Abstract
Insecticide resistance in insects severely threatens both human health and agriculture, making insecticides less compelling and valuable, leading to frequent pest management failures, rising input costs, lowering crop yields, and disastrous public health. Insecticide resistance results from multiple factors, mainly indiscriminate insecticide usage and mounted selection pressure on insect populations. Insects respond to insecticide stress at the cellular level by modest yet significant genetic propagations. Transcriptional, co-transcriptional, and post-transcriptional regulatory signals of cells in organisms regulate the intricate processes in gene expressions churning the genetic information in transcriptional units into proteins and non-coding transcripts. Upregulation of detoxification enzymes, notably cytochrome P450s (CYPs), glutathione S-transferases (GSTs), esterases [carboxyl choline esterase (CCE), carboxyl esterase (CarE)] and ATP Binding Cassettes (ABC) at the transcriptional level, modification of target sites, decreased penetration, or higher excretion of insecticides are the noted insect physiological responses. The transcriptional regulatory pathways such as AhR/ARNT, Nuclear receptors, CncC/Keap1, MAPK/CREB, and GPCR/cAMP/PKA were found to regulate the detoxification genes at the transcriptional level. Post-transcriptional changes of non-coding RNAs (ncRNAs) such as microRNAs (miRNA), long non-coding RNAs (lncRNA), and epitranscriptomics, including RNA methylation, are reported in resistant insects. Additionally, genetic modifications such as mutations in the target sites and copy number variations (CNV) are also influencing insecticide resistance. Therefore, these cellular intricacies may decrease insecticide sensitivity, altering the concentrations or activities of proteins involved in insecticide interactions or detoxification. The cellular episodes at the transcriptional and post-transcriptional levels pertinent to insecticide resistance responses in insects are extensively covered in this review. An overview of molecular mechanisms underlying these biological rhythms allows for developing alternative pest control methods to focus on insect vulnerabilities, employing reverse genetics approaches like RNA interference (RNAi) technology to silence particular resistance-related genes for sustained insect management.
Keywords: insects, insecticide resistance, detoxification enzymes, pathways, ncRNAs, RNA methylation
Introduction
Insects are the most common species on the planet, inhabiting and interacting with fauna and flora in ecological systems, including humans. Few insect species serve humans and their wellbeing by pollinating crops, scavenging garbage, and performing other tasks. On the other hand, many insect species adversely affect public health, crops, hygiene, and other sectors (Eggleton, 2020). When a biological equilibrium is disturbed, insects expand uncontrollably, wreaking havoc on humans, threatening food production, spreading human diseases, and requiring pest control treatments. The principal pest-reduction strategy employs chemicals that challenge insects and their reproduction. Insecticides are chemical or biological molecules used to kill or otherwise inhibit insects from engaging in damaging behaviors (Manyilizu, 2019).
According to the Insecticide Resistance Action Committee (IRAC) Mode of Action (MoA) Classification Version 10.5 March 2023, 36 insecticide groups are available, each of which contains a sub-group, class, or exemplifying active ingredient of the main groups (IRAC, 2023), indicating that these insecticides differ in structure, synthesis, and mode of action. The diversity of these compounds can be seen in their availability, which includes chlorinated hydrocarbons, organophosphates, carbamates, pyrethroids, neonicotinoids, formamidines, phenylpyrazoles, sulfoximines, spinosyns, juvenile hormone analogs, benzoylureas, buprofezin, cyromazine, and other molecules, as well as botanical and microbial agents (Simon, 2014; IRAC, 2023).
Long-term insecticide efficacy is crucial for successful and sustainable food and fiber production and public health. However, given numerous reports of chemical agents’ ineffectiveness in suppressing insect populations, prolonged use of insecticides has had unexpected consequences, most notably the emergence of insecticide-resistant insect pests. Insecticide resistance (IR) is common and widespread, nearly nine decades after synthetic pesticides proved popular in pest management. IR is defined as a reduction in an insect population’s susceptibility to a previously effective insecticide caused by the continued use and/or possible cross-selection with other chemical substances, which occurs through genetic, physiological, or behavioral changes and is also a hereditary trait (WHO, 2016; Oppold and Müller, 2017; IRAC, 2023).
The San Jose scale, Comstockaspis perniciosa Comstock (Hemiptera: Diaspididae), demonstrated the first known insecticide resistance to lime sulfur in 1914 (Melander, 1914). Since the discovery and widespread application of DDT and other synthetic insecticides in the late 1940s, the number of resistant insect species has steadily expanded. Insecticide resistance to 339 insecticides and five insecticidal characteristics expressed in genetically modified plants has been documented for 602 insect species as of 2019 (Sparks and Nauen, 2015; Sparks et al., 2019). The most notorious insect at the top of the list of resistant insects is Plutella xylostella (Linnaeus) (Lepidoptera: Plutellidae), which has evolved resistance to 101 different active ingredients of insecticides (APRD, 2023). Pests like the Colorado potato beetle, Leptinotarsa decimlineata (Say) (Coleoptera: Chrysomelidae), the two-spotted spider mite, Tetranychus urticae Koch (Trombidiformes: Tetranychidae), and the green peach aphid, Myzus persicae (Sulzer) (Hemiptera: Aphididae) are each resistant to 56, 84, and 96 different insecticides, respectively (APRD, 2023). Unsurprisingly, worldwide reports of insecticide resistance to the majority of WHO-approved public health insecticides have been made (Ranson et al., 2011; Moyes et al., 2017). Human disease vectors like the malaria mosquito Anopheles sacharovi Favre (Diptera: Culicidae), Anopheles albimanus Wiedemann (Diptera: Culicidae), and the house fly Musca domestica Linnaeus (Muscidae: Diptera) have also been resistant to 20, 21, and 65 different insecticidal compounds, respectively (APRD, 2023). Ninety percent of malaria-endemic countries have documented resistance in Anopheles mosquitoes to at least one class of insecticide, with 32% reporting resistance to pyrethroids, carbamates, organophosphates, and organochlorines that were recommended until 2016 (WHO, 2012). IR eliminates pest management alternatives and may lower agricultural profitability. The availability of new insect-challenging chemicals is becoming increasingly difficult because of rising costs for discovery, development, and registration, fueled partly by public concerns about environmental safety and human health (Van Leeuwen et al., 2020).
Resistance is an evolutionary phenomenon characterized by toxicodynamic and toxicokinetic changes in the physiology and biochemistry of resistant strains, resulting in shifts in penetration, activation, metabolism, transport, and excretion - altering the amount of toxin that reaches the target site (toxicokinetic mechanisms), and alterations to the pesticide target-site due to structural changes, knock-out, and amplify mechanisms (toxicodynamic mechanisms) (Kennedy and Tierney, 2012; Feyereisen et al., 2015).
Insecticide resistance mechanisms can be broadly classified as follows: 1) behavioral resistance, 2) fitness cost, 3) penetration resistance, 4) target-site resistance, 5) metabolic resistance, and 6) resistance-inducing operational parameters (Siddiqui et al., 2023). Insect pests overcome both the host plant defenses and the toxicity of insecticides to adapt and survive. Detoxification genes are essential for pests to withstand plant poisons and insecticides at the molecular level. Many research studies in arthropods have shown that microRNAs (miRNAs/miRs) play critical roles in physiological and developmental pathways such as metamorphosis, embryogenesis, molting, reproduction, immunity, wing development, and metabolism of plant toxins and insecticide resistance (Qiao et al., 2019). MiRNAs are abundant in the insects’ genomes and are important regulators of gene expression in response to xenobiotic stressors.
Non-coding RNAs (ncRNAs) are crucial for managing insecticide resistance and pest control (Etebari et al., 2015). Recently, miRNAs associated with resistance have been found to target detoxification genes. Along with their verified target genes, their regulatory roles in insecticide resistance and detoxification in various pests have also been established (Zhang et al., 2021). The growing body of information suggests that oxidative and other cellular stress influence miRNA expression. This review majorly summarised miRNAs associated with insecticide-resistant pests and their potential relevance in insect pest management. Information on the involvement of epitranscriptomic regulation, long non-coding RNAs, and the xenobiotic pathways regulating detoxification genes has also been reviewed.
