 3 in remyelination condition.
(G) Volcano plot showing subcluster 7 marker genes.
(H) Representative IF images showing colocalization of SERPINA3N and GFAP in lateral CC in each condition. SERPINA3N, red; GFAP, green; DAPI, blue. Scalebar, 50 μm.
(I) Quantification of SERPINA3N+GFAP+/GFAP+ ratio across different conditions. Data are presented as mean ± SD. n = 3–4 mice/condition.
(J) Volcano plot showing subcluster 8 marker genes.
(K) Representative IF images showing colocalization of STAT1 and GFAP in LCC in each condition. STAT1, red; GFAP, green; DAPI, blue. Scale bar, 50 μm.
(L) Quantification of STAT1+GFAP+/GFAP+ ratio across different conditions. Data are presented as mean ± SD. n = 3–5 mice/condition.
(M) Volcano plot showing DEGs of subcluster 2 vs. subclusters 0 and 1.
Adjusted p values by non-parametric Wilcoxon rank-sum test, Bonferroni correction (G, J, and M). p values by one-way ANOVA with Tukey’s multiple comparisons test (I and L). See also Figure S4 and Table S2.
To decipher whether stress-induced responses involved all astrocytes or distinct subsets, astrocyte nuclei were further subclustered into nine subsets (Figure 3D). At steady state, homeostatic astrocytes were heterogeneous: in addition to clusters 0 and 1 that expressed pan-astrocyte genes, cluster 3 expressed genes controlling ECM production, including Agt, Sparc, Igsf1, Itih3, Spon1, and Slc6a11.52 Cluster 4 was enriched for mitochondrial genes, indicating dying cells induced by CPZ or tissue processing. Cluster 5 expressed the synaptogenic gene Thbs4 and the neurogenesis-related gene Igfbp5, which may reflect astrocytes in the subventricular zone niche.53 Cluster 6 expressed high levels of Gfap, as well as Myoc, Nrp2, Disp3, and Serpinf1 that may have neuroprotective functions54 (Figure S4G). In demyelination, stress-response and mTOR pathway genes (Ephx1, Slc7a11, Slc7a5, and Slc3a2) were upregulated in all subsets, whereas classical astrocyte activation genes and IFN-R genes were enriched in two distinctive subsets, clusters 7 and 8, which were absent in steady state (Figures 3E and 3F). Cluster 7 was featured by Serpina3n, Gfap, Vim, and C3 (Figure 3G); immunostaining of lateral CC confirmed the expression of SERPINA3N, VIM, and GFAP (Figures 3H, 3I, S4H, and S4I). Cluster 8 expressed IFN-R genes (Figure 3J) and was distinguished by STAT1 accumulation within GFAP+ astrocytes in CC from mice undergoing both de- and remyelination (Figures 3K and 3L). During remyelination, abundance of clusters 7 and 8 abated (Figures 3E and 3F). In addition, homeostatic Gfap− astrocyte clusters 0 and 1 were not restored, but rather replaced by cluster 2 expressing NF-κB-related genes (Irak2 and Il18), circadian clock genes (Cry2), genes controlling astrocyte adhesion (Ezr and Igsf11), metabolism (Glud1, Nrros, and Slc39a14), and stress response (Paxx and Neat1) (Figures 3F and 3M). Together, these data demonstrate that astrocytes follow stress- or IFN-response trajectories during demyelination while acquiring a distinct response state demarcated by detoxifying metabolic pathways and activation of the NF-κB pathway during remyelination.
To compare astrocyte signatures in the CPZ model with those in human MS, we overlaid astrocyte DEGs of the CPZ model with featured genes of the “astrocytes inflamed in MS” characterized from MS CA lesions.8 We found that human MS shared GFAP, VIM, and NUPR1 with mouse demyelination and CST3, CD81, GJA1, CSRP1, and FAM107A with mouse remyelination (Figures S4J and S4K; Tables S2 and S3). Some of the shared genes, such as the stress-inducible transcription regulator NUPR1, suggested commonality of essential pathways controlling astrocyte reactive responses.
MAFBhi microglia emerge during remyelination
Microglia expanded during de- and remyelination and displayed abundant DEGs (Figures S5A–S5D). Subclustering of microglia identified nine subsets with distinct cluster markers (Figures 4A and S5E). In steady state, microglia were mainly composed of cluster 0 enriched for homeostatic genes (Figures 4B, 4C, and S5E). Demyelination induced the emergence of several microglia subsets (1, 2, 4–8) (Figures 4B and 4C). Cluster 2 (DAM-like, DAM-L) exhibited a composite signature similar to that of disease-associated microglia (DAM)55 (Figures 4D–4F) and white matter associated microglia (WAM)56 (Figures 4G–4I), demarcated by expression of Apoe, Lpl, and Spp1 (Table S3). However, variegated expression of DAM genes, exemplified by non-overlapping expression patterns of Ccl4, Itgax, Gpnmb, and Ch25h, suggested graded polarization and functional heterogeneity within this cluster (Figure 4J). Moreover, the DAM-L cluster highly expressed genes not previously defined as DAM genes, including the transcription factor Mitf, which has been reported to induce GPNMB in response to lysosomal stress57; and Plau, the urokinase plasminogen activator (uPA) gene that is associated with microglial migration58 and is upregulated in MS lesions59 (Figure 4K). Clusters 1 and 5 (transition microglia) bridged the gamut between homeostatic and DAM-L microglia (Figures S5F and S5G). Demyelination-induced clusters 6, 7, and 8 selectively expressed major histocompatibility complex (MHC) class II (Cd74 and H2-Aa), IFN-R (Ifi204 and Oasl2), and proliferation genes (Top2a and Mki67), respectively (Figure S5E) and evoked similar subsets reported in mouse models of AD.60,61 Co-staining markers for microglia (IBA1), DAM (CD11c), and MHC class II (CD74) corroborated the presence of these microglia subsets (Figures S5H and S5I). Another subset (cluster 4) was enriched for mitochondrial genes, which indicated stressed cells possibly induced by CPZ or tissue processing. CPZ withdrawal correlated with a decline in demyelination-induced clusters (1, 5, and 8), paralleled by reconstitution of homeostatic microglia (cluster 0) (Figures 4B and 4C). In addition, a remyelination-specific subset that selectively expressed Mafb (cluster 3) also emerged (Figures 4L and 4M). Since Mafb is required for maintenance of the microglia homeostatic signature,62 this subset may encompass microglia that transition from reactive states back to homeostasis after CPZ withdrawal. Remarkably, MAFB expression was enriched in microglia from CA lesions8 and acute lesions22 of human MS (Figures 4N and 4O; Table S3), implicating MAFB is crucial for driving microglia responses to myelin alterations. MAFBhi microglia were validated by immunostaining in the CPZ model (Figure 4P). Pseudo-time analysis of microglia corroborated that DAM-L, IFN-R, and MHCII trajectories persisted throughout de- and remyelination, whereas MAFBhi microglia peaked during remyelination (Figures S5J–S5L).
Figure 4. MAFBhi microglia emerge in remyelination.
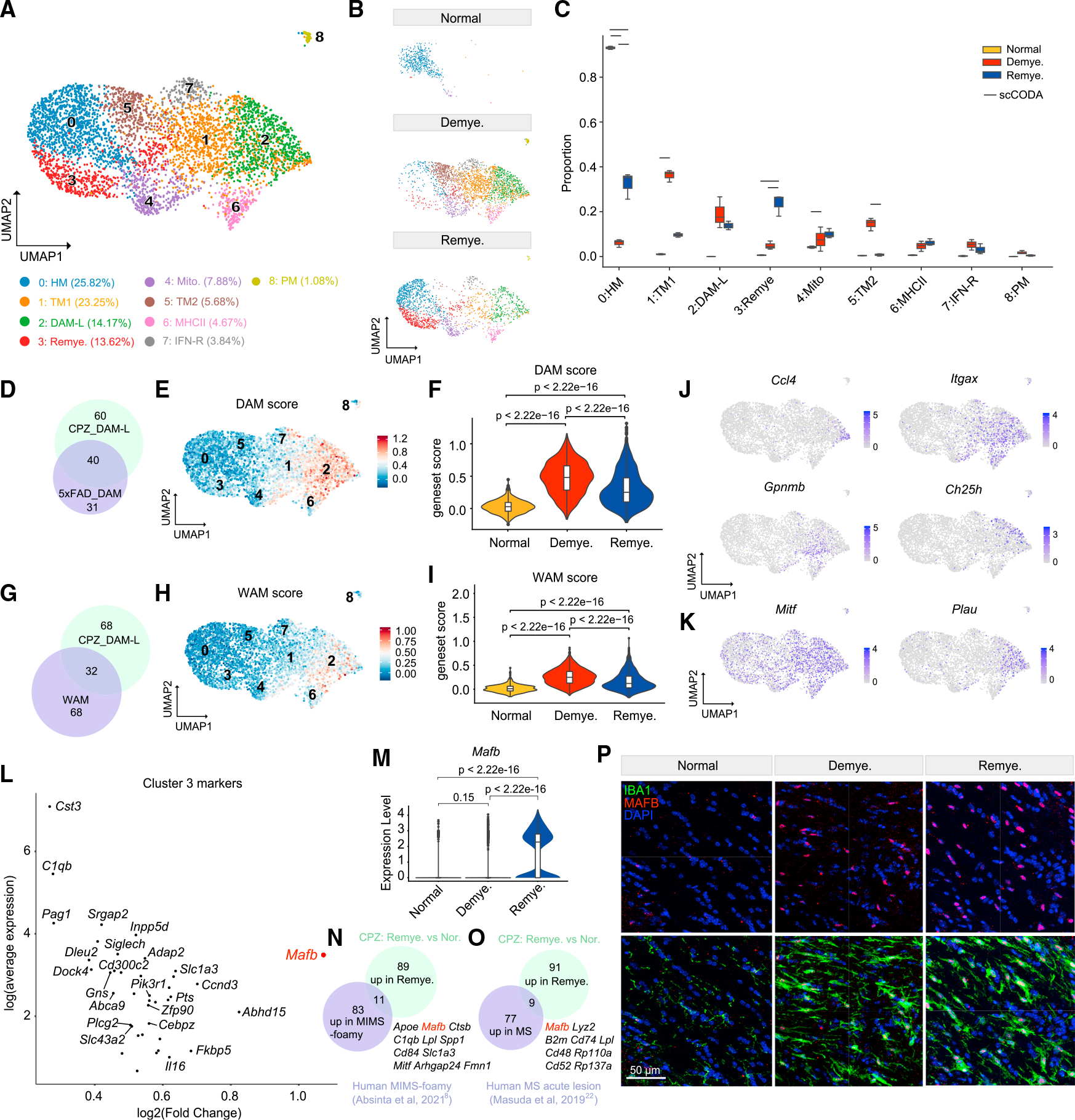
(A) UMAP plot of reclustered microglia identifying nine subclusters. n = 4,736 microglial nuclei.
(B) UMAP plot of microglia subclusters split by condition. n = 510 nuclei in normal, n = 2,450 nuclei in demyelination, and n = 1,776 nuclei in remyelination.
(C) Subcluster compositional analysis across different conditions. Statistically credible changes, as tested by scCODA, are denoted with bars on top. n = 2 mice in normal, n = 3 in demyelination, and n = 3 in remyelination condition.
(D) Venn diagram depicting overlap between the top 100 markers of microglia subcluster 2 (DAM-L) and the top 71 signature genes of DAM identified in the 5XFADmodel.55 Gene sets used are listed in Table S3.
(E) UMAP plot showing the DAM signature score calculated using the top 71 signature genes of DAM.
(F) Violin plot showing DAM gene set score in each condition. p value by Wilcoxon test.
(G) Venn diagram depicting overlap between the top 100 markers of microglia subcluster 2 (DAM-L) and the top 100 signature genes of white matter associatedmicroglia (WAM) from Safaiyan et al.56 Gene sets used are listed in Table S3.
(H) UMAP plot showing the WAM score calculated using the top 100 signature genes of WAM.
(I) Violin plot showing WAM gene set score in each condition. p value by Wilcoxon test.
(J) UMAP plots showing expression pattern of Ccl4, Itgax, Gpnmb, and Ch25h in microglia subclusters.
(K) UMAP plots showing expression pattern of Mitf and Plau in microglia subclusters.
(L) Scatter plot showing subcluster 3 marker genes. Adjusted p value by non-parametric Wilcoxon rank-sum test, Bonferroni correction.
(M) Violin plot of microglial expression of Mafb in each condition. p value by Wilcoxon test.
(N) Venn diagram of overlap between top 94 markers of “microglia inflamed in multiple sclerosis”-foamy (MIMS-foamy) from Absinta et al.8 and top 100 genes upregulated in microglia in remyelination vs. normal in the mouse CPZ model. Overlapping genes are listed on the side. Gene sets used are listed in Table S3.
(O) Venn diagram of overlap between top 86 genes upregulated in microglia from patients with early active MS from Masuda et al.22 and top 100 genes upregulated in microglia in remyelination vs. normal in the mouse CPZ model. Overlapping genes are listed on the side. Gene sets used are listed in Table S3.