Non-coding RNAs
The ncRNAs, diverse RNA molecules, including ribosomal RNA (rRNA), transfer RNA (tRNA), small ncRNAs (sncRNAs), and long ncRNAs (lncRNAs), are categorized based on their length and intended function. Small interfering RNA (siRNA), small nuclear RNA (snRNA), and PIWI-interacting RNAs (piRNA) with fewer than 200 nucleotides are classed as sncRNAs. Their sizes range from 18 to 25 nucleotides for small RNAs like siRNAs and miRNAs and from 20 to 200 nucleotides for other small RNAs. On the other hand, ncRNAs of more than 200 nucleotides are classified as long non-coding RNAs (lncRNAs), which are found in practically all eukaryotic creatures (Mercer et al., 2009). All of these RNAs typically operate as transcriptional and translational regulators.
MicroRNAs
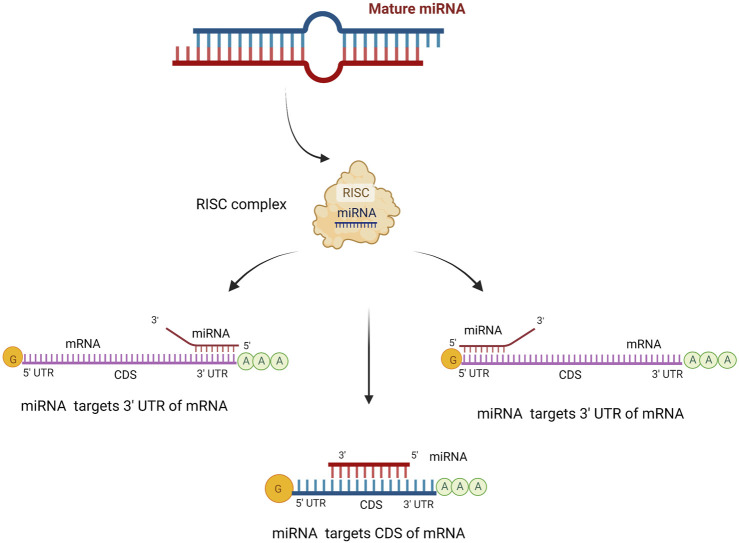
MicroRNAs bind to the 3′-untranslated regions (UTR) of the messenger RNA of target genes via imperfect base pairing between the miRNA’s “seed” sequence (nucleotides 2-8 at its 5′end) and its complementary seed match sequence, causing post-transcriptional gene expression regulation. Initially, it was assumed that miRNA target sequences could only be found in the 3′UTR of target mRNAs. However, leads from research imply that target sequences may reside in the open reading frame, 5′UTR, and 3′UTR (Bartel, 2009) (Figure 1).
FIGURE 1.
The target site of miRNA. MiRNAs can silence the expression of target genes by binding to 5′or 3′untranslated regions (UTRs) or coding sequence (CDS) of mRNA. MiRNA can bind with different complementarities, such as incomplete pairing in UTR regions and complete pairing in the CDS region. Created with BioRender.com.
In a search for genes essential for post-embryonic development in the nematode Caenorhabditis elegans (Rhabditida: Rhabditidae), the first miRNA, lin-4, was found (Lee et al., 1993). Though, at the time, it was practically regarded as a genetic anomaly, the discovery of the lin-4 locus and its regulatory mechanism through the 3′UTR of lin-14 mRNA was intriguing. However, the identification of another miRNA, let-7, first in C. elegans (Reinhart et al., 2000) and then in many bilaterian species (Pasquinelli et al., 2000), proved that the interactions between lin-4 and lin-14 were not at all unusual but rather a new and fundamental layer of the mechanisms governing gene expression (Lai et al., 2003).
MicroRNA biogenesis
MiRNAs may be encoded from non-coding transcripts, introns, or coding regions. MiRNA genes, mostly independent transcriptional units, are predominantly transcribed by RNA polymerase II as a primary miRNA (pri-miRNA), which may contain one or more stem loops. Like mRNAs, pri-miRNA transcripts are 5′capped and polyadenylated (Bracht et al., 2004). Drosha, associated with Pasha (equivalent to DGCR8 in mammals), further processes the stem loop into a short hairpin of around 70 bases known as the precursor miRNA (Bartel, 2009). Pre-miRNA hairpins may also be directly processed from primary mRNA transcripts by splicing and debranching of short introns, referred to as mirtrons. After being carried into the cytoplasm by Exportin-5, the terminal loop of pre-miRNA is removed by ribonuclease enzyme Dicer-1 (Dcr-1), resulting in a miRNA:miRNA* duplex with two nucleotide overhangs on both ends (Hutvagner et al., 2001). The duplex is integrated into the RNA Inducing Silencing Complex (RISC), which is mainly made up of the Argonaute-1 (Ago-1) protein (Miyoshi et al., 2009). The miRNA strand (guide strand) subsequently directs the RISC complex to the target mRNA once the miRNA* strand (passenger strand) has been torn and degraded.
MicroRNA classification
MiRNA classification is based on the “seed sequence” region because selection pressures appear to regulate the nucleotide substitution pattern in miRNA genes and because it is a functionally important region (Brennecke et al., 2003; Bartel, 2009). In the case of metazoans, 858 miRNA families have been deposited in the miRBase database (v21.0) (Griffiths-Jones et al., 2007), with 254 (30%) of these families found in at least five species. These records may vary as more high-throughput sequencing experiments are performed; however, current statistics show that most miRNA families (562) are discovered in vertebrates, followed by insects (178 families recorded). Different miRNA families may have varying degrees of conservation in the seed region. The miRNAs miR-100, miR-125, and let-7 illustrate a well-conserved seed region (Behura, 2007). There are insect-specific miRNA families, including bantam, miR-2, and miR-3, whose seed region is likewise highly preserved.
MicroRNA in insects
According to the InsectBase version 2.0 database, 112,162 miRNAs in 807 insects have been discovered (Mei et al., 2022). The insect with the highest number of miRNAs (576) is Aedes albopictus (Skuse) (Diptera: Culicidae). Insect development, including the formation of the germ cell, the wing, and the muscle, the neurogenesis, the apoptosis, and phenotypic plasticity, is greatly influenced by miRNA (Xu et al., 2003; Hilgers et al., 2010; Truscott et al., 2011; Ge et al., 2012; Li and Padgett, 2012). Specific miRNAs, including miR-263, miR-14, bantam, and the miR-2 family, have been discovered to affect apoptosis in Drosophila melanogaster Meigen (Diptera: Drosophilidae). Apoptosis is controlled by miR-263a/b as well. miR-8, a highly conserved miRNA, has been linked to the insulin signaling system in the fat body of Drosophila larvae (Hyun et al., 2009). The conserved miRNA miR-14 has been demonstrated to play a role in 20-hydroxyecdysone (20E) signaling pathway of Drosophila. The miR-14 mutant has a shortened lifespan because miR-14 participates in the 20E signaling pathway (Varghese and Cohen, 2007).
In insects, carbon dioxide (CO2) receptors are structured differently, resulting in various olfactory behaviors. Feeding-related behavior was correlated with olfactory detection of CO2 through neurons present in the mouthparts of an insect, such as maxillary palps (MPs) and labial palps. In the absence of miR-279 in Drosophila, a CO2 sensing system is developed in the maxillary palps, similar to that reported in mosquitoes (Cayirlioglu et al., 2008). miR-14, discovered in insulin-producing cells in the fly brain, regulates insulin production and metabolism in Drosophila, which explains why miR-14 mutant flies are metabolically deficient (Varghese et al., 2010). miR-34 expression increases with age in the Drosophila brain (Liu N. et al., 2012), and miR-34 deficiency results in accelerated brain aging, late-onset brain degeneration, and lower survival, whereas miR-34 over-expression extends the median lifespan and reduces neurodegeneration. In Drosophila, miR-8 has been associated with neurodegeneration prevention (Karres et al., 2007). miR-7 functions within genes implicated in photoreceptor and proprioceptor determination in Drosophila to protect these networks from environmental changes and other pressures (Li et al., 2009).
MiRNAs miR-31a, let-7, miR-279, and miR-275 were over-expressed in the honey bee, Apis mellifera Linnaeus (Hymenoptera: Apidae) nurses than foragers, but miR-13b, miR-133, miR-210, miR-278, and miR-92a were downregulated in nurses compared to foragers (Liu F. et al., 2012). Putative miRNAs have been experimentally identified and mapped to the pea aphid genome. Two parthenogenetic pea aphid morphs, sexuparae and virginoparae, differed in their expression of five miRNAs: miR-34, miR-X47 and miR-X103, miR-307*, and miR-X52* (Legeai et al., 2010).
Although insects lack adaptive immunity, parts of their innate immunity involved in cellular and humoral responses can identify foreign things and then express the appropriate reaction to the foreign intruder in their presence. These include melanization, phagocytosis, nodule/capsule development, antimicrobial peptide synthesis, wound healing, and nodule/capsule formation (Lemaitre and Hoffmann, 2007). Due to their capacity to control gene expression at the post-transcriptional level, miRNAs may be essential for preserving the homeostasis and plasticity of immunity. For instance, it has been demonstrated that miR-8 negatively regulates the expression of antimicrobial peptides, such as Drosomycin and Diptericin, in Drosophila to keep their expression level low during typical non-infection settings, facilitating the homeostasis of immunity (Choi and Hyun, 2012).
Databases related to microRNAs
In the post-genomic era, biological data are being created at an increasing rate with the development of high-throughput sequencing technologies (Katz et al., 2022). Using next-generation sequencing (NGS) technology, researchers could predict miRNAs from target insects more quickly and inexpensively. Experts worldwide established databases to globalize biological data, which might aid novice researchers in making quick and accurate discoveries on miRNAs. The primary databases with the bulk of information on insect miRNAs are InsectBase 2.0 and miRBase; additional useful databases are given in Table 1.
TABLE 1.
Different databases related to miRNAs in insects.
| Database | Information packed | References |
|---|---|---|
| miRBase | This database contains miRNA sequences, both mature and precursors of 31 insect species | Kozomara et al. (2019) |
| InsectBase2.0 | This database contains many miRNAs (112,162) from 801 insect species | Mei et al. (2022) |
| ENA (European Nucleotide Archive) | It contains 2,661 miRNA sequences of more than 30 insects | Burgin et al. (2023) |
| MirGeneDB 2.1 | 9 insect species from four insect orders contains 1372 miRNAs | Fromm et al. (2020) |
| FlyBase | A database exclusively for Drosophila Genes and Genomes where it contains 440 miRNA sequences | Larkin et al. (2021) |
| BmncRNAdb | Database dedicated to bombyx mori, which contains 1,986 miRNAs | Zhou et al. (2016) |
MicroRNA regulation in insecticide metabolism
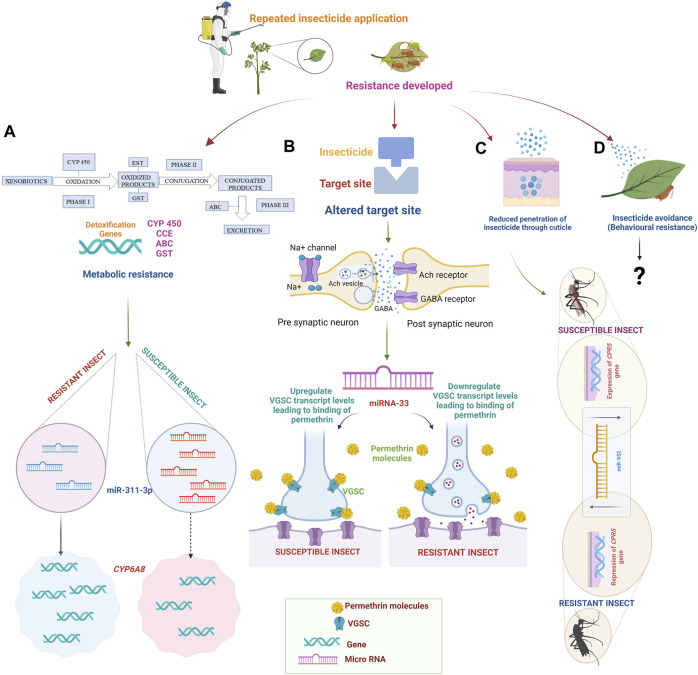
Four factors are primarily involved in the evolution of insect resistance to insecticides: increased metabolic capacity for detoxification, target insensitivity (Troczka et al., 2012), delayed cuticular penetration, and behavioral modification (Liu, 2007) (Figure 2).
FIGURE 2.
Mechanism of insecticide resistance. Due to the repeated application of insecticides, insects develop resistance. (A) Target site insensitivity: Mutations in the target receptors of insecticides. Nicotinic acetylcholine receptors (nAChRs), Gamma-aminobutyric acid (GABA), Voltage-gated sodium channel (VGSC), and Acetylcholine esterase (Ach) receptors may face resistance due to alterations in the structure or expression levels of these receptors. In permethrin susceptible insects, miR-33 upregulates the transcript level of VGSC, leading to increased binding of permethrin to VGSC. In the case of permethrin-resistant insects, the binding of permethrin to VGSC is less due to the downregulation of VGSC by miR-33. (B) Metabolic resistance: Insects possess a wide range of metabolic enzymes, such as cytochrome P450 monooxygenases (P450s), esterases, and glutathione S-transferases (GSTs), which can detoxify insecticides. miRNAs can bind to target detoxification gene mRNA molecules, leading to mRNA translational repression or degradation. The upregulation of miR-311-3p degrades the mRNA of the CYP6AB gene resulting in lower expression of CYP6AB in susceptible insects. In contrast, the resistant insects had overexpression of CYP6AB due to downregulation of miR-311-3p. (C) Reduced cuticular penetration: Insects possess a cuticle that acts as a barrier to the penetration of insecticides. Resistance can occur through the thickening or modification of the cuticle, reducing insecticide uptake. The CPR5 gene has been highly expressed in susceptible insects due to the downregulation of miR-932. However, the CPR5 gene is repressed in resistant insects due to the upregulation of miR-932. (D) Behavioral resistance: Some insects can develop behavioral changes that reduce their insecticide exposure. They may avoid treated areas, alter feeding or breeding behaviors, or exhibit reduced contact with insecticides. The involvement of microRNA in behavioral resistance has not been reported. Created with BioRender.com.
According to most researchers, metabolic resistance is the primary mechanism underlying the early emergence of resistance (Hyun et al., 2009). Metabolic resistance is also intimately linked to the differential expression of genes encoding detoxification enzymes (Feng et al., 2018). Primary phase I, which entails hydrolysis or oxidation, and secondary phase II, which entails conjugating phase I products, are the two detoxification steps (Berenbaum and Johnson, 2015). Esterases (EST), monooxygenases (like those found in cytochrome P-450 (CYP)), and transferases (like glutathione-S-transferase (GST)) are only a few of the many enzymes that are crucial to its function. The overproduction of specific enzymes causes insecticides to degrade before binding to their target sites, and with these excessively generated enzymes, pests can develop resistance to insecticides. Notably, these enzymes degrade xenobiotics into non-toxic molecules. A list of studies describing insect microRNAs involved in detoxification of and resistance to different insecticides is reported in Supplementary Table S1.