(P) Representative IF images showing staining of MAFB and IBA1 in the MCC in each condition. IBA1, green; MAFB, red; DAPI, blue. Scale bar, 50 μm.
Remyelination is associated with reprogramming of vascular cells
In addition to glial cells, CPZ treatment induced evident changes in the signatures of vascular and mesenchymal brain populations. Analysis of DEGs in these cell compartments revealed substantial changes during remyelination vs. demyelination (Figures S6A and S6B). Pathway analysis of theseDEGs highlighted tissue development, cell adhesion, and migration pathways (Figure S6C). We further distinguished the vascular-mesenchymal cells into endothelial cells (Endo.1 and Endo.2), pericytes, smooth muscle cells (SMCs), meningeal fibroblasts (Fibro.1), and perivascular fibroblasts (Fibro.2–4)63,64 (Figures 5A and S6D) and examined the expression of DEGs (Figure 5B). Perivascular fibroblasts (Fibro.4) and pericytes upregulated Cd248, a marker for angiogenesis.65 Pericytes amplified expression of Il34, a ligand of CSF1R that may sustain perivascular macrophages during vascular remodeling.66 Fibroblasts highly expressed Loxl2, encoding a hypoxia-induced amine oxidase that promotes ECM remodeling by catalyzing crosslinking of collagen and elastin.67 SMCs upregulated Pdlim5, which promotes hypoxia-mediated vascular remodeling68 (Figure 5B).
Figure 5. Remyelination is associated with reprogramming of vascular cells.
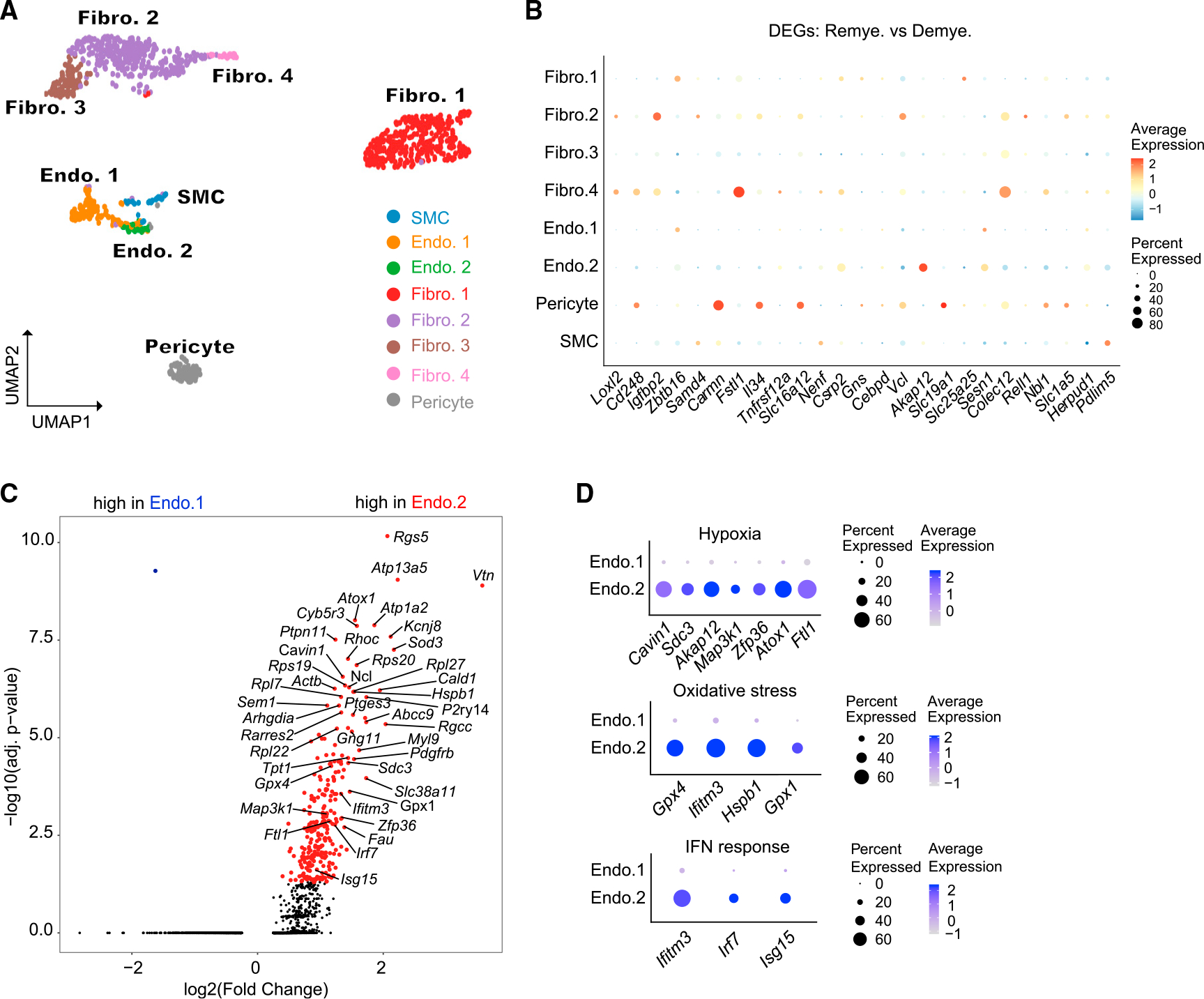
(A) UMAP plot of reclustered vascular cells identifying fibroblasts (Fibro.1–4), endothelial cells (Endo.1 and Endo.2), smooth muscle cells (SMC), and pericytes. n = 1,062 vascular nuclei. n = 2 mice in normal, n = 3 in demyelination, and n = 3 in remyelination condition.
(B) Dot plot showing expression of top DEGs between remyelination and demyelination in each subcluster.
(C) Volcano plot showing significant DEGs between Endo.2 and Endo.1. Log2(fold change) > 0.5, adjusted p < 0.05, non-parametric Wilcoxon rank-sum test, Bonferroni correction.
(D) Dot plot showing expression of DEGs (from C) involved in hypoxia, oxidative stress, and IFN-response pathways across Endo.1 and Endo.2.
We examined the relative abundance of vascular subclusters throughout CPZ treatment: pericytes trended to decline, while Endo.2 expanded during demyelination (Figure S6E). Pericyte loss may result in increased permeability of the blood-brain barrier.69,70 Compared with Endo.1, the Endo.2 cluster expressed the hypoxia-induced gene Rgs5,71 together with a signature indicative of oxidative stress and exposure to IFN (Figures 5C and 5D). Interestingly, during remyelination, perivascular fibroblasts (Fibro.2) upregulated genes that impact endothelial biology (Figure S6F): for example, Ccn1 that promotes angiogenic activities in endothelial cells72 and Timp3 that promotes endothelial apoptosis.73 Altogether, these results document a response of vascular cells to hypoxia and vascular injury.
Transcriptional changes reflect neuronal functional defects only in part
snRNA-seq analysis of neurons revealed a slight reduction in the proportion of neuronal nuclei after remyelination (Figure 1D), which may be a delayed consequence of axonal degeneration after demyelination. Analysis of DEGs across neuronal subclusters revealed that remyelination was associated with an upregulation of genes involved in neurogenesis, suggesting an attempt to compensate for neuronal damage (Figures S7A and S7B). One of the top genes increased in neurons was Epha5, encoding an Eph receptor involved in neuronal synapse formation.74 These modest transcriptional changes were somewhat in contrast to the markedly reduced neuronal activity previously reported in CPZ-induced demyelination.75 Moreover, immunostaining for the presynaptic marker synaptophysin in CPZ-treated mice did corroborate diminished synapses during demyelination and remyelination (Figure S7C). We conclude that transcriptional changes only partially reflect the neuronal defects induced by CPZ.
snRNA-seq reveals cell-cell interactions elicited by demyelination
Leveraging the comprehensive transcriptomic information provided by snRNA-seq, we explored potential cellular interactions between all brain cell types using NicheNet analysis, which predicts which ligands in the sender cell potentially regulate gene expression changes in the receiver cell.76 Specifically, we exploited NicheNet to rank ligand-receptor pairs that most likely account for differential gene expression during demyelination vs. normal in the receiver cells (Figures 6A–6C), and mapped gene expression of these ligands and receptors to the various cell types (Figures S7D and S7E). One of the top ligands was neuron/glia-derived neurofascin (NFASC), predicted to engage Contactin1 (CNTN1) expressed in oligodendrocyte lineage cells (Figures 6A, 6B, S7D, and S7E). NFASC-CNTN1 interaction between glia and axons impacts axo-glial junction formation, myelin maintenance, and axon segregation.77,78 NFASC-CNTN1 interaction also induces differentiation of OPCs into mature oligodendrocytes, promoting myelination during neurodevelopment.79 Thus, NFASC-CNTN1 interaction during CPZ treatment may underline a process of oligodendrocyte differentiation similar to that occurring during postnatal brain development.
Figure 6. NicheNet analysis reveals cell-cell interactions elicited by demyelination.
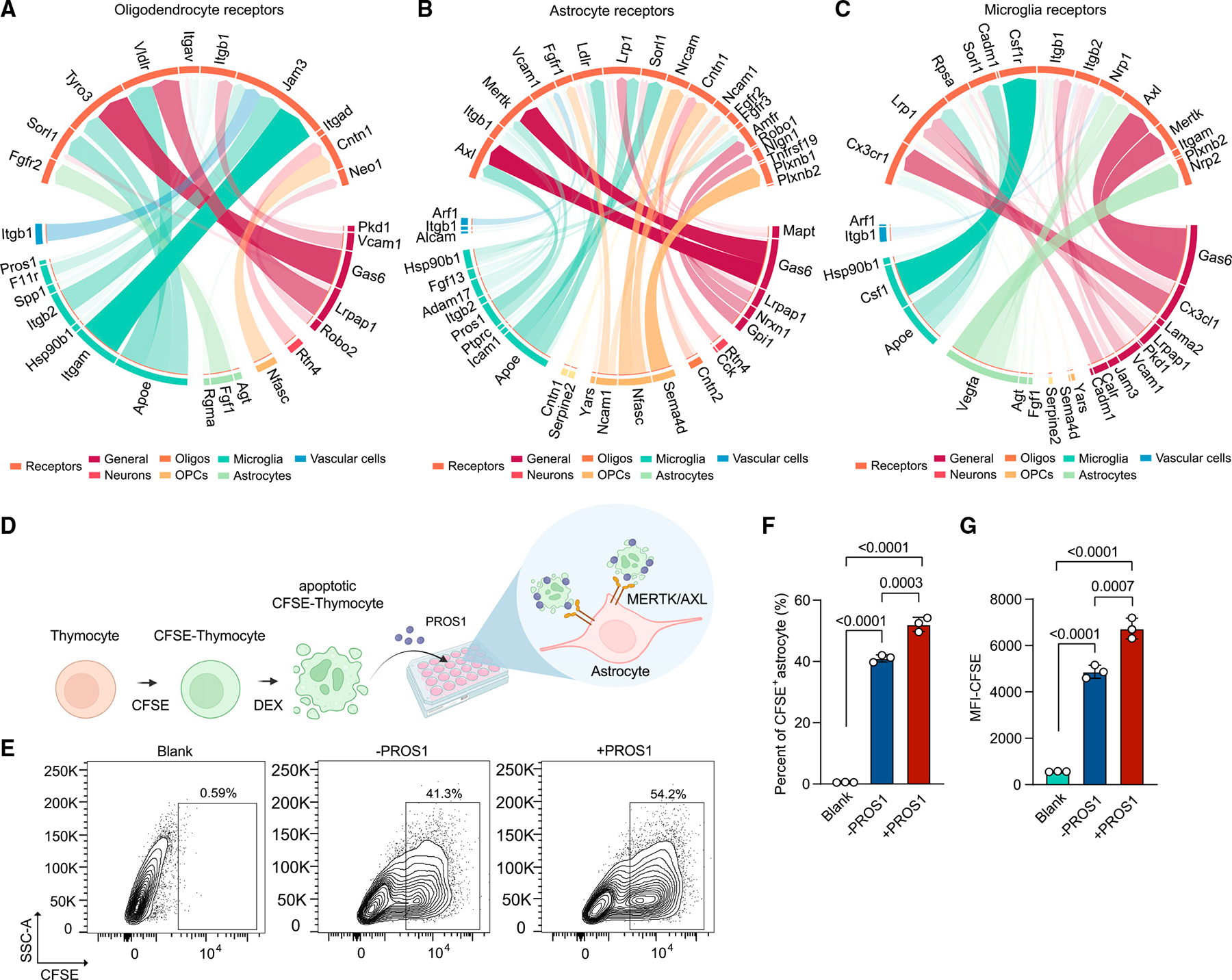
(A–C) Circos plot showing links between predicted ligands from sender cells (bottom) with their associated receptors on receiver cells (top, tomato). Width and transparency of the blocks represent the prior interaction weight of the ligand-receptor interactions. Colors on the lower part of the plots represent cell types where ligands are mostly derived from: dark red, ligands expressed in multiple cell types (general); dark orange, neurons; orange, oligodendrocytes; light orange, OPCs; dark green, microglia; light green, astrocytes; dark blue, vascular cells.