Lepidoptera
Diamondback moth, Plutella xylostella
The diamondback moth, P. xylostella, is a severe pest of crucifer crops such as cauliflower, mustard, radish, turnip, Chinese cabbage, broccoli, rape, and kale (Ganehiarachchi and Knodel, 2008). It has become one of the most resistant pests in the world due to the repeated application of insecticides. It has developed resistance against organophosphates, pyrethroids, new molecules, and microbial-derived pesticides (Furlong et al., 2013). To date, 1010 cases of insecticide resistance to more than 101 insecticides have been recorded due to dependence on insecticides to control diamondback moth (APRD, 2023). In P. xylostella, enhanced detoxifying enzyme activity (56%) and altered target sites (44%) are the two leading causes of different pesticide resistance (Banazeer et al., 2021). The pesticide resistance in P. xylostella has been demonstrated to be significantly influenced by miRNA. Recently, chlorantraniliprole has been one of the pesticides most frequently employed on P. xylostella. It is an anthranilic diamide insecticide that opens up the muscles’ ryanodine receptor (RyR) (Lahm et al., 2009). Plutella xylostella has developed resistance to chlorantraniliprole due to point mutations in the RyR gene and increased activity of detoxifying enzymes like CYPs, carboxylesterase (CarE), and GSTs (Liu et al., 2015).
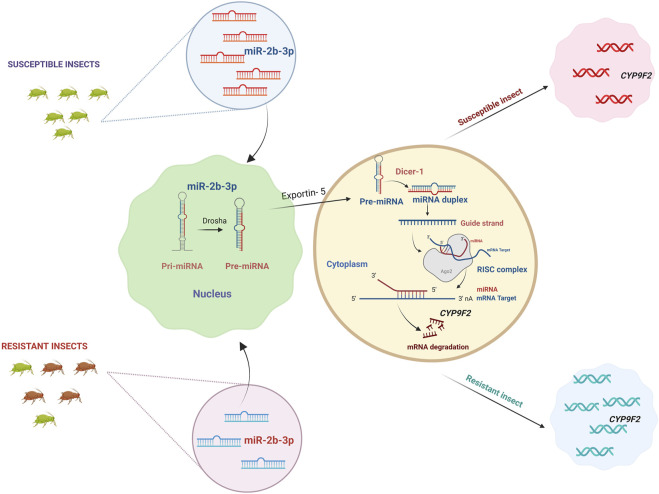
Detoxifying enzyme CYP9F2 has a binding site for miR-2b-3p in its 3′UTR, but the binding sites for the other two miRNAs, miR-14b-5p and let-7-5p, were CYP9F2 and CYP307a1, and GST and CYP9F2, respectively (Etebari et al., 2018) (Figure 3). Four common differentially expressed miRNAs (miR-8491-5p, miR-4969-5p, mir-8488-5p, and novel-13_1575) in chlorantraniliprole exposed P. xylostella two resistant and one susceptible strain. At the same time, miR-8533-3p, miR-8534-5p, and miR-375-5p were downregulated, and their corresponding targets included larval cuticle protein LCP-30, CYP6B6, and CYP4G15, respectively were upregulated after chlorantraniliprole exposure (Zhu et al., 2017) (Figure 4).
FIGURE 3.
miRNAs regulating detoxification genes. miRNA genes are transcribed by RNA polymerase II to primary miRNA (pri-miRNA) with one or more stem-loops. Drosha further processes the stem loop into precursor miRNA. Pre-miRNA is carried into the cytoplasm by Exportin-5, and the terminal loop of pre-miRNA is removed by ribonuclease enzyme Dicer-1 (Dcr-1), resulting in a miRNA:miRNA* duplex. The duplex is integrated into the RNA Inducing Silencing Complex (RISC), mainly made up of the Argonaute-1 (Ago-1) protein. The miRNA strand (guide strand) subsequently directs the RISC complex to the target mRNA, and the miRNA* strand (passenger strand) will be degraded. miRNAs primarily bind to the mRNA of target molecules, leading to mRNA translational repression or degradation. They are involved in the post-transcriptional regulation of detoxification gene expression involved in insecticide metabolism, and the expression is negatively correlated between microRNAs and detoxification genes. In the case of susceptible insects, the microRNA miR-2b-3p is upregulated, where these microRNAs bind to the mRNA of detoxification gene CYP9F2 and degrade them. The CYP9F2 genes produced are fewer in number, which is not sufficient for insecticide detoxification, making the insect susceptible. In contrast to susceptible insects, miR-2b-3p is downregulated, which upregulates CYP9F2 genes, and the insect creates resistance. Created with BioRender.com.
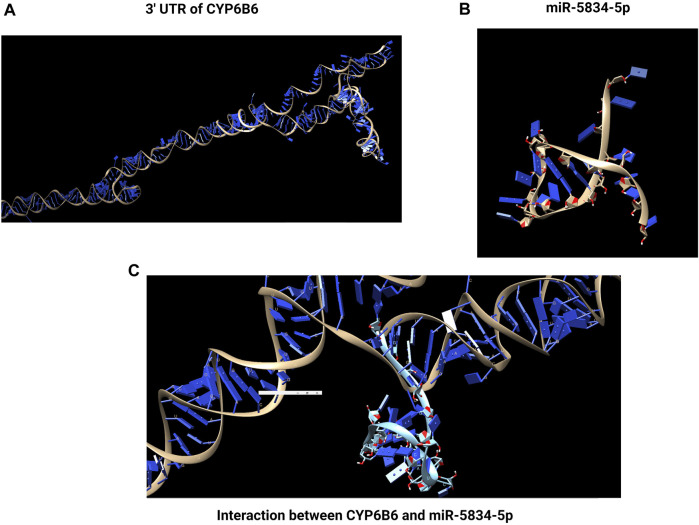
FIGURE 4.
Interaction between miRNA and detoxification gene. The binding of a miRNA to its target site typically occurs in the 3′untranslated region (UTR) of the mRNA. In chlorantraniliprole exposed to P. xylostella, miR-8534-5p was downregulated, and its corresponding target CYP6B6 was upregulated. (A) 3′ UTR of CYP6B6. (B) miR-5834-5p. (C) Interaction of CYP6B6 and miR-5834-5p. Created with BioRender.com.
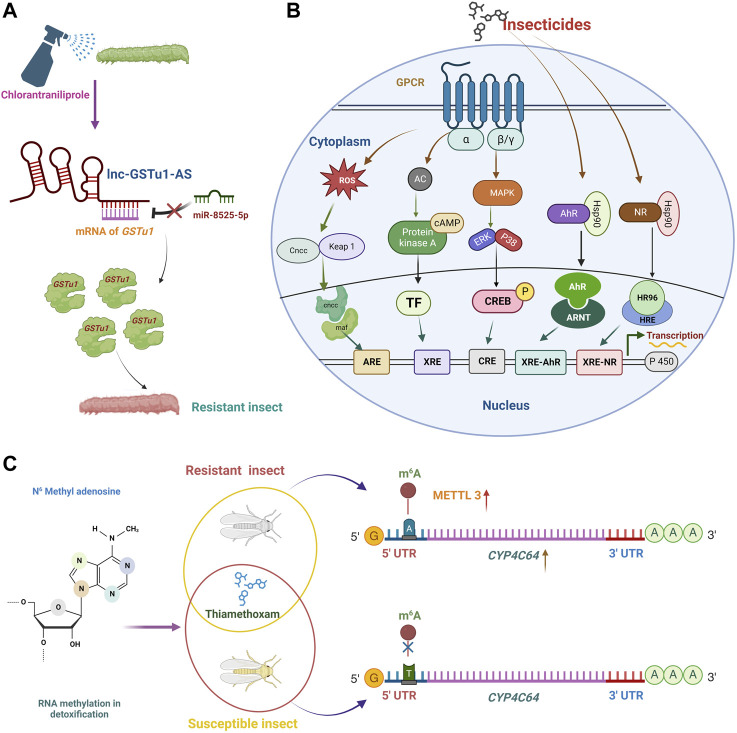
Seven possible miRNAs were predicted to target the PxEcR-B following fufenozide treatment. Plutella xylostella showed a 2.28-fold increase in the expression of miR-189942 and a 29% decrease in the expression of PxEcR-B (Li et al., 2020). The ATP-binding cassette transporter (ABC transporter) proteins have been recognized as essential receptors for several Cry toxins in insects. By interacting with the ABCC2 gene’ coding sequence (CDS) in opposition to the Cry1Ac toxin, miR-998-3p enhanced Cry toxins resistance in P. xylostella (Zhu et al., 2020). Besides CYPs, GSTs also significantly impact P. xylostella ability to withstand chlorantraniliprole. For instance, lnc-GSTu1-AS, antisense transcript formed an RNA duplex with GSTu1, preventing miR-8525-5p from binding at the GSTu1-3′ UTR and therefore masked GSTu1 degradation that could have been induced by miR-8525-5p and thus increased the resistance of P. xylostella to chlorantraniliprole (Zhu et al., 2021) (Figure 5A).