(D) Schematic of the assessment of astrocyte phagocytosis. DEX, dexamethasone. Created with BioRender.com.
(E) Representative flow-cytometry plots showing CFSE+ phagocytic astrocytes. Numbers indicate percentage of cells in the indicated gates.
(F and G) Quantification of the phagocytic astrocytes by percentage of CFSE+ astrocytes (F) and CFSE mean fluorescence intensity (MFI) (G). Astrocytes without thymocyte incubation were included as blank control. Data are presented as mean ± SD. p value by one-way ANOVA with Tukey’s multiple comparisons test. n = 3 cell culture wells per group. Experiment was repeated three times.
See also Figure S7.
NicheNet analysis also revealed that fibroblast growth factor 13 (FGF13) expressed by microglia may stimulate astrocytes through fibroblast growth factor receptor 3 (FGFR3). Additionally, SEMA4D, expressed by oligodendrocytes and microglia, may impact astrocytes by interacting with Plexinb1 (PLXNB1) (Figures 6B, S7D, and S7E). PLXNB1 is associated with cognitive decline in patients with AD, and knockdown of astrocyte PLXNB1 led to lower extracellular levels of Aβ42 in astrocyte cultures.80 Moreover, microglial-astrocyte SEMA4D-PLXNB1 interaction exacerbates CNS inflammation during EAE.81 Likewise, SEMA4D may aggravate pathology in the CPZ model by interacting with astrocyte PLXNB1.
Vascular endothelial growth factor A (VEGFA) produced by astrocytes was among the top ligands predicted to affect gene expression changes in microglia during demyelination via the VEGFA co-receptors Neuropilin 1/2 (NRP1/2)82 (Figures 6C, S7D, and S7E). Microglial NRP1 promotes remyelination by interacting with platelet-derived growth factor α (PDGFRα) on OPCs.83 Thus, astrocyte-derived VEGFA, microglia NRP1, and OPC PDGFRα may anchor a circuitry that controls myelination. In addition to aforementioned interactions, we corroborated the well-known interactions of TAM receptors with GAS6 and PROS1, which recognize phosphatidylserine on the surface of apoptotic cells and have been shown to impact myelination.20,84,85 Interestingly, we found that distinct cell types utilize disparate TAM receptors for binding GAS6 and PROS1: microglia and astrocytes expressed AXL and MERTK, whereas oligodendrocytes and neurons mainly expressed TYRO3 (Figures 6A–6C, S7D, and S7E). In the CPZ model, microglia predominantly expressed Pros1 (Figure S7D), which we validated at the protein level in cell lysates of cultured primary microglia (Figure S7F). To further demonstrate the impact of PROS1 on the uptake of apoptotic cells by astrocytes, carboxyfluorescein succinimidyl ester (CFSE)-labeled apoptotic thymocytes were cocultured with astrocytes with or without mouse PROS1, and phagocytosis was assessed by flow cytometry (Figure 6D). PROS1 protein enhanced astrocyte phagocytosis of apoptotic thymocytes (Figures 6E–6G), supporting the importance of PROS1-AXL/MERTK interaction in astrocyte-mediated clearance of apoptotic cells in the CPZ model.
TREM2 deficiency indirectly impairs induction of DOLs
Since our previous work suggested that reactive oligodendrocytes are partially TREM2 dependent in the 5XFAD mouse model,30 we examined the impact of TREM2 deficiency on DOL induction. TREM2 sustains the capacity of microglia to clear damaged myelin, enabling subsequent remyelination.21,86–88 However, does TREM2 indirectly affect other cells in response to myelin alterations in a cell-extrinsic manner? To address this question, we performed snRNA-seq of brain specimens from WT and Trem2−/− mice treated with CPZ (Figure S8A). Demyelination was delayed and axon myelination curtailed in Trem2−/− vs. WT mice, as evidenced by EM analysis of myelin in the CC (Figures S8B–S8D). Consistently, Trem2−/− mice had fewer OLIG2+ oligodendrocyte lineage cells than did WT mice during remyelination (Figures S8E and S8F). Unbiased clustering of single nuclei sequenced identified major cell types in the brain (Figures 7A, S8G, and S8H; Table S4). Differential expression analysis of WT and Trem2−/− mice showed that microglia and oligodendrocytes bore the greatest number of DEGs (Figure 7B and Table S5).
Figure 7. TREM2 deficiency indirectly impairs DOL induction.
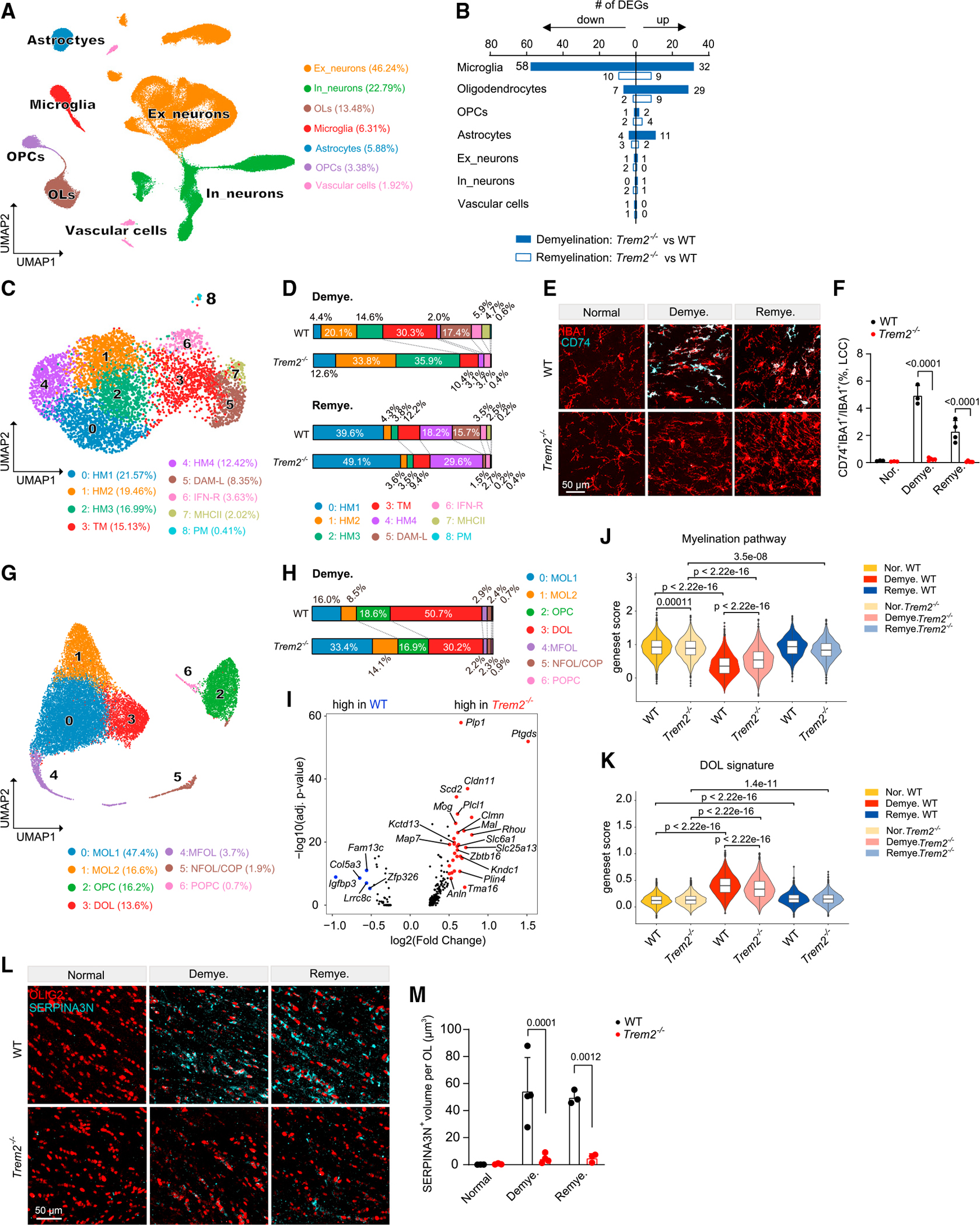
(A) UMAP plot of 117,729 nuclei showing seven distinguished cell types of Trem2−/− mice and littermate WT controls. n = 3 mice per genotype per condition.
(B) Number of DEGs between Trem2−/− and WT mice in demyelination and remyelination. Log2(fold change) > 0.5, adjusted p < 0.05, non-parametric Wilcoxon rank-sum test, Bonferroni correction.
(C) UMAP plot of reclustered microglia. n = 5,567 total nuclei. n = 619 nuclei in normal WT, n = 594 nuclei in normal Trem2−/−, n = 1,848 nuclei in demyelination WT, n = 1,121 nuclei in demyelination Trem2−/−, n = 866 nuclei in remyelination WT, n = 519 nuclei in remyelination Trem2−/−.
(D) Distribution of microglia subsets in demyelination and remyelination across WT and Trem2−/− mice.
(E) Representative IF images of CD74 and IBA1 staining in the LCC of Trem2−/− and WT mice in each condition. CD74, cyan; IBA1, red. Scale bar, 50 μm.
(F) Quantification of CD74+IBA1+/IBA1+ ratio in LCC across different conditions. n = 3–5 mice per genotype per condition. Data are presented as mean ± SD. p value by two-way ANOVA with Tukey’s multiple comparisons test.
(G) UMAP plot of reclustered oligodendrocyte lineage cells. n = 19,523 total nuclei. n = 3,939 nuclei in normal WT, n = 4,013 nuclei in normal Trem2−/−, n = 2,966 nuclei in demyelination WT, n = 2,964 nuclei in demyelination Trem2−/−, n = 3,468 nuclei in remyelination WT, n = 2,173 nuclei in remyelination Trem2−/−.
(H) Distribution of oligodendrocyte lineage subsets in demyelination across WT and Trem2−/−.
(I) Volcano plot showing DEGs of mature oligodendrocytes in Trem2−/− vs. WT during demyelination. Log2(fold change) > 0.5, adjusted p < 0.05, non-parametric Wilcoxon rank-sum test, Bonferroni correction.
(J and K) Violin plots showing myelination pathway gene set score (J) and DOL signature score (K) in the oligodendrocytes in each condition. p value by Wilcoxon test. Gene sets used are listed in Table S3.
(L) Representative IF images of SERPINA3N and OLIG2 staining in the MCC of Trem2−/− and WT mice in each condition. SERPINA3N, cyan; OLIG2, red. Scale bar, 50 μm.
(M) Quantification of SERPINA3N+ volume per OLIG2+ oligodendrocyte in MCC across different conditions. Data are presented as mean ± SD. p value by two-way ANOVA with Tukey’s multiple comparisons test. n = 3–4 mice per genotype per condition.
See also Figures S8 and S9; Table S3, Tables S4 and S5.
Subclustering of microglia delineated distinct demyelination-induced populations, including DAM-L, MHCII, and transition microglia, all of which were severely contracted in the absence of TREM2 (Figures 7C, 7D, S9A, and S9B). The abatement of Trem2−/− DAM-L and MHCII microglia was sustained through remyelination. The demyelination-induced IFN-R and proliferating subclusters were not evidently TREM2 dependent (Figures 7D and S9B). We validated the loss of DAM-L and MHCII microglia in Trem2−/− mice by co-staining for IBA1, CD74, APOE, and CD11c (Figures 7E, 7F, and S9C–S9F).
We next analyzed the relative abundance of oligodendrocyte subclusters in WT and Trem2−/− mice (Figures 7G, 7H, S9G, and S9H). During demyelination, MOLs (MOL1 and MOL2) were more abundant in Trem2−/− than in WT mice, which was paralleled by a reduction of DOLs (Figures 7H and S9H). Consistent with this, myelination-associated genes, such as Plp1 and Ptgds, were more abundant, whereas genes associated with demyelination, such as Col5a3, were more weakly expressed in Trem2−/− than in WT oligodendrocytes (Figures 7I–7K). Double immunolabeling for SERPINA3N and OLIG2 corroborated a significant diminution of DOLs in Trem2−/− mice (Figures 7L and 7M). Microglia are necessary for removal of damaged myelin,89 and TREM2 is required to mount a microglial response to myelin breakdown.86,87,90 Hence, we speculate that the defects emanating from Trem2 deficiency hamper removal of myelin debris, which causes a secondary delay in the stress response of oligodendrocytes, preserving their homeostatic configuration.
DISCUSSION
The CPZ mouse model is widely used to simulate demyelination occurring in MS and neurodegenerative diseases. While gene expression changes in microglia at the single-cell level have been delineated in this model recently,21,22 less is known about those occurring in other brain cell types. Here, we present a comprehensive snRNA-seq analysis of gene expression changes in the CPZ model, defining cell-type-specific signatures for demyelination and remyelination. Furthermore, snRNA-seq data provide insights into ligand-receptor interaction networks, enabling construction of complex cellular circuitries involved in de- and remyelination.