FIGURE 5.
The regulation of insecticide resistance mechanisms involves epi-transcriptome signals, detoxification signaling pathways, and long non-coding RNAs (lncRNAs). (A) lncRNAs influence miRNA-mediated insecticide resistance regulation. Regulation mechanisms include lncRNA-miRNA-mRNA interaction and lncRNA-mediated regulation of miRNA expression. GSTu1, a detoxifying gene, is involved in the chlorantraniliprole resistance of P. xylostella. A long non-coding RNA, lnc-GSTu1-AS, interacted with GSTu1 by forming an RNA duplex, masking the binding site of microRNA, miR-8525-5p, at the GSTu1-3′ UTR. lnc-GSTu1-AS maintained the mRNA stability of GSTu1 by preventing its degradation, which could have been induced by miR-8525-5p and thus resulting in increased production of detoxification gene (GSTu1), increased the resistance of P. xylostella to chlorantraniliprole. (B) Detoxification enzyme induction pathway. This pathway involves upregulating detoxification enzymes, such as cytochrome P450 monooxygenases, esterases, and glutathione S-transferases. They are the CncC/Keap1, NR, PKA, MAPK/CREB, and AhR/ARNT pathways. The activated molecules of these pathways, such as Cncc/maf, TF, CREB, Ahr/AHRNT, and HR96/HRE, respectively, in the cytoplasm, interact with their corresponding response element in the nucleus to regulate the expression of detoxification genes through transcription. The arrows indicate the cascade of effectors in the signaling pathway. (C) N6-methyladenosine (m6A) is a modified form of adenosine widely involved in gene expression regulation. Mutation (T to A at position-206 bp) was observed in the 5′UTR of CYP4C64 that was observed at a much greater frequency in the thiamethoxam-resistant strains compared with the susceptible strain. The T at 206 bp helps bind m6A, and the overexpression of the enzyme METTL (methyltransferase) led to the development of thiamethoxam-resistant insects. CncC, Cap‘n’ Collar isoform C; Maf, Musculoaponeurotic fibrosarcoma; ARE, Antioxidant responsive element; Gas, G protein alpha unit which stimulates adenyl cyclase; AC, adenyl cyclase; cAMP, cyclic adenosine monophosphate; PKA, protein kinase A; TF, Transcription factor; XRE, xenobiotic response element; MAPK, mitogen-activated protein kinase; ERK, extracellular regulated protein kinase; P38, P38 mitogen-activated protein kinase; CREB, c-AMP response element binding protein; -P, phosphorylation; CRE, cAMP response element; NR, nuclear receptor; AhR, aryl hydrocarbon receptor; Hsp90, heat shock protein 90; ARNT, aryl hydrocarbon receptor nuclear translocator; XRE-NR, xenobiotic response element-nuclear receptor; XRE-AhR, xenobiotic response element-aryl hydrocarbon receptor. Created with BioRender.com.
ABCG20, a member of the ABCG subfamily, was discovered to be substantially expressed in Cry1Ac-susceptible P. xylostella population, and its associated miRNA, a novel-miR-310, was predicted. Thirty-four miRNAs were discovered to have at least one binding site that targets the CDS of ABCG20. In contrast to the ABCG20 expression pattern, novel-miR-310 was much more abundant in the Cry1S1000 strain (resistant) than the G88 strain (susceptible). A high-throughput sequencing analysis of small RNA libraries constructed from the midguts of the P. xylostella Cry1Ac-resistant strain and the Cry1Ac-susceptible strain revealed 12 differentially expressed miRNAs between the strains, with specific, nine miRNAs downregulated and three upregulated in the resistant strain. The discovered mRNA targets were genes involved in the cellular process, metabolism, membrane and catalytic activity, and the Hippo, MAPK signaling pathway (Yang et al., 2022).
The juvenile hormone esterase (JHE) gene PxJHE, whose inhibition increases Cry1Ac protoxin resistance, was differently expressed in the Cry1Ac-resistant and Cry1Ac-susceptible strains. Two novel miRNAs (miR-108 and miR-234) that were in inverse connection with the degree of PxJHE expression were predicted to target the PxJHE CDS (Yang et al., 2023).
Fall armyworm, Spodoptera frugiperda
The fall armyworm, Spodoptera frugiperda (Smith) (Lepidoptera: Noctuidae), is a critical migratory pest worldwide (Zhao et al., 2020). Spodoptera frugiperda is a severe maize pest with over 80 distinct crop hosts. Excessive pesticide use has resulted in resistance to 29 active insecticidal chemicals across six modes of action (Chao et al., 2019; APRD, 2023). Target site insensitivity is one of the reported mechanisms of insecticide resistance in S. frugiperda, such as the RyR that confers resistance to diamide insecticides (Boaventura et al., 2020), acetylcholinesterase (AChE) that confers resistance to carbamates and organophosphates, and voltage-gated sodium channel (VGSC) imparts resistance to synthetic pyrethroids (Carvalho et al., 2013). Resistance to pyrethroids, organophosphates, and carbamates is caused by metabolic detoxification, wherein chromosomal changes result in GST, CYP, and CarE gene amplification, over-expression, and alteration. Chlorantraniliprole significantly increased and decreased the expression of CYP6K2 and miR-190-5p in S. frugiperda by 2.96-fold and 56.5%, respectively (Zhang M. Y. et al., 2022).
Other Lepidopterans
The Asian spongy moth, Lymantria dispar (Linnaeus) (Lepidoptera: Erebidae), is a global forest pest that kills over 500 plant species (Zhang et al., 2023a). examined miRNA and mRNA levels in L. dispar larvae treated with cyantraniliprole, the anthranilic diamide insecticide of the second generation, used to combat Lepidopteran, Coleopteran, Dipteran, and Hemipteran pests (Liu, 2007; Foster et al., 2012). Eleven differently expressed miRNAs predicted twenty-one genes relevant to insecticide resistance, with 25 miRNA-mRNA interactions discovered. CYP4C1 was the only differentially expressed gene in the miRNA-mRNA network influenced by novel-miR-4 upregulation (Zhang et al., 2023a). A lncRNA in cadherin allele intron 20 was recently found to modulate cadherin 1 transcription in the pink bollworm Pectinophora gossypiella (Saunders) (Lepidoptera: Gelechiidae). The lncRNA promotes PgCad1 transcription and pink bollworm sensitivity to Cry1Ac (Li S. et al., 2019).
Planthoppers
The brown planthopper (BPH), Nilaparvata lugens (Stal) (Hemiptera: Delphacidae), is a highly harmful pest in rice-growing parts of Asian countries (Hereward et al., 2020). Because of the extensive use of insecticides, BPH has developed high levels of resistance to the major classes of insecticides, covering 34 active components of pesticides, with 453 recorded resistance cases worldwide, including neonicotinoids, phenylpyrazoles, carbamates, pyridine azomethine derivatives, and inhibitors of chitin biosynthesis (Liao et al., 2021; APRD, 2023). Resistance mechanisms in BPH have included target-site mutation and upregulation of detoxifying enzyme genes (Bao and Zhang, 2019).
The ABC transporter was involved in N. lugens resisting nitenpyram and clothianidin. Fourteen and four ABC genes were considerably increased in nitenpyram- and clothianidin-resistant N. lugens strains, respectively, with ABCD3 and ABCG3 highly over-expressed in both nitenpyram and clothianidin-resistant strains. The novel_268 miRNA has been predicted to target the ABCD3 and ABCG3 CDS (Li et al., 2022). Furthermore (Mao et al., 2022), discovered 72 differently expressed miRNAs in N. lugens, with 29 miRNAs being over-expressed and 28 miRNAs being downregulated on exposure to nitenpyram. The bioinformatics study showed that novel 85 and novel 191 have been predicted to target the CDS of CYP6ER1 and CarE1, respectively.
Aphids
The Aphididae family contains around 4,000 species of aphids, although just 20 are highly polyphagous and infested plants from 50 families (Blackman and Eastop, 2000). Aphids are almost resistant to most commonly used insecticides such as organophosphates, carbamates, pyrethroids, neonicotinoids, and newer insecticides (Guo et al., 2017). miRNA influences aphid resistance development activities through i) post-translational modification of CYP genes, ii) downregulation of insecticide receptors, and iii) causing the over-expression of acetyl co A carboxylase, a crucial enzyme in fatty acid biosynthesis. Recent advances in next-generation sequencing (NGS) technologies have facilitated the identification of aphid miRNAs. Acyrthosiphon pisum (Harris) (Hemiptera: Aphididae) was the first aphid to have its miRNA sequenced and used as a reference database (Legeai et al., 2010).
MiRNA profiling of M. persicae revealed 22 miRNAs that actively bind to the target gene CYP6CY3, which modulates resistance to nicotine, the plant’s secondary metabolite. Among these 22 miRNAs, let-7 and miR-100 are linked to post-translational alteration of the CYP6CY3 gene, which leads to enhanced nicotinic compound breakdown (Peng et al., 2016). Similarly, Aphis gossypii (Glover) (Hemiptera: Aphididae) adapts to gossypol, a plant toxin, and other secondary metabolites such as tannic acid by overexpressing CYP genes (Du et al., 2004; Peng et al., 2017). A novel CYP450 gene, CYP4CJ1, discovered in A. gossypii, was associated with the mechanism of gossypol resistance, and CYP4CJ1 gene over-expression was governed by miR-4133-3p via reduced expression (Ma et al., 2019). Spirotetramat is a cyclic keto-enol insecticide targeting the enzyme acetyl-CoA carboxylase (ACC) essential for lipid biosynthesis (Nauen et al., 2008). Two miRNAs, miR-276 and miR-3016, were downregulated in the spirotetramat-resistant strain of A. gossypii, involving post-translational modification of the ACC gene (Wei et al., 2016).
In Sitobion miscanthi (Takahashi) (Hemiptera: Aphididae), downregulation of miR-278 and miR-263b is essential in targeting the nicotinic acetylcholine receptors nAchRα1 and nAchRβ1, resulting in imidacloprid insensitivity. Also, the downregulation of miR-316 targets the over-expression of the CYP4CJ6 gene, resulting in imidacloprid resistance (Zhang B. Z. et al., 2022).
Mite, Tetranychus cinnabarinus
Over 100 crops, including cotton and beans, are fed by the carmine spider mite, Tetranychus cinnabarinus (Boisduval) (Trombidiformes: Tetranychidae) (Jia et al., 2011). The primary method of mite management involves using insecticides and acaricides, which inevitably results in the development of resistance. Due to their high fecundity, short generation period, and high inbreeding tendency, mites are more predisposed to pesticide resistance problems than other crop pests. In New York in 1949, there was a first-ever report of parathion resistance in T. cinnabarinus. Variations in the activity of several detoxification enzymes, including MFO, GST, and CarE, are linked to acarid resistance. Tetranychus cinnabarinus developed cyflumetofen resistance in 2014, with significantly greater activity levels of the detoxifying enzymes CarE, CYPs, and GSTs (Wang et al., 2014).
The miR-1-3p (miR-1 family) levels were lower in cyflumetofen-resistant T. cinnabarinus and that the detoxifying enzyme gene TCGSTM4 (mu class GST gene) was a target of miR-1-3p (Zhang et al., 2018). It was the first report of a miRNA and its target implicated in acaricide (cyflumetofen) resistance in T. cinnabarinus. Fenpropathrin is a broad-spectrum insecticide widely employed against mites and pests of many crops (Solomon et al., 2001). Tetranychus cinnabarinus resistance mechanism to fenpropathrin comprises a mutation in the sodium channel gene (F1538I), target site resistance, and increased enzyme activity of GST, CYP, and carboxyl choline esterase (CCEs). Using transcriptome sequencing, 4,454 lncRNAs in the carmine spider mite T. cinnabarinus were discovered (Feng et al., 2020). Among these, the detoxifying enzyme gene TcGSTm02 and lincRNA_Tc13743.2 each had a miRNA (miR-133-5p) response element. A cyflumetofen-resistant strain of T. cinnabarinus (CyR) was found to over-express lincRNA_Tc13743.2 and TcGSTm02, while miR-133-5p was downregulated.
Mosquito, Culex pipiens pallens
Mosquitoes are vectors for various illnesses that can significantly affect human health. Mosquitoes must be managed effectively to prevent and control mosquito-borne illnesses. Excessive usage of pesticides against mosquitoes in recent decades has resulted in insecticide resistance, which impedes efficient control. Studies show that the targeted areas’ decreasing sensitivity has led to complex multiple insecticide resistance. miRNAs degrade target mRNAs and regulate host-pathogen interactions, metabolism, development, and pesticide resistance.
miRDeep prediction methods assisted in discovering nine novel miRNAs from C. pipiens pallens (Linnaeus) (Diptera: Culicidae). The most abundant among them is miR-13664, which was identified to target the 3′-UTR of CYP314A1 (Sun et al., 2019). miR-932 was found to be 1.8-fold over-expressed, resulting in a 2.8-fold decrease of its target CPR5 (Liu et al., 2016). In another instance, miR-4448 was 6.49-fold more abundant while its predicted target gene, CYP4H31, was 2.77-fold less abundant in the deltamethrin-susceptible strain (Li et al., 2021a). CYP6N23, CYP6AG11, CYP9J35, CYP325BG3, and CYP6Cp1 were shown to be inversely linked with their respective miRNAs (miR-285, miR-278-3P, miR2, miR-71). CYP9J35, on the other hand, had a positive connection with its regulator miR-13 (Fahmy et al., 2020).
Fruit fly, Drosophila melanogaster
The dichloro diphenyl trichloroethane (DDT) resistance in the fruit fly, D. melanogaster, was due to a single CYP gene, CYP6G1 (monogenic) in low-level DDT resistance phenotype (Daborn et al., 2001; Le Goff and Hilliou, 2017) but moderate to high-level DDT resistance was found to be polygenic due to multiple resistance genes (Kim et al., 2018). Drosophila metabolic resistance to DDT has been linked to constitutively over-expressed genes and higher levels of CYPs (P450s), GSTs, and ESTs (Tu and Akgül, 2005). The 3′UTR sequences of DDT-resistant D. melanogaster over-expressed genes CYP6G1, CYP6G2, CYP6A8, and CYP4G1 had a target site for their respective regulator miRNAs miR-310-3p, miR-311-3p, miR-312-3p, miR-313-3p, and miR-92a-3p, which exhibited downregulation following DDT exposure (Seong et al., 2019).
Long non-coding RNAs
Although lncRNAs are not translated into proteins, they have a similar structure to mRNA. The lncRNAs, previously considered insignificant, are increasingly garnering attention due to their regulatory involvement in various biological processes in animals and plants. The lncRNAs have a role in the stability and translation of mRNAs, pre-mRNA splicing, and protein activities and also serve as precursors of siRNA and miRNA in post-transcriptional control. Based on InsectBase version 2.0 database, 1,293,430 lncRNAs in 376 insects have been discovered (Mei et al., 2022). According to the region of the genome that was transcribed, lncRNAs are divided into four categories: 1) sense lncRNAs overlap exonic regions of another transcript made from the same strand; 2) antisense lncRNAs are present on the complementary strand of the sense strand; 3) intergenic lncRNAs (lincRNAs), which are made from the DNA between two genes (intergenic regions); 4) bidirectional lncRNAs are concurrently transcribed at the opposing strands from coding transcripts (Quinn and Chang, 2016). Recent research has demonstrated the importance of lncRNAs in all aspects of insect development, reproduction, and genetic plasticity (Choudhary et al., 2021). According to recent studies, lncRNAs have been linked to enhanced fitness, xenobiotic sensitivity, and pesticide resistance (Lawrie et al., 2022).