The oligodendrocyte response to CPZ-induced demyelination was remarkably complex and multipronged. We noted that oligodendrocytes spared by demyelination acquired a DOL signature, marked by inflammatory mediators, stress-response factors, anti-oxidant enzymes, cholesterol metabolism, and growth factors. Comparison of DOLs with oligodendrocytes in mouse models of amyloid pathology revealed common expression of Serpina3n and C4b genes,30,91 while Syt4, Moxd1, and Gdf15 were demyelination specific. DOLs largely overlapped with the disease-associated oligodendrocytes recently reported in multiple neurodegenerative models.92 Moreover, we demonstrated that the emergence of DOLs is affected by TREM2 deficiency, suggesting that DOLs are partly a secondary ramification of microglia activation. A further interesting aspect of the oligodendrocyte response was the release of nuclear IL-33, which attenuated CPZ-mediated demyelination. The response of oligodendrocytes to demyelination was paralleled by compensatory expansion of OPCs expressing transcription factors that drive oligodendrocyte maturation and myelination. Comparing CPZ oligodendrocytes with those in juvenile mice revealed that oligodendrocyte recovery during remyelination follows a trajectory echoing that of developmental myelination. Thus, remyelination in the CPZ model partially recapitulates oligodendrocyte development.
Astrocytes also evinced unique transcriptional signatures in response to CPZ. During demyelination, astrocytes adopted a gene expression program encompassing wound healing (Vim, Gfap, and Thbs4), complement and coagulation cascades (C4b and Serpina3n), cellular growth (Slc7a5, Igfbp2, and Igfbp5), and anti-oxidative metabolism (Slc7a11/Slc3a2 and Ephx1). This program probably depends on STAT3 among other transcription factors. Another demyelination-induced subset expressed genes indicating type I IFN signaling, mirroring a subset found in a model of lipopolysaccharide-induced inflammation.93 Demyelination-induced astrocyte subsets declined during remyelination, conceding to an emerging subset with a transcriptional profile indicative of cholesterol and fatty acid metabolism (Ldlr and Sqle), detoxifying and stress-response pathways (Paxx, Nrros, and Neat1), cell adhesion (Ezr and Igsf11), and activation of NF-κB (Irak2, Il18, and Nfkbia).
In addition to reported signatures of DAM, IFN-R, and MHCII in microglia responses to demyelination,20,22 our snRNA-seq study identified MAFB as a major transcription factor underlying microglia responses during remyelination. MAFB represses macrophage self-renewal and promotes differentiation.94–96 Expression of Mafb is elevated in adult microglia and required for maintenance of the homeostatic signature during adulthood by inhibiting IFN pathway genes.62 Thus, MAFB may be crucial for microglial recovery of homeostasis phenotype during remyelination. Our data also revealed unprecedented alterations in the vascular and mesenchymal compartment during remyelination that affect endothelial cells, pericytes, and perivascular fibroblasts. These changes reflected endothelial responses to hypoxia and reactive oxygen species, pericyte supply of IL-34 for perivascular macrophages, and fibroblast-mediated remodeling of ECM. While CPZ impairs CNS functions,75,97 its impact on the neuronal transcriptional profile was not dramatic and contrasted with the marked synaptic loss detected by immunostaining. Thus, snRNA-seq may provide a partial proxy for the impaired neuronal connectivity caused by CPZ-induced demyelination.
NicheNet analysis unveiled a rich network of interactions among oligodendrocytes, astrocytes, and microglia that may be altered during CPZ-induced demyelination. These interactions mediate oligodendrocyte regeneration and formation of myelin sheaths,79 microglial phagocytosis of damaged myelin and apoptotic neurons,20 and astrocyte repair functions.81 Our study also provides a benchmark for comparing mouse signatures of de- and remyelination with human diseases. We identified key genes governing glial responses to myelin alterations shared by mouse CPZ and human MS, particularly HMGCS1, NUPR1, and MAFB. Moreover, we noted co-existence of gene expression profiles related to both CPZ de- and remyelination in human MS, implying that MS involves demyelination and simultaneous attempts to reconstitute myelin sheaths. Altogether, this study provides a rich resource that can be leveraged to generate new mechanistic hypotheses and tools to fully delineate demyelination and remyelination.
Limitations of the study
While CPZ selectively lowers the mitochondrial transmembrane potential of oligodendrocytes, with negligible effects on microglia, astrocytes, or neurons in vitro,98 we could not exclude that CPZ subtly affects these cells. Yet the similarity of DOL signatures in CPZ and mouse models of AD suggests that the responses identified here may be general and not limited to CPZ-induced damage. Although our analysis of germline Il1rl1−/− mice revealed a role for IL-33 in delaying demyelination, oligodendrocyte-specific deletion of Il1rl1 is necessary to precisely define the impact of oligodendrocyte-secreted IL-33.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Marco Colonna (mcolonna@wustl.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Single-nucleus RNA-seq data have been deposited at the Gene Expression Omnibus (GEO) database and are publicly available as of the date of publication under accession number GSE204770.
Original code has been deposited at Github at: https://github.com/marcocolonnalab/CPZ.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
WT C57BL/6J mice were purchased from Jackson Laboratory (#000664). Trem2−/− mice were generated as previously described.102 Il1rl1−/− mice were generated in the lab of Dr. S. Akira (Osaka University)103 and were a gift from Steve Van Dyken (Washington University in St. Louis). WT littermates were used as controls. All mice were housed in specific-pathogen-free conditions. The Institutional Animal Care and Use Committee at Washington University in St. Louis approved all protocols used in this study. 8-week-old mice were fed with 0.2% (w/w) cuprizone (bis-cyclohexanone oxaldihydrazone) (Sigma, #c9012) in ground breeder chow for 5 weeks to induce demyelination.13 For WT C57BL/6J snRNA-seq dataset, tissues from 2 WT C57BL/6J mice in normal condition, 3 in demyelination and 3 in remyelination were processed. For Trem2−/− snRNA-seq dataset, tissues from 3 mice per genotype per condition were processed. The normal control mice were sacrificed at the same time with the demyelination group (5 weeks after cuprizone). All mice applied in this study were male.
Primary cell culture
Primary mixed glia were cultured from neonatal brains as described previously with minor modifications.104 Superficial cortex dissected from P2 or P3 WT mice were dissociated with 300 μg/mL DNase I (#11284932001, Sigma) and 0.15% trypsin (#T1426, Sigma) in 37°C water bath for 10 min with gentle agitation every 2 min. After removal of undispersed tissue using a 70-μm cell strainer, cells were suspended in 5 mL complete DMEM medium (containing 10% heat-inactivated fetal bovine serum, 1X kanamycin sulfate, 1mM sodium pyruvate, 1X MEM nonessential amino acids, and 1X GlutaMAX) and seeded in poly-L-lysine-coated 25-cm2 culture flasks (#P4707, Sigma). The culture was maintained by changing the medium every 3 days. After 14 days in culture, astrocytes were at the bottom layer, and microglia grew on top of the astrocyte layer. Microglia were then detached from the bottom of the flasks by vigorous shaking. Adhered cells, which mainly consisted of astrocytes, were detached using 0.05% trypsin, seeded on a poly-L-lysine-coated 24-well plate, and maintained in complete DMEM medium until use.
METHOD DETAILS
Isolation of nuclei from frozen brain tissue
Isolation of nuclei was performed similarly as previously described.30 Briefly, frozen brain cortex and white matter were Dounce homogenized in 5 mL of lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.025% NP-40) for 15 min. Following initial Dounce homogenization, the solution was then filtered through a 30-μm cell filter and pelleted at 500g for 5 min at 4°C. Nuclei were washed and filtered twice with nuclei wash buffer (1% BSA in PBS with 0.2 U μl−1 RNasin (Promega)). Nuclei pellets were resuspended in 500 μL nuclei wash and 900 μL 1.8 M sucrose. This 1,400 μL mixture was carefully layered on top of 500 μL 1.8 M sucrose and centrifuged at 13,000g for 45 min at 4°C to separate the nuclei from myelin debris. The nuclei pellet was resuspended in nuclei wash buffer at 1,200 nuclei μl−1 and filtered through a 40-μm FlowMi Cell Strainer.
Single-nucleus RNA sequencing and data processing
Droplet-based snRNA-seq was performed using the Chromium Single Cell 5’ Reagent Kits (v1 for WT C57BL/6J snRNA-seq dataset and v2 for Trem2−/− snRNA-seq dataset) per the manufacturer’s instructions (10x Genomics). Nuclei were resuspended to a concentration of 1,200 nuclei per μl before loading according to the manufacturer’s protocol. The libraries were sequenced using Illumina sequencers (NovaSeq instrument) with 150bp paired-end sequencing at the McDonnell Genome Institute. Sample demultiplexing, barcode processing and single-cell counting were performed using the Cell Ranger Single-Cell Software Suite (10x Genomics). To include Clec7a gene, we built a custom reference genome by adding Clec7a cDNA to the pre-built mouse reference genome (mm10). Cellranger count (v.3.0.2) was used to align reads to the custom reference genome, quantify reads and filter reads with a quality score below 30.
Processing data with seurat package and quality control
The Seurat package in R was used for subsequent analysis.105 For quality control, nuclei with mitochondrial content >5% were removed. Nuclei that are doublets or multiplets were filtered out by two steps. First, nuclei with high UMI and gene number per nucleus were filtered out. Cutoffs for UMI and gene number were determined on the basis of histograms showing cell density as a function of UMI per gene counts. Then nuclei with more than one cell type marker gene expressed were removed. For WT C57BL/6J mouse brain snRNA-seq analysis, a cutoff of 500–10,000 UMI and 500–4,000 genes was applied. After filtering, a total of 58,079 individual nuclei across all conditions remained, with a median of 1,808 UMIs and 1,250 genes per nucleus for downstream analysis. For Trem2−/− and littermate WT mouse brain snRNA-seq analysis, a cutoff of 500–15,000 UMI and 400–5,000 genes was applied. After filtering, a total of 117,729 individual nuclei across all genotypes remained, with a median of 3,082 UMIs and 1,951 genes per nucleus for downstream analysis. Data were log normalized and regressed on mitochondrial gene percentage during data scaling, using the mitochondria ratio as the argument for ‘vars.to.regress’. Samples were batch corrected using FindIntegrationAnchors function and Canonical Correlation Analysis (CCA). Principal component analysis was performed using the top 3,000 most variable genes and UMAP analysis was performed with the top 20 PCAs. Clustering was performed using a resolution of 0.3. For identifying markers for each cluster, we performed differential expression of each cluster against all other clusters, identifying positive markers for that cluster, using FindAllMarkers() function in the Seurat package with default parameters. Cluster marker genes are listed in Tables S1 and S4. Nuclei from broad cell types (neurons, oligodendrocytes, astrocytes, microglia and vascular cells) were taken and re-clustered to further analyze the sub-clusters in each cell type. For data visualization, BBrowser (version 2) was also used.
Sub-clustering analysis
To identify subsets in each cell type, nuclei from all samples belonging to a given major cell type were extracted for downstream analysis. Doublets were filtered out as described above. Re-integration was applied to microglia sub-clustering (both C57BL/6J and Trem2−/− datasets), and oligodendrocyte sub-clustering (Trem2−/− dataset). Principal component analysis was performed prior to clustering and the first ten principal components were used based on the ElbowPlot. Clustering was performed using the FindClusters() function. For C57BL/6J dataset, clustering resolution at 0.3 was used for oligodendrocyte lineage, 0.6 for astrocytes, and 0.6 for microglia. For vascular cells, the first 15 principal components were used, and clustering was done with resolution 1.1. For Trem2−/− dataset, clustering resolution at 0.3 was used for oligodendrocyte lineage, and 0.6 for microglia.
Identification of DEGs across conditions
Differential expression of genes between conditions was done using FindMarkers() function with default parameters of the Seurat package in R,106 which is based on the non-parametric Wilcoxon rank-sum test. Log2(fold change) of average expression and the percentage of cells (pct) expressing the genes in each condition were generated. The adjusted p value was calculated using Bonferroni correction. Genes with log2(Fold Change) > 0.5, adjusted p < 0.05 were considered as significant DEGs. Lists of DEGs were generated by filtering all genes for log2 (fold change) > 0.25 (Tables S2 and S5). These gene lists were used as inputs for downstream pathway analyses.
Heatmap
Average expression per cluster per sample was calculated using the AverageExpression() function in the Seurat package. For heat maps of relative gene expression across cell types, average expression of each gene was Z score transformed across samples and plotted using heat.map2 function in the gplots packge (v3.1.1) in R.
NicheNet analysis
Ligand-receptor interactions were mined from the snRNA-seq data using the NicheNet algorithm (nichenetr 1.0.0).76 Briefly, sender and receiver cells were defined and indicated in Figure 6. NicheNet analysis was performed using upregulated DEGs in the defined receiver cells between demyelination versus normal (log2(fold change) > 0.5, adjusted p value <0.05) as input with default settings. Circos plot was computed based on calculation of interaction scores between possible ligand-receptor combinations. Interaction pairs were filtered for those bona fide interactions that were documented in the literature and publicly available datasets.