Researchers discovered 6,171 lncRNA transcripts from Bactrocera dorsalis (Hendel) (Diptera: Tephritidae) malathion-resistant (MR1) and susceptible (MS) strains, including 3,728 lincRNAs, 653 antisense lncRNAs, 1,402 intronic lncRNAs, and 388 sense lncRNAs. Twenty-seven of these lncRNAa were expressed similarly in both males and females of the MR1 strain, with only 15 lncRNAs upregulated and 12 downregulated. Among them, the MR1 strain cuticle showed significant levels of expression of the lncRNAs lnc15010.10 and lnc3774.2, indicating that these two lncRNAs may be related to malathion resistance (Meng et al., 2021).
Differential gene expression patterns of lncRNAs in A. gossypii revealed 6,059 lncRNAs in spirotetramat-resistant (SR) and susceptible (SS) strains. Among them, 874 lncRNAs were differentially expressed, of which MSTRG.28822.1, MSTRG.28822.2, MSTRG.28822.3, MSTRG.28822.4, and MSTRG.28822.5, were predicted to be acetyl-CoA carboxylase (ACC) targeting. A combined study of reverse transcription real-time quantitative PCR (RT-qPCR) and RNA interference (RNAi) confirmed that the selected ACC lncRNA was related to the ACC expression it was predicted found that the transcription factors, C/EBP and C/EBPzeta were regulating ACC lncRNA (Peng et al., 2021).
Furthermore, when the CYP380C6, CYP4CJ1, CYP6DA2, CYP6CY7, and CYP6CY21 genes which were discovered to be important for spirotetramat resistance in A. gossypii SR strain, when were ectopically expressed in Drosophila resulted in significantly decreased mortality after spirotetramat exposure. Silencing investigations revealed that lncRNAs MSTRG.36649.2/5 and MSTRG.71880.1 influence CYP6CY21 and CYP380C6 expression, affecting the sensitivity of the SR strain to spirotetramat (Peng et al., 2022).
Totally 11,978 lncRNAs, including 3,136 intergenic lncRNAs, 7,393 intronic lncRNAs, and 1,449 antisense lncRNAs, were identified from indoxacarb susceptible (SS) and resistant strains (Lab-InRS and Field-FInRS) of Spodoptera litura (Fabricius) (Lepidoptera: Noctuidae). Compared with the SS, 51 and 134 lncRNAs were upregulated and downregulated in the two resistant strains, respectively, and 908 differentially expressed mRNAs were their target genes. Expression of 14 P450s, seven CCEs, one GST, six UGTs, five ABC transporters, and 24 cuticle protein genes was under 112 differentially expressed lncRNAs. By sponging 10 miRNAs, 79 differentially expressed lncRNAs controlled the expression of 14 detoxifying and 19 cuticle protein genes that lead to indoxacarb resistance. The regulatory pathways of lncRNA-mRNA and lncRNA-miRNA-mRNA resulted in indoxacarb resistance with 47 differentially expressed lncRNAs. The involvement of LNC_004867 and LNC_006576 in S. litura indoxacarb resistance was confirmed through molecular and bioassay studies (Shi et al., 2022).
Mutations
In nitenpyram-resistant (NitR) and imidacloprid-resistant (ImiR) strains of N. lugens, the transcription factor FoxO controlled CYP4CE1 expression. In the differential influence of FoxO on CYP4CE1 expression, CYP4CE1 promoter sequence variations between susceptible and resistant insects were discovered. Single-nucleotide polymorphisms (SNPs) in six FoxO response locations predicted in the CYP4CE1 promoter were found in more than 50% of NitR and ImiR strains. The active control of CYP4CE1 expression by FoxO was primarily caused by two mutations, −650T/G and −2205T/A, in two response elements located at −648 bp and −2,200 bp, respectively (Zhang H. et al., 2023).
The GSTe2 polymorphism analysis discovered a strong link between the point mutation and DDT resistance in the malaria vector Anopheles funestus, Giles (Diptera: Culicidae) with a single amino acid substitution L119F in the upregulated GSTe gene provided significant levels of metabolic resistance (Riveron et al., 2014).
PxABCG1 (Pxwhite), an ABC transporter gene, is implicated in the downregulation of P. xylostella Bt Cry1Ac toxin-functional midgut receptor, and the gene expression has been shown to be transcriptionally controlled. It was found that Antennapedia (Antp), a Hox family transcription factor, interacted with a cis-response element (CRE) in the PxABCG1 promoter of the susceptible strain to stimulate gene expression. A cis-acting mutation, on the other hand, inhibited Antp from binding to the CRE and regulating the PxABCG1 gene creating Cry1Ac resistance (Qin et al., 2021).
There were seven base alterations (M1-M7) in the CYP6B7 promoter associated with resistance between fenvalerate-resistant (HDTJFR) and susceptible (HDTJ) strains of H. armigera (Hubner) (Lepidoptera: Noctuidae). pGL3-CYP6B7 reporter genes with varied mutation sites revealed that genes with M3, M4, and M7 mutations had significantly lower fenvalerate-induced activity. The transcription factors Ubx and Br were over-expressed in HDTJFR, with binding sites containing M3 and M7, respectively, confirming their role in fenvalerate resistance in Helicoverpa armigera (Huang et al., 2023).
Cloning and characterization of the CYP332A1 gene linked with fenvalerate resistance in H. armigera revealed that the gene was more expressed in the resistant (BJ) strain than in the susceptible (HDS) strain. The resistant strain’s sequence had five amino acid changes (F21C, G28R, K64Q, A290S, and V477I). The 5′-flanking region of CYP332A1 had potential binding sites for the transcription factors Dr, Ubx, Cf2, caup, ara, Antp, ftz, eve, and otp, and the resistant BJ strain had three significant deletions of 386–392 bp, 547–554 bp, and 965–971 bp (Huang et al., 2022).
ACC gene was significantly over-expressed in the spirotetramat-resistant strain compared to the laboratory-selected resistant and susceptible strains of A. gossypii. The full-length ACC gene sequenced from resistant and susceptible cotton aphids indicated a substantial relationship between spirotetramat resistance and 14 amino acid alterations in the ACC gene’s biotin carboxylase domain and carboxyl transferase domain (Pan et al., 2017).
Copy number variation of detoxification genes and insecticide resistance
A specific DNA sequence copies vary in number among individual genomes, which is referred to as copy number variation (CNV). CNV in insect genomes is a rich source of potentially adaptive polymorphism, which may assist in overcoming the restrictions of purifying selection on conserved genes and allow for increased transcription (Weetman et al., 2018). CNV of detoxification genes such as GSTs, CYPs, ESTs, UGTs, and oxidative stress genes have been found in insects such as mosquitoes, tobacco cutworm, and fall armyworm (S. frugiperda). Adaptive evolution of multi-copy detoxifying genes has been implicated in insecticide resistance. Considerable allelic differentiation of genomic copy number changes between fall armyworm S. frugiperda regional populations but not among host-plant-based strains.