Pseudotime trajectory analysis
Single-cell pseudotime trajectories were inferred using the R package slingshot (v2.1.0).100 All C57BL/6J microglia nuclei except for the proliferating microglia were used for the analysis.
Estimation of RNA velocity
RNA velocity analysis was performed using the package scVelo (v.0.2.4) with stochastic modeling, as instructed.38 Briefly, we used the.bam and.bam.bai files from the CellRanger output and generated the.loom files for the representative samples in each condition (demyelination: A3, and remyelination: B7) using the velocyto.py version 0.17 with the velocyto run10x command. Next, the.loom files were loaded into Python (v3.9.13) using scv.read function to generate count tables in JupyterLab interface. The Seurat objects from each genotype were converted into anndata files as described (https://smorabit.github.io/tutorials/8_velocyto/) and then loaded separately into JupterLab interface with Python via the function sc.read.h5ad. RNA velocity was computed via scv.tl.velocity(), and the final velocity streams and pseudotime were generated using the commands scv.pl.velocity_embedding_stream() and scv.pl.velocity_pseudotime(), respectively. For visualization, the RNA velocity stream was projected onto each predefined UMAPs.
Dataset integration
To compare oligodendrocyte gene signatures in the cuprizone model to that observed in mouse juvenile CNS26 as well as DOL signature characterized in the 5XFAD model,39 we re-analyzed the raw39 or filtered26 matrix data of the two aforementioned datasets. Filtering parameters were the same as in the original manuscript. After filtering, each dataset was processed using functions implemented in the Seurat package (SCTransform, RunPCA, RunUMAP, FindNeighbors, FindClusters). Oligodendrocyte lineage cells were extracted using subset() function in the Seurat package. Samples were integrated using FindIntegrationAnchors() function and CCA.107 CPZ oligodendrocyte dataset was integrated with Marques et al.26 dataset and Kenigsbuch et al.39 dataset, respectively. Principal component analysis was performed using the top 3,000 most variable genes and UMAP analysis was performed with the top 20 PCAs. Annotation of subclusters were based on original reference or clusters characterized in the current manuscript. Dot plot of marker gene expression was generated using scanpy (v1.9.3) and dendrogram was built by performing hierarchical clustering on predefined subclusters.
Re-analysis of public datasets
To reanalyze publicly available datasets on human patients with MS, raw or filtered matrix of UMI counts and associated metadata were downloaded from GEO under accession number GSE124335, GSE118257 and GSE180759, and analyzed using Seurat package. For oligodendrocyte signature comparisons, oligodendrocyte lineage cells were extracted from the two datasets, respectively.6,8 Differential expression analysis was performed between each pathological state and the control as described above. DEGs were considered as genes with log2 (fold change) > 0.25, adj. p value <0.05 for GSE124335 dataset, with log2 (fold change) > 0.25, adj. p value <0.05 for GSE118257 dataset, and with log2 (fold change) > 0.5, adj. p value <0.05 for GSE180759 dataset. For astrocyte signature comparisons, top 100 marker genes of the AIMS were extracted from Absinta et al., 2021.8 For microglia comparisons, top 94 marker genes of the MIMS-foamy isolated from the chronic lesions of patients with MS,8 or top 100 feature genes of microglia from acute lesions of patients with MS22 were extracted. Gene sets listed in Table S3 were used as inputs for this analysis. Human genes were converted to mouse genes using biomaRt package (2.48.3) in R. Venn diagrams were plotted using BioVenn package (v1.1.3) in R.
Gene set score analysis
Gene set scores were calculated using the AddModuleScore() function in Seurat package. Briefly, for each cell, the log-transformed average expression of all genes in each gene set was calculated and subtracted by the log-transformed expression of control features sets. The white matter-associated microglia (WAM) score was calculated by using the top 100 signature genes of WAM identified from aged mice.56 The disease-associated microglia (DAM) score was determined by using the top 71 signature genes of DAM identified in the 5XFAD model.55 The reactive oligodendrocyte signature was defined by the cluster markers of DOLs cluster. Gene signatures of myelination was obtained from the “Myelination” pathway from Metascape pathway analysis. Gene sets listed in Table S3 were used as inputs for this analysis.
Pathway enrichment analysis
For pathway enrichment analysis, enrichment for Gene Ontology terms was obtained through Metascape.30 Gene set enrichment analysis (GSEA) was performed using the hallmark pathways from the Molecular Signature Database.
Immunofluorescence
Animals were anesthetized by 10 mg/mL ketamine and 1 mg/mL xylazine solution i.p. and perfused transcardially with PBS containing 1U/ml heparin. Fixation of brain samples was done in 4% paraformaldehyde (PFA) overnight. The brain tissues were then incubated in 30% sucrose in PBS for 24 h. After freezing the tissue on dry ice using Tissue-Tek O.C.T and 30% sucrose at a ratio of 1:2, 20-μm coronal sections were cut by a Leica CM 1900 cryostat. Floating mouse brain sections were blocked with 3% BSA and 0.25% Triton X-100 in PBS for 1h at room temperature, and stained with anti-IBA1 (1:500; rabbit, #17198S, Cell Signaling Technology), anti-IBA1 (1:500; goat, #5076, Abcam), anti-CD74 (1:200; Alexa Fluor 647-labeled rat IgG2b, #151004, Biolegend), anti-OLIG2 (1:500; Rabbit, #AB9610, EMB Millipore), anti-CA2 (1:200, rat, #MAB2184, R&D), anti-CC1 (1:50, Mouse, #OP80, Calbiochem), anti-SER-PINA3N (1:300; Goat, #AF4709, R&D), anti-IL33 (5 μg/mL; Goat, #AF3626, R&D), anti-GFAP (1:1000; Alexa Fluor 488-labbeled mouse IgG1, #53–9892-82, ThermoFisher Scientific), anti-APOE (1:300; biotinylated, mouse monoclonal: HJ6.3, a gift from Dr. David M. Holtzman), anti-CD11c (1:500; Rabbit, #97585, Cell Signaling Technology), anti-MAFB (1:400; Rabbit, #A700–046, BETHYL), anti-Vimentin (1:500; Mouse, #sc-373717, Santa Cruz Biotechnology), anti-STAT1 (1:300; Rabbit, #14994, Cell Signaling Technology), anti-HMGCS1 (1:200; Rabbit, #PA5–29488, Invitrogen) and anti-Synaptophysin (1:200; Rabbit, #36406, Cell Signaling Technology) overnight at 4°C, followed by staining with anti-rabbit IgG Alexa Fluor 555 (1:2,000; #ab150074, Abcam), anti-rat IgG Alexa Fluor 647 (1:2,000; #A21472, Invitrogen), anti-mouse IgG2b Alexa Fluor 488 (1:2,000; #A21141, Invitrogen), anti-goat IgG Alexa Fluor 488 (1:2,000; #ab150129, Abcam), anti-Streptavidin, conjugate (1:2,000; #s868, ThermoFisher Scientific), and DAPI (1 μg/ml; #D9542, Sigma) for 1 h at room temperature. All antibodies were used in blocking buffer, and between all incubations, sections were washed for 10 min in PBS 3 times. Images were collected using a Nikon A1Rsi confocal microscope or Zeiss LSM880 microscope. z-Stacks with 1-μm steps in the z direction, 1,024 × 1,024-pixel resolution, were recorded. Extraction of parameters were performed in Imaris (version 9.8.0), and further processing was performed using automated scripts in MATLAB.88
To quantify staining signals in the confocal images by volume in MCC or LCC, we created surfaces using the “skip automatic function, edit manually” option in Imaris and manually selected the MCC or LCC area on the top and bottom sections; we then used the “masked channel” function to create new channels that selectively contain signals of protein of interest inside the selected regions. Then “batch colocalization” of MATLAB function was run on the newly generated channels and colocalization channels were created. The volumes of signal of interest in each image were determined using “Surface” function in Imaris. Ratios of double-positive volumes over single positive volumes (IBA1+CD74+/IBA1+, IBA1+CD11C/IBA1+, IBA1+APOE+/IBA1+ and SERPINA3N+GFAP+/GFAP+) were then calculated. To analyze SERPINA3N+ volume in Trem2−/− and WT mice, total SERPINA3N+ volume was measured within 2-μm radius of OLIG2, then ratios of total SERPINA3N+ volume over OLIG2+ number were calculated. To quantify VIM+ and Synaptophysin+ area in the LCC and cortex, images were first converted to a 2D image and then were converted to a binary image with the ImageJ “Make Binary” tool. The area of LCC or cortex were selected manually on the binary image using the ImageJ “Freehand Selections” tool. The mean signal intensity value was measured by the ImageJ “Measure” tool, using “mean gray value” as the readout.
Flow cytometry
Brains from 4 mice in each group were dissected and homogenized by a Dounce homogenizer in the dissociation buffer (1%BSA, 1mM EDTA). Myelin debris was removed by 30% Percoll density gradient centrifugation, followed by the labeling of microglia with CD45-APC-Cy7 (1:200, #103116, BioLegend) and IL-33R (ST2)-PE (1:200; #12-9333-80, Invitrogen). Dead cells were excluded using DAPI (1 μg/ml; D9542, Sigma). Cells were acquired using BD FACSCantoII flow cytometer. Microglia were gated on CD45low cells and analyzed using Flowjo (v10.8.1).
Transmission electron microscopy
Brains were processed for transmission electron microscopy (TEM) analysis as previously described.108 Briefly, animals were perfused with the 4% PFA (Electron Microscopy Sciences, #15714-S), and post-fixed in the fixative containing 2.5% glutaraldehyde/2% paraformaldehyde/0.15M cacodylate buffer with 2mM CaCl2 (Electron Microscopy Sciences, #16300, #15710) at 4°C for 24 h before further dissection. Post fixation, brain samples were cut in sagittal direction near the midline into 100 μm thick sections with a vibratome (Leica VT1200S, Vienna, Austria). Whole brain sections containing CC were rinsed in 0.15 M cacodylate buffer containing 2 mM calcium chloride 3 times for 10 min each followed by a secondary fixation in 1% osmium tetroxide/1.5% potassium ferrocyanide in 0.15 M cacodylate buffer containing 2 mM calcium chloride for 1 h in the dark. The samples were then rinsed 3 times in ultrapure water for 10 min each and en bloc stained with 2% aqueous uranyl acetate overnight at 4°C in the dark. After another 4 washes in ultrapure water, the samples were dehydrated in a graded ethanol series (30%, 50%, 70%, 90%, 100% x3) for 10 min each step. Once dehydrated, samples were infiltrated with LX112 resin (Electron Microscopy Sciences) and flat embedded and polymerized at 60°C for 48 h. Post curing, medial regions of the CC were excised and mounted on blank epoxy stubs for cross sectioning. 70 nm sections were then cut, post-stained with 2% aqueous uranyl acetate and Sato’s lead and imaged on a TEM (JEOL JEM-1400 Plus) at an operating voltage of 120 Kv.109
For each TEM analysis, five to ten randomly selected CC regions in each mouse were imaged. Data analysis was performed using ImageJ 1.53a software. The myelinated ratios and g-ratios were measured with ImageJ on transverse electron micrographs at 3,000× magnification. The perimeters of each axon and the myelin sheath were measured with the freehand tool by tracing the outer surfaces of each structure. The g-ratio was calculated as the perimeter of the axon over the perimeter of its myelin.
LFB staining
Myelin sheaths were stained with luxol fast blue (Electron microscopy sciences, Luxol Fast Blue, 0.1% in 95% alcohol, #26056–15). For the measurement of LFB intensity, the micrograph was first converted to a binary image with the ImageJ “Make Binary” tool. The area of MCC was then selected manually on the binary image using the ImageJ “Freehand Selections” tool. The mean LFB intensity value was measured by the ImageJ “Measure” tool, using “mean gray value” as the readout. For the quantification of LFB intensity, the mean intensity values in 3 adjacent brain sections from one animal were calculated and used for further statistical analysis. At least 3 animals per genotype were analyzed and the n was specified in the figure legends.
Microglial PROS1 ELISA
Microglia detached from the mixed glia culture were seeded in 24-well plate at a density of 100,000 cells/well in complete RPMI 1640 medium containing 10% LCCM. After one day, microglia were stimulated with myelin debris (500 μg/ml) for 48hrs, and the microglial lysis was harvested for PROS1 ELISA (#AE25758MO, Abebio). Total protein concentration was quantified by DC protein assay (#5000112, BIO-RAD).
Assessment of astrocytic phagocytosis
For fluorescent labeling of thymocyte membranes, WT thymocytes were incubated with CFSE dye (5μM; C1157, ThermoFisher Scientific) for 8 min with constant agitation at room temperature. CFSE-labeled thymocytes were then incubated with 2μM Dexamethasone (D1756, Sigma) for 6 h in 37°C incubator to induce apoptosis. Our preliminary experiment confirmed that around 70% CFSE-labeled thymocytes became apoptotic after dexamethasone incubation by Annexin V and 7AAD staining (#559763, BD Biosciences). The CFSE-labeled apoptotic thymocytes were then incubated for 10 min with or without 10nM recombinant mouse PROS1 (#9740-PS-050, R&D systems), added to the astrocyte culture at a ratio of 20:1 (apoptotic thymocytes: astrocytes), and incubated for 12 h at 37°C. Astrocytes were then briefly washed in PBS for 3 times, incubated for 3 min at 37°C in 0.25% trypsin and detached by vigorous pipetting. Phagocytic astrocytes were assessed using LSR Fortessa (BD Biosciences). Dead cells were excluded by PI staining. The post-acquisition data was analyzed by FlowJo software (v10.8.1).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical tests used are indicated in figure legends. No data were excluded from the analysis. Data are presented as mean ± sd unless specifically described. All statistical analyses for validation assay were performed using Prism 8.0 software with one-way or two-way ANOVA with Tukey’s multiple comparisons test, and p values less than 0.05 were considered statistically significant. Statistics on cell type compositional analysis were calculated using scCODA package (v.0.1.8) in Python (v3.9.2).101 p values in snRNA-seq analysis were adjusted based on Bonferroni correction.106 p values in violin plots were calculated using stat_compare_means() function of ggpubr package (0.4.0) in R, using Wilcox test.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| anti-Ibal (rabbit monoclonal) | Cell Signaling Technology | Cat#17198; RRID: AB_2820254 |
| anti-Ibal (goat polyclonal) | Abcam | Cat#5076; RRID: AB_2224402 |
| anti-CD11c (rabbit monoclonal) | Cell Signaling Technology | Cat#97585; RRID: AB_2800282 |
| anti-CD74-AF647 (rat monoclonal) | BioLegend | Cat# 151004; RRID: AB_2632609 |
| anti-APOE (HJ6.3, biotinylated, mouse monoclonal) | Holtzman Lab (Kim et al., 20 1 299) | N/A |
| anti-SERPINA3N (goat polyclonal) | R&D Systems | Cat#AF4709; RRID: AB_2270116 |
| anti-APC (CC-1, mouse monoclonal) | MilliporeSigma | Cat#OP80; RRID: AB_2057371 |
| anti-IL33 (goat polyclonal) | R&D Systems | Cat#AF3626; RRID: AB_884269 |
| anti-GFAP (Alexa Fluor 488-labbeled, mouse monoclonal) | ThermoFisher Scientific | Cat #53–9892-82; RRID: AB_10598515 |
| anti-Vimentin (E-5, mouse monoclonal) | Santa Cruz Biotechnology | Cat#sc-373717; RRID: AB_10917747 |
| anti-MAFB (rabbit monoclonal) | BETHYL | Cat#A700–046; RRID: AB_2891845 |
| anti-STATI (rabbit monoclonal) | Cell Signaling Technology | Cat#14994; RRID: AB_2737027 |
| anti-HMGCSI (rabbit Polyclonal) | Invitrogen | Cat#PA5–29488; RRID: AB_2546964 |
| anti-Synaptophysin (Rabbit monoclonal) | Cell Signaling Technology | Cat#364065; RRID: AB_2799098 |
| anti-OLIG2 (rabbit polyclonal) | EMB Millipore | Cat#AB9610; RRID: AB_570666 |
| anti-IL33R(ST2)-PE (rat monoclonal) | Invitrogen | Cat#12–9333-80; RRID: AB_2572705 |
| CD45-APC/Cyanine7 (30-F11, rat monoclonal) | BioLegend | Cat#103116; RRID: AB_312981 |
| anti-Carbonic Anhydrase II/CA2 (rat monoclonal) | R&D Systems | Cat#MAB2184; RRID: AB_2065858 |
| anti-rabbit IgG AF555 (donkey polyclonal) | Abcam | Cat#150074; RRID: AB_2636997 |
| anti-goat IgG AF647 (donkey polyclonal) | Abcam | Cat#150131; RRID: AB_2732857 |
| anti-goat IgG AF488 (donkey polyclonal) | Abcam | Cat#150129; RRID: AB_2687506 |
| anti-rat IgG AF647 (chicken polyclonal) | Invitrogen | Cat# A-21472; RRID: AB_2535875 |
| anti-mouse IgG2b AF488 (goat polyclonal) | Invitrogen | Cat#A21141; RRID: AB_2535778 |
| anti-Streptavidin, APC | ThermoFisher Scientific | Cat#s868 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Luxol Fast Blue Solution, 0.1% in 95% Alcohol | Electron microscopy sciences | Cat#26056–15 |
| CFDA SE (CFSE) dye | ThermoFisher Scientific | Cat# C1157 |
| Poly-L-lysine solution | MilliporeSigma | Cat#P4707 |
| 4’,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) | MilliporeSigma | Cat# D9542 |
| DNase I | MilliporeSigma | Cat#11284932001 |
| Trypsin from bovine pancreas | MilliporeSigma | Cat#T1426 |
| Recombinant Mouse Protein S/PROS1 | R&D systems | Cat#9740-PS-050 |
| Paraformaldehyde 32% | Electron Microscopy Sciences | Cat#15714-S |
| Formaldehyde Aqueous Solution, EM Grade | Electron Microscopy Sciences | Cat#15710 |
| Dexamethasone | MilliporeSigma | Cat#D1756 |
| Cuprizone (bis-cyclohexanone oxaldihydrazone) | MilliporeSigma | Cat#c9012 |
| EDTA | Corning | Cat#46–034-Cl |
| Triton X-100 | MilliporeSigma | Cat#T8787 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Chromium Single Cell 5’ Library & Gel Bead Kit, 16 rxns | 10x Genomics | Cat#PN-1000006 |
| Chromium Next GEM Single Cell 5’ Kit v2, 16 rxns | 10x Genomics | Cat#PN-1000263 |
| Mouse Vitamin K-dependent protein S (PROS1) ELISA Kit | Abebio | Cat#AE25758MO |
| PE Annexin V Apoptosis Detection Kit I | BD Biosciences | Cat#559763 RRID: AB_2869265 |
| DC protein assay | BIO-RAD | Cat#5000112 |
|
| ||
| Deposited data | ||
|
| ||
| 10x Single Nucleus RNA-seq | This paper | GEO: GSE204770 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Primary murine microglia | This paper | N/A |
| Primary murine astrocytes | This paper | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6J | Jackson Laboratory | Cat #000664 |
| Mouse: wildtype | In house | N/A |
| Mouse: Trem2−/− | In house | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| MATLAB | MathWorks |
http://www.mathworks.com/products/ matlab/;RRID:SCR_001622 |
| ImageJ | National Institutes of Health | https://imagej.net/;RRID:SCR_003070 |
| FlowJo (v10.8.1) | FlowJo | https://www.flowjo.com/solutions/flowjo/;RRID:SCR_008520 |
| Imaris (v9.8.0) | Bitplane | http://www.bitplane.com/imaris/imaris/;RRID:SCR_007370 |
| R(v4.1.0) | R Core Team (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. | http://www.r-project.org/; RRID:SCR_001905 |
| RStudio (v1.4.1717) | https://posit.co | RRID: SCR_000432 |
| Seurat (v4.0) | http://seurat.r-forge.r-project.org/ | RRID: SCR_007322 |
| Slingshot (v2.1.0) | Street etal.,2018100 | N/A |
| scCODA (v0.1.8) | Büttner et al., 2021101 | N/A |
| scVelo (v0.2.4) | La Manno et al., 201838 | N/A |
| NicheNet (v1.0.0) | Browaeys et al., 201976 | N/A |
|
| ||
| Python (v3.9.13) | Python Software Foundation |
https://www.python.org/; RRID:SCR_008394 |
| BBrowser (v2.7.5) | BioTuring Inc. | www.bioturing.com/bbrowser |
| Cell Ranger (v3.0.2) | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome |
| JupyterLab (v3.4.7) | https://jupyter.org/ | RRID:SCR_022695 |
| GraphPad Prism (v8.0.2) | http://www.graphpad.com/ | RRID:SCR_002798 |
| BioRender | https://biorender.com/ | RRID: SCR_018361 |
| Metascape | http://metascape.org/gp/index.html#/main/step1 | RRID:SCR_016620 |
| Code | This paper | https://github.com/marcocolonnalab/CPZ |
|
| ||
| Other | ||
|
| ||
| Cuprizone Diet (2018, 0.2%) | Envigo | TD.06172 |
Highlights.
Cuprizone-induced de- and remyelination are analyzed by single-nucleus RNA-seq
Oligodendrocytes exhibit a demyelination-specific subset dependent on microglia TREM2
Astrocytes and vascular cells show metabolic, anti-oxidative, and interferon signatures
Microglia acquire a transcription factor MAFB-induced state during remyelination
ACKNOWLEDGMENTS
We thank Tristan Q. Li, Simone Brioschi, and Vincent Peng for scientific discussions. We thank David M. Holtzman for providing the anti-APOE antibody. We acknowledge the Washington University Center for Cellular Imaging for assistance in imaging studies, supported by Washington University School of Medicine, the Children’s Discovery Institute of University and St. Louis Children’s Hospital (CDI-CORE-2015-505 and CDI-CORE-2019-813), the Foundation for Barnes-Jewish Hospital (3770), the Washington University Diabetes Research Center (DK020579), and the Alvin J. Siteman Cancer Center of Washington University School of Medicine and Barnes-Jewish Hospital (CA091842). The Zeiss LSM880 microscope was purchased with support from the Office of Research Infrastructure Programs, National Institutes of Health, a part of the NIH Office of the Director (OD021629). J.A.J.F. is supported by a Chan Zuckerberg Initiative Imaging Scientist award (2020-225726). R.M.G. is supported by the Sao Paulo Research Foundation (FAPESP, 2021/10477-5). M.C. is supported by the NIH (RF1 AG051485, R21 AG059176, and RF1 AG059082) and Cure Alzheimer’s Fund. Graphical abstract and schematics were created with BioRender.com.
Footnotes
DECLARATION OF INTERESTS
M.C. is a member of the scientific advisory board of Vigil, receives research support from Vigil, and is a consultant for CST.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112293.
REFERENCES
- 1.Franklin RJM, and Ffrench-Constant C (2017). Regenerating CNSmyelin - from mechanisms to experimental medicines. Nat. Rev. Neurosci. 18, 753–769. 10.1038/nrn.2017.136. [DOI] [PubMed] [Google Scholar]
- 2.Lassmann H (2018). Multiple sclerosis pathology. Cold Spring Harb. Perspect. Med. 8, a028936. 10.1101/cshperspect.a028936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarbu N, Shih RY, Jones RV, Horkayne-Szakaly I, Oleaga L, and Smirniotopoulos JG (2016). White matter diseases with radiologic-pathologic correlation. Radiographics 36, 1426–1447. 10.1148/rg.2016160031. [DOI] [PubMed] [Google Scholar]
- 4.Nave KA (2010). Myelination and support of axonal integrity by glia. Nature 468, 244–252. 10.1038/nature09614. [DOI] [PubMed] [Google Scholar]
- 5.Ihara M, Polvikoski TM, Hall R, Slade JY, Perry RH, Oakley AE, Englund E, O’Brien JT, Ince PG, and Kalaria RN (2010). Quantification of myelin loss in frontal lobe white matter in vascular dementia, Alzheimer’s disease, and dementia with Lewy bodies. Acta Neuropathol. 119, 579–589. 10.1007/s00401-009-0635-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jäkel S, Agirre E, Mendanha Falcão A, van Bruggen D, Lee KW, Knuesel I, Malhotra D, Ffrench-Constant C, Williams A, and Castelo-Branco G (2019). Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566, 543–547. 10.1038/s41586-019-0903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schirmer L, Velmeshev D, Holmqvist S, Kaufmann M, Werneburg S, Jung D, Vistnes S, Stockley JH, Young A, Steindel M, et al. (2019). Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature 573, 75–82. 10.1038/s41586-019-1404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Absinta M, Maric D, Gharagozloo M, Garton T, Smith MD, Jin J, Fitzgerald KC, Song A, Liu P, Lin JP, et al. (2021). A lymphocyte-microglia-astrocyte axis in chronic active multiple sclerosis. Nature 597, 709–714. 10.1038/s41586-021-03892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falcão AM, van Bruggen D, Marques S, Meijer M, Jäkel S, Agirre E, Samudyata Floriddia EM, Floriddia EM, Vanichkina DP, Ffrench-Constant C, et al. (2018). Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nat. Med. 24, 1837–1844. 10.1038/s41591-018-0236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordão MJC, Sankowski R, Brendecke SM, Sagar Locatelli G, Lo-catelli G, Tai YH, Tay TL, Schramm E, Armbruster S, Hagemeyer N, et al. (2019). Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363, eaat7554. 10.1126/science.aat7554. [DOI] [PubMed] [Google Scholar]
- 11.Zirngibl M, Assinck P, Sizov A, Caprariello AV, and Plemel JR (2022). Oligodendrocyte death and myelin loss in the cuprizone model: an updated overview of the intrinsic and extrinsic causes of cuprizone demyelination. Mol. Neurodegener. 17, 34. 10.1186/s13024-022-00538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan J, Mann T, Joost S, Behrangi N, Frank M, and Kipp M (2020). The cuprizone model: dos and do nots. Cells 9. 10.3390/cells9040843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsushima GK, and Morell P (2001). The neurotoxicant, cuprizone, as a model to study demyelination and remyelination in the central nervous system. Brain Pathol. 11, 107–116. 10.1111/j.1750-3639.2001.tb00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skripuletz T, Lindner M, Kotsiari A, Garde N, Fokuhl J, Linsmeier F, Trebst C, and Stangel M (2008). Cortical demyelination is prominent in the murine cuprizone model and is strain-dependent. Am. J. Pathol. 172, 1053–1061. 10.2353/ajpath.2008.070850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gudi V, Moharregh-Khiabani D, Skripuletz T, Koutsoudaki PN, Kotsiari A, Skuljec J, Trebst C, and Stangel M(2009).Regional differences between grey and white matter in cuprizone induced demyelination. Brain Res. 1283, 127–138. 10.1016/j.brainres.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Gudi V, Gingele S, Skripuletz T, and Stangel M (2014). Glial response during cuprizone-induced de- and remyelination in the CNS: lessons learned. Front. Cell. Neurosci. 8, 73. 10.3389/fncel.2014.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marzan DE, Brügger-Verdon V, West BL, Liddelow S, Samanta J, and Salzer JL (2021). Activated microglia drive demyelination via CSF1R signaling. Glia 69, 1583–1604. 10.1002/glia.23980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lloyd AF, and Miron VE (2019). The pro-remyelination properties of microglia in the central nervous system. Nat. Rev. Neurol. 15, 447–458. 10.1038/s41582-019-0184-2. [DOI] [PubMed] [Google Scholar]
- 19.Skripuletz T, Hackstette D, Bauer K, Gudi V, Pul R, Voss E, Berger K, Kipp M, Baumgärtner W, and Stangel M (2013). Astrocytes regulate myelin clearance through recruitment of microglia during cuprizone-induced demyelination. Brain 136, 147–167. 10.1093/brain/aws262. [DOI] [PubMed] [Google Scholar]
- 20.Shen K, Reichelt M, Kyauk RV, Ngu H, Shen YAA, Foreman O, Modrusan Z, Friedman BA, Sheng M, and Yuen TJ (2021). Multiple sclerosis risk gene Mertk is required for microglial activation and subsequent remyelination. Cell Rep. 34, 108835. 10.1016/j.celrep.2021.108835. [DOI] [PubMed] [Google Scholar]
- 21.Nugent AA, Lin K, van Lengerich B, Lianoglou S, Przybyla L, Davis SS, Llapashtica C, Wang J, Kim DJ, Xia D, et al. (2020). TREM2 regulates microglial cholesterol metabolism upon chronic phagocytic challenge. Neuron 105, 837–854.e9. 10.1016/j.neuron.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Masuda T, Sankowski R, Staszewski O, Böttcher C, Amann L, Sagar Scheiwe C, Scheiwe C, Nessler S, Kunz P, van Loo G, et al. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392. 10.1038/s41586-019-0924-x. [DOI] [PubMed] [Google Scholar]
- 23.Voskuhl RR, Itoh N, Tassoni A, Matsukawa MA, Ren E, Tse V, Jang E, Suen TT, and Itoh Y (2019). Gene expression in oligodendrocytes during remyelination reveals cholesterol homeostasis as a therapeutic target in multiple sclerosis. Proc. Natl. Acad. Sci. USA 116, 10130–10139. 10.1073/pnas.1821306116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berghoff SA, Spieth L, and Saher G (2022). Local cholesterol metabolism orchestrates remyelination. Trends Neurosci. 45, 272–283. 10.1016/j.tins.2022.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947. 10.1523/jneurosci.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcão A, Xiao L, Li H, Häring M, Hochgerner H, Romanov RA, et al. (2016). Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329. 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trimarco A, Forese MG, Alfieri V, Lucente A, Brambilla P, Dina G, Pieragostino D, Sacchetta P, Urade Y, Boizet-Bonhoure B, et al. (2014). Prostaglandin D2 synthase/GPR44: a signaling axis in PNS myelination. Nat. Neurosci. 17, 1682–1692. 10.1038/nn.3857. [DOI] [PubMed] [Google Scholar]
- 28.Ljunggren-Rose Å, Natarajan C, Matta P, Pandey A, Upender I, and Sriram S (2020). Anacardic acid induces IL-33 and promotes remyelination in CNS. Proc. Natl. Acad. Sci. USA 117, 21527–21535. 10.1073/pnas.2006566117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allan D, Fairlie-Clarke KJ, Elliott C, Schuh C, Barnett SC, Lassmann H, Linnington C, and Jiang HR (2016). Role of IL-33 and ST2 signalling pathway in multiple sclerosis: expression by oligodendrocytes and inhibition of myelination in central nervous system. Acta Neuropathol. Commun. 4, 75. 10.1186/s40478-016-0344-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Miller KR, Poliani PL, Cominelli M, Grover S, Gilfillan S, et al. (2020). Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142. 10.1038/s41591-019-0695-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strelau J, Strzelczyk A, Rusu P, Bendner G, Wiese S, Diella F, Altick AL, von Bartheld CS, Klein R, Sendtner M, and Unsicker K (2009). Progressive postnatal motoneuron loss in mice lacking GDF-15. J. Neurosci. 29, 13640–13648. 10.1523/jneurosci.1133-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amstad A, Coray M, Frick C, Barro C, Oechtering J, Amann M, Wischhusen J, Kappos L, Naegelin Y, Kuhle J, and Mehling M (2020). Growth differentiation factor 15 is increased in stable MS. Neurol. Neuroimmunol. Neuroinflamm. 7, e675. 10.1212/nxi.0000000000000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harlow DE, and Macklin WB (2014). Inhibitors of myelination: ECMchanges, CSPGs and PTPs. Exp. Neurol. 251, 39–46. 10.1016/j.expneurol.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.González-Castillo C, Ortuño-Sahagún D, Guzmán-Brambila C, Pallàs M, and Rojas-Mayorquín AE (2014). Pleiotrophin as a central nervous system neuromodulator, evidences from the hippocampus. Front. Cell. Neurosci. 8, 443. 10.3389/fncel.2014.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuboyama K, Fujikawa A, Suzuki R, and Noda M (2015). Inactivation of protein tyrosine phosphatase receptor type Z by pleiotrophin promotes remyelination through activation of differentiation of oligodendrocyte precursor cells. J. Neurosci. 35, 12162–12171. 10.1523/jneurosci.2127-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emery B (2010). Regulation of oligodendrocyte differentiation and myelination. Science 330, 779–782. 10.1126/science.1190927. [DOI] [PubMed] [Google Scholar]
- 37.Sock E, and Wegner M (2019). Transcriptional control of myelination and remyelination. Glia 67, 2153–2165. 10.1002/glia.23636. [DOI] [PubMed] [Google Scholar]
- 38.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kenigsbuch M, Bost P, Halevi S, Chang Y, Chen S, Ma Q, Hajbi R, Schwikowski B, Bodenmiller B, Fu H, et al. (2022). A shared disease-associated oligodendrocyte signature among multiple CNS pathologies. Nat.Neurosci.25,876–886. 10.1038/s41593-022-01104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christophi GP, Gruber RC, Panos M, Christophi RL, Jubelt B, and Massa PT (2012). Interleukin-33 upregulation in peripheral leukocytes and CNS of multiple sclerosis patients. Clin. Immunol. 142, 308–319. 10.1016/j.clim.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jafarzadeh A, Mahdavi R, Jamali M, Hajghani H, Nemati M, and Ebrahimi HA (2016). Increased concentrations of interleukin-33 in the serum and cerebrospinal fluid of patients with multiple sclerosis. Oman Med. J. 31, 40–45. 10.5001/omj.2016.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gadani SP, Walsh JT, Smirnov I, Zheng J, and Kipnis J (2015). The glia-derived alarm in IL-33 orchestrates the immune response and promotes recovery following CNS injury. Neuron 85, 703–709. 10.1016/j.neuron.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Vainchtein ID, Chin G, Cho FS, Kelley KW, Miller JG, Chien EC, Liddelow SA, Nguyen PT, Nakao-Inoue H, Dorman LC, et al. (2018). Astrocyte-derived interleukin-33 promotes microglial synapse engulfment and neural circuit development. Science 359, 1269–1273. 10.1126/science.aal3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cormerais Y, Giuliano S, LeFloch R, Front B, Durivault J, Tambutté E, Massard PA, de la Ballina LR, Endou H, Wempe MF, et al. (2016). Genetic disruption of the multifunctional CD98/LAT1 complex demonstrates the key role of essential amino acid transport in the control of mTORC1 and tumor growth. Cancer Res. 76, 4481–4492. 10.1158/0008-5472.can-15-3376. [DOI] [PubMed] [Google Scholar]
- 45.Bridges RJ, Natale NR, and Patel SA (2012). System xc− cystine/glutamate antiporter: an update on molecular pharmacology and roles within the CNS. Br. J. Pharmacol. 165, 20–34. 10.1111/j.1476-5381.2011.01480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kandasamy P, Gyimesi G, Kanai Y, and Hediger MA (2018). Amino acid transporters revisited: new views in health and disease. Trends Biochem. Sci. 43, 752–789. 10.1016/j.tibs.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Mahmoud S, Gharagozloo M, Simard C, and Gris D (2019). Astrocytes maintain glutamate homeostasis in the CNS by controlling the balance between glutamate uptake and release. Cells 8. 10.3390/cells8020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gautheron J, Morisseau C, Chung WK, Zammouri J, Auclair M, Baujat G, Capel E, Moulin C, Wang Y, Yang J, et al. (2021). EPHX1 mutations cause a lipoatrophic diabetes syndrome due to impaired epoxide hydrolysis and increased cellular senescence. Elife 10, e68445. 10.7554/eLife.68445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Routhe LJ, Andersen IK, Hauerslev LV, Issa II, Moos T, and Thomsen MS (2020). Astrocytic expression of ZIP14 (SLC39A14) is part of the inflammatory reaction in chronic neurodegeneration with iron overload. Glia 68, 1810–1823. 10.1002/glia.23806. [DOI] [PubMed] [Google Scholar]
- 50.Karaca M, Frigerio F, Migrenne S, Martin-Levilain J, Skytt DM, Pajecka K, Martin-del-Rio R, Gruetter R, Tamarit-Rodriguez J, Waage-petersen HS, et al. (2015). GDH-dependent glutamate oxidation in the brain dictates peripheral energy substrate distribution. Cell Rep. 13, 365–375. 10.1016/j.celrep.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 51.Rawji KS, Gonzalez Martinez GA, Sharma A, and Franklin RJM (2020). The role of astrocytes in remyelination. Trends Neurosci. 43, 596–607. 10.1016/j.tins.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 52.Wiese S, Karus M, and Faissner A (2012). Astrocytes as a source for extracellular matrix molecules and cytokines. Front. Pharmacol. 3, 120. 10.3389/fphar.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benner EJ, Luciano D, Jo R, Abdi K, Paez-Gonzalez P, Sheng H, Warner DS, Liu C, Eroglu C, and Kuo CT (2013). Protective astrogenesis from the SVZ niche after injury is controlled by Notch modulator Thbs4. Nature 497, 369–373. 10.1038/nature12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu YE, Pan L, Zuo Y, Li X, and Hong W (2017). Detecting activated cell populations using single-cell RNA-seq. Neuron 96, 313–329.e6. 10.1016/j.neuron.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 55.Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, et al. (2017). A unique microglia type associated with restricting development of alzheimer’s disease. Cell 169, 1276–1290.e17. 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 56.Safaiyan S, Besson-Girard S, Kaya T, Cantuti-Castelvetri L, Liu L, Ji H, Schifferer M, Gouna G, Usifo F, Kannaiyan N, et al. (2021). White matter aging drives microglial diversity. Neuron 109, 1100–1117.e10. 10.1016/j.neuron.2021.01.027. [DOI] [PubMed] [Google Scholar]
- 57.Gabriel TL, Tol MJ, Ottenhof R, van Roomen C, Aten J, Claessen N, Hooibrink B, de Weijer B, Serlie MJ, Argmann C, et al. (2014). Lysosomal stress in obese adipose tissue macrophages contributes to MITF-dependent Gpnmb induction. Diabetes 63, 3310–3323. 10.2337/db13-1720. [DOI] [PubMed] [Google Scholar]
- 58.Shin SM, Cho KS, Choi MS, Lee SH, Han SH, Kang YS, Kim HJ, Cheong JH, Shin CY, and Ko KH (2010). Urokinase-type plasminogen activator induces BV-2 microglial cell migration through activation of matrix metalloproteinase-9. Neurochem. Res. 35, 976–985. 10.1007/s11064-010-0141-3. [DOI] [PubMed] [Google Scholar]
- 59.Gveric D, Hanemaaijer R, Newcombe J, van Lent NA, Sier CF, and Cuzner ML (2001). Plasminogen activators in multiple sclerosis lesions: implications for the inflammatory response and axonal damage. Brain 124, 1978–1988. 10.1093/brain/124.10.1978. [DOI] [PubMed] [Google Scholar]
- 60.Ellwanger DC, Wang S, Brioschi S, Shao Z, Green L, Case R, Yoo D, Weishuhn D, Rathanaswami P, Bradley J, et al. (2021). Prior activation state shapes the microglia response to antihuman TREM2 in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 118, e2017742118. 10.1073/pnas.2017742118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mathys H, Adaikkan C, Gao F, Young JZ, Manet E, Hemberg M, De Jager PL, Ransohoff RM, Regev A, and Tsai LH (2017). Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 21, 366–380. 10.1016/j.celrep.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada González F, Perrin P, et al. (2016). Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670. 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- 63.Yang AC, Vest RT, Kern F, Lee DP, Agam M, Maat CA, Losada PM, Chen MB, Schaum N, Khoury N, et al. (2022). A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature 603, 885–892. 10.1038/s41586-021-04369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, et al. (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480. 10.1038/nature25739. [DOI] [PubMed] [Google Scholar]
- 65.Bagley RG, Honma N, Weber W, Boutin P, Rouleau C, Shankara S, Kataoka S, Ishida I, Roberts BL, and Teicher BA (2008). Endosialin/TEM 1/CD248 is a pericyte marker of embryonic and tumor neovascularization. Microvasc. Res. 76, 180–188. 10.1016/j.mvr.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 66.Delaney C, Farrell M, Doherty CP, Brennan K, O’Keeffe E, Greene C, Byrne K, Kelly E, Birmingham N, Hickey P, et al. (2021). Attenuated CSF-1R signalling drives cerebrovascular pathology. EMBO Mol. Med. 13, e12889. 10.15252/emmm.202012889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kagan HM, and Li W (2003). Lysyl oxidase: properties, specificity, and biological roles inside and outside of the cell. J. Cell. Biochem. 88, 660–672. 10.1002/jcb.10413. [DOI] [PubMed] [Google Scholar]
- 68.Huang X, Qu R, Ouyang J, Zhong S, and Dai J (2020). An overview of the cytoskeleton-associated role of PDLIM5. Front. Physiol. 11, 975. 10.3389/fphys.2020.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Armulik A, Genové G, Mäe M, Nisancioglu MH, Wallgard E, Niaudet C, He L, Norlin J, Lindblom P, Strittmatter K, et al. (2010). Pericytes regulate the blood-brain barrier. Nature 468, 557–561. 10.1038/nature09522. [DOI] [PubMed] [Google Scholar]
- 70.Daneman R, Zhou L, Kebede AA, and Barres BA (2010). Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 468, 562–566. 10.1038/nature09513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin Y, An X, Ye Z, Cully B, Wu J, and Li J (2009). RGS5, a hypoxia-inducible apoptotic stimulator in endothelial cells. J. Biol. Chem. 284, 23436–23443. 10.1074/jbc.M109.032664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Athanasopoulos AN, Schneider D, Keiper T, Alt V, Pendurthi UR, Liegibel UM, Sommer U, Nawroth PP, Kasperk C, and Chavakis T (2007). Vascular endothelial growth factor (VEGF)-induced up-regulation of CCN1 in osteoblasts mediates proangiogenic activities in endothelial cells and promotes fracture healing. J. Biol. Chem. 282, 26746–26753. 10.1074/jbc.M705200200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qi JH, and Anand-Apte B (2015). Tissue inhibitor of metalloproteinase-3 (TIMP3) promotes endothelial apoptosis via a caspase-independent mechanism. Apoptosis 20, 523–534. 10.1007/s10495-014-1076-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murai KK, and Pasquale EB (2011). Eph receptors and ephrins in neuron-astrocyte communication at synapses. Glia 59, 1567–1578. 10.1002/glia.21226. [DOI] [PubMed] [Google Scholar]
- 75.Wellman SM, Guzman K, Stieger KC, Brink LE, Sridhar S, Dubaniewicz MT, Li L, Cambi F, and Kozai TDY (2020). Cuprizone-induced oligodendrocyte loss and demyelination impairs recording performance of chronically implanted neural interfaces. Biomaterials 239, 119842. 10.1016/j.biomaterials.2020.119842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Browaeys R, Saelens W, and Saeys Y (2020). NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162. 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 77.Pillai AM, Thaxton C, Pribisko AL, Cheng JG, Dupree JL, and Bhat MA (2009). Spatiotemporal ablation of myelinating glia-specific neurofascin (Nfasc NF155) in mice reveals gradual loss of paranodal axoglial junctions and concomitant disorganization of axonal domains. J. Neurosci. Res. 87, 1773–1793. 10.1002/jnr.22015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boyle ME, Berglund EO, Murai KK, Weber L, Peles E, and Ranscht B (2001). Contactin orchestrates assembly of the septate-like junctions at the paranode in myelinated peripheral nerve. Neuron 30, 385–397. 10.1016/s0896-6273(01)00296-3. [DOI] [PubMed] [Google Scholar]
- 79.Çolakoglu G, Bergstrom-Tyrberg U, Berglund EO, and Ranscht B (2014). Contactin-1 regulates myelination and nodal/paranodal domain organization in the central nervous system. Proc. Natl. Acad. Sci. USA 111, E394–E403. 10.1073/pnas.1313769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mostafavi S, Gaiteri C, Sullivan SE, White CC, Tasaki S, Xu J, Taga M, Klein HU, Patrick E, Komashko V, et al. (2018). A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nat. Neurosci. 21, 811–819. 10.1038/s41593-018-0154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clark IC, Gutiérrez-Vázquez C, Wheeler MA, Li Z, Rothhammer V, Linnerbauer M, Sanmarco LM, Guo L, Blain M, Zandee SEJ, et al. (2021). Barcoded viral tracing of single-cell interactions in central nervous system inflammation. Science 372, eabf1230. 10.1126/science.abf1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Argaw AT, Asp L, Zhang J,Navrazhina K, Pham T, Mariani JN, Mahase S, Dutta DJ, Seto J, Kramer EG, et al. (2012). Astrocyte-derived VEGF-A drives blood-brain barrier disruption in CNS inflammatory disease. J. Clin. Invest. 122, 2454–2468. 10.1172/jci60842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sherafat A, Pfeiffer F, Reiss AM, Wood WM, and Nishiyama A (2021). Microglial neuropilin-1 promotes oligodendrocyte expansion during development and remyelination by trans-activating platelet-derived growth factor receptor. Nat. Commun. 12, 2265. 10.1038/s41467-021-22532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goudarzi S, Rivera A, Butt AM, and Hafizi S (2016). Gas6 promotes oligodendrogenesis and myelination in the adult central nervous system and after lysolecithin-induced demyelination. ASN neuro 8, 1759091416668430. 10.1177/1759091416668430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ye P, Li L, Richards RG, DiAugustine RP, and D’Ercole AJ (2002). Myelination is altered in insulin-like growth factor-I null mutant mice. J. Neurosci. 22, 6041–6051. 10.1523/jneurosci.22-14-06041.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Poliani PL, Wang Y, Fontana E, Robinette ML, Yamanishi Y, Gilfillan S, and Colonna M (2015). TREM2 sustains microglial expansion during aging and response to demyelination. J. Clin. Invest. 125, 2161–2170. 10.1172/jci77983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cantoni C, Bollman B, Licastro D, Xie M, Mikesell R, Schmidt R, Yuede CM, Galimberti D, Olivecrona G, Klein RS, et al. (2015). TREM2 regulates microglial cell activation in response to demyelination in vivo. Acta Neuropathol. 129, 429–447. 10.1007/s00401-015-1388-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gouna G, Klose C, Bosch-Queralt M, Liu L, Gokce O, Schifferer M, Cantuti-Castelvetri L, and Simons M (2021). TREM2-dependent lipid droplet biogenesis in phagocytes is required for remyelination. J. Exp. Med. 218, e20210227. 10.1084/jem.20210227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lampron A, Larochelle A, Laflamme N, Préfontaine P, Plante MM, Sánchez MG, Yong VW, Stys PK, Tremblay MÈ, and Rivest S (2015). Inefficient clearance of myelin debris by microglia impairs remyelinating processes. J. Exp. Med. 212, 481–495. 10.1084/jem.20141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cignarella F, Filipello F, Bollman B, Cantoni C, Locca A, Mikesell R, Manis M, Ibrahim A, Deng L, Benitez BA, et al. (2020). TREM2 activation on microglia promotes myelin debris clearance and remyelination in a model of multiple sclerosis. Acta Neuropathol. 140, 513–534. 10.1007/s00401-020-02193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee SH, Rezzonico MG, Friedman BA, Huntley MH, Meilandt WJ, Pandey S, Chen YJJ, Easton A, Modrusan Z, Hansen DV, et al. (2021). TREM2-independent oligodendrocyte, astrocyte, and T cell responses to tau and amyloid pathology in mouse models of Alzheimer disease. Cell Rep. 37, 110158. 10.1016/j.celrep.2021.110158. [DOI] [PubMed] [Google Scholar]
- 92.Pandey S, Shen K, Lee SH, Shen YAA, Wang Y, Otero-García M, Kotova N,Vito ST,Laufer BI,Newton DF,et al.(2022).Disease-associated oligodendrocyte responses across neurodegenerative diseases. Cell Rep. 40, 111189. 10.1016/j.celrep.2022.111189. [DOI] [PubMed] [Google Scholar]
- 93.Hasel P, Rose IVL, Sadick JS, Kim RD, and Liddelow SA (2021). Neuroinflammatory astrocyte subtypes in the mouse brain. Nat. Neurosci. 24, 1475–1487. 10.1038/s41593-021-00905-6. [DOI] [PubMed] [Google Scholar]
- 94.Soucie EL, Weng Z, Geirsdóttir L, Molawi K, Maurizio J, Fenouil R, Mossadegh-Keller N, Gimenez G, VanHille L, Beniazza M, et al. (2016). Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Science 351, aad5510. 10.1126/science.aad5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aziz A, Soucie E, Sarrazin S, and Sieweke MH (2009). MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science 326, 867–871. 10.1126/science.1176056. [DOI] [PubMed] [Google Scholar]
- 96.Kelly LM, Englmeier U, Lafon I, Sieweke MH, and Graf T (2000). MafB is an inducer of monocytic differentiation. EMBO J. 19, 1987–1997. 10.1093/emboj/19.9.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Das A, Bastian C, Trestan L, Suh J, Dey T, Trapp BD, Baltan S, and Dana H (2019). Reversible loss of hippocampal function in a mouse model of demyelination/remyelination. Front. Cell. Neurosci. 13, 588. 10.3389/fncel.2019.00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bénardais K, Kotsiari A, Skuljec J, Koutsoudaki PN, Gudi V, Singh V, Vulinovic F, Skripuletz T, and Stangel M (2013). Cuprizone [bis(cyclohexylidenehydrazide)] is selectively toxic for mature oligodendrocytes. Neurotox. Res. 24, 244–250. 10.1007/s12640-013-9380-9. [DOI] [PubMed] [Google Scholar]
- 99.Kim J, Eltorai AEM, Jiang H, Liao F, Verghese PB, Kim J, Stewart FR, Basak JM, and Holtzman DM (2012). Anti-apoE immunotherapy inhibits amyloid accumulation in a transgenic mouse model of Aβ amyloidosis. J. Exp. Med. 209, 2149–2156. 10.1084/jem.20121274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, and Dudoit S (2018). Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 19, 477. 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Büttner M, Ostner J, Müller CL, Theis FJ, and Schubert B (2021). scCODA is a Bayesian model for compositional single-cell data analysis. Nat. Commun. 12, 6876. 10.1038/s41467-021-27150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M, Piccio L, Hernandez M, and Colonna M (2006). Cutting edge: TREM-2 attenuates macrophage activation. J. Immunol. 177, 3520–3524. 10.4049/jimmunol.177.6.3520. [DOI] [PubMed] [Google Scholar]
- 103.Hoshino K, Kashiwamura S, Kuribayashi K, Kodama T, Tsujimura T, Nakanishi K, Matsuyama T, Takeda K, and Akira S (1999). The absence of interleukin 1 receptor-related T1/ST2 does not affect T helper cell type 2 development and its effector function. J. Exp. Med. 190, 1541–1548. 10.1084/jem.190.10.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Konishi H, Okamoto T, Hara Y, Komine O, Tamada H, Maeda M, Osako F, Kobayashi M, Nishiyama A, Kataoka Y, et al. (2020). Astrocytic phagocytosis is a compensatory mechanism for microglial dysfunction. EMBO J. 39, e104464. 10.15252/embj.2020104464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278. 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun LO, Mulinyawe SB, Collins HY, Ibrahim A, Li Q, Simon DJ, Tessier-Lavigne M, and Barres BA (2018). Spatiotemporal control of CNS myelination by oligodendrocyte programmed cell death through the TFEB-PUMA Axis. Cell 175, 1811–1826.e21. 10.1016/j.cell.2018.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hanaichi T, Sato T, Iwamoto T, Malavasi-Yamashiro J, Hoshino M, and Mizuno N (1986). A stable lead by modification of Sato’s method. J. Electron. Microsc. 35, 304–306. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-nucleus RNA-seq data have been deposited at the Gene Expression Omnibus (GEO) database and are publicly available as of the date of publication under accession number GSE204770.
Original code has been deposited at Github at: https://github.com/marcocolonnalab/CPZ.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