Insecticides containing organophosphates and carbamates, widely utilized to reduce mosquito populations worldwide, resulted in resistance to both insecticides and are conferred by the identical amino acid alteration (G119S) in the ace-1 gene. G119S mutation is part of homogenous duplications that associate multiple resistant copies of the ace-1 gene in Anopheles gambiae and C. pipiens (Gimenez et al., 2020). Multiple copies provided higher degrees of resistance, demonstrating the adaptability of the genetic architecture of resistance to organophosphate and carbamate insecticides surrounding the ace-1 locus (Milesi et al., 2022). Because of an increased gene copy number, overexpression of the CYP6CY3 gene gives neonicotinoid resistance in the aphid M. persicae (Kirkland et al., 2023). Copy number variation increased CYP6G1 gene expression in D. melanogaster (Schmidt et al., 2010). In brown planthopper N. lugens, the CYP6ER1 gene had been duplicated, and allelic variations of some of the duplicated genes encoded enzymes that may metabolize imidacloprid (Heckel, 2021).
Regulation of the expression of detoxification genes in insects
As inducing agents and substrates, xenobiotics are known to cause the over-expression of broad groups of genes involved in detoxification. Transcriptional regulation is frequently driven by cis-regulatory elements (cis-acting), which are short sequences within the promoter region that certain transcription factors bind (trans-acting) to and further recruit the transcriptional machinery (Guo et al., 2018). Several insect transcription regulatory pathways, such as AhR/ARNT, HR96, ROS/CncC/Keap1, GPCR/PKA, and MAPK/CREB pathways, govern insecticide and phytochemical detoxification (Figure 5B). A list of studies describing different transcription factors regulated by these pathways is reported in Supplementary Table S2.
AhR/ARNT pathway
The xanthotoxin cascade is one of the first regulatory networks for detoxifying genes revealed in insects (Sogawa et al., 1995). They are found in insects and mammals and have bHLH DNA binding domains, Per-ARNT-Sim (PAS) protein-protein interaction domains, and ligand-binding domains (Denison et al., 1988; Hahn, 2002). In the cytoplasm, the molecular chaperone heat shock protein 90 (Hsp90) detects the inactivated forms of the Aryl hydrocarbon Receptor (AhR), the xenobiotic sensor. Following exposure to and binding to a variety of ligands, such as toxic substances, AhR is activated and translocated to the nucleus, where it heterodimerizes with ARNT (Aryl Hydrocarbon Receptor Nuclear Translocator) to affect the expression of numerous genes interacting with Xenobiotic Response Elements to AhR (XRE-Ahr) (Nakata et al., 2006).
NlAhR and NlARNT bound the NlCarE7 promoter, significantly enhancing the transcriptional activity in N. lugens resistant to imidacloprid, etofenprox, and sporocarp. In Locusta migratoria (Orthoptera: Acrididae), AhR is associated with chlorpyrifos susceptibility by regulating LmGSTd7 expression (Zhang et al., 2019).
In the cotton aphid A. gossypii, CYP6DA2 is linked to gossypol and spirotetramat tolerance. In resistant strains of A. gossypii, AhR transcript levels were 9-fold more significant than in susceptible strains (Peng et al., 2017). Similarly, AhR/ARNT pathway was also engaged in nicotine tolerance in M. persicae via over-expression of CYP6CY3 and CYP6CY4 (Pan et al., 2019). It was fascinating to learn that CYP6CY3 is controlled by microRNAs, namely, let-7 and miR-100 (Peng et al., 2016). Transcription beginning point in the CYP6B6a promoter contained a short region of 138 bp that was highly connected to promoter activity in 2-tridecanone exposed H. armigera larvae (Li F. et al., 2014). This sequence (5′ -CATGACACCTG-3′) was comparable to Xenobiotic Response Element (XRE), suggesting that 2-tridecanone regulation of CYP6B6 was mediated through the AhR/ARNT pathway. In chlorantraniliprole-exposed P. xylostella, the gene CYP6B6, was discovered to be controlled by miR-8534-5p (Zhu et al., 2017). Different pathways regulating detoxification genes and miRNAs are represented in Table 2.
TABLE 2.
Pathways mediating in regulation of insect xenobiotic detoxification genes and microRNAs.
| Detoxification enzyme | Reported insecticide | Reported insect | Pathway | miRNA associated | Study source | |
|---|---|---|---|---|---|---|
| Target enzyme | miRNA | |||||
| CYP6CY3 | Nicotine | Myzus persicae | AhR/ARNT | let 7 miR 100 | Pan et al. (2019) | Peng et al. (2016) |
| CYP6B6 | 2-tridecanone | Helicoverpa armigera Plutella xylostella | AhR/ARNT | miR-8534-5p | Li et al. (2014a) | Zhu et al. (2017) |
| CYP6ER1 | Imidacloprid | Nilaparvata lugens | CncC Pathway | Novel_85 | Tang et al. (2020) | Mao et al. (2022) |
| Nitenpyram | Novel_191 | |||||
| CYP6G1 | DDT | Drosophila melanogaster | HR96 pathway | miR-310-3p | Cheesman et al. (2013) | Seong et al. (2019) |
| CYP6A8 | DDT | D. melanogaster | CncC Pathway | miR-312-3p | Misra et al. (2013) | Seong et al. (2019) |
NR (Nuclear receptors) pathway
Three insect NRs, including HR96 (hormone receptor-like in 96), Hnf4 (Hepatocyte nuclear factor 4), and Ftz-f1 (Ftz transcription factor 1), have been implicated in phytochemical and insecticide regulation of P450s (Li et al., 2021b).
HR96 (Hormone receptor-like in 96)
Hormone receptor-like in 96 (HR96) is the nuclear receptor superfamily (NR) transcription factor. The DNA binding domain (DBD), which consists of two zinc fingers, is a highly conserved functional domain in nuclear receptors. The ligand binding domain (LBD), which forms a ligand binding pocket, dimerization unit, and transactivation domain, is a less conserved functional domain (Germain et al., 2006; Markov and Laudet, 2011).
When activated, the two primary xenobiotic-binding nuclear receptors (NRs), constitutive androstane receptor (CAR), and Steroid and xenobiotic receptor/Pregnane X Receptor (SXR/PXR), translocate to the nucleus and dimerize with the retinoid-X receptor (RXR) to promote detoxification gene transcription. The ortholog HR96 gene in invertebrates represents CAR/PXR. Most arthropod genomes have it, including T. urticae (Grbic et al., 2011), D. melanogaster (King-Jones et al., 2006), Tribolium castaneum (Herbst) (Coleoptera: Tenebrionidae) (Xu et al., 2010), A. mellifera (Velarde et al., 2006), and S. frugiperda (Giraudo et al., 2015). Furthermore, it has been proposed that HR96 acts as a dimerizing partner of ultraspiracle protein (USP), the closest insect RXR orthologue, and as a dimerizing partner of insect ecdysone receptors (EcR) in mediating transcriptional regulation of the insect detoxification process (Giraudo et al., 2015).
In Drosophila, DHR96 is a crucial mediator of xenobiotic tolerance. The binding motif of DHR96 was found multiple times in the promoter region of two detoxification genes, GSTe1 and CYP6G1. DDT is metabolized by CYP6G1 via the HR96 route (Cheesman et al., 2013). Surprisingly, miR-310-3p affected the gene CYP6G1 in D. melanogaster (Seong et al., 2019). TcHR96 (Tribolium HR96) over-expression caused by imidacloprid detoxification considerably boosted the promoter activity of genes such as CYP4Q4, CYP4G7, CYP4BR3, and CYP345A1 (Kim et al., 2021).
HNF4 (Hepatocyte nuclear factor)
HNF4a, a member of the NR2A subfamily, acts as an important transactivator of P450 genes involved in drug metabolism and clearance (Jover et al., 2001). A single HNF4a ortholog called HNF4 is widely expressed in insect species and plays a role in diverse processes, including lipid mobilization, b-oxidation, insulin signaling, glucose homeostasis, and metabolic control. An imidacloprid-resistant strain (Res) exhibited a 251.69-fold resistance to imidacloprid compared to the susceptible counterpart (Sus). The expression level of Hepatocyte nuclear factor 4 (HNF4) in the Res strain was lower than that in Sus. When the HNF4 was silenced, UGT-1-7, UGT-2B10, and CYP6ER1 were significantly higher in the Res strain than in the Sus strain by 1.73-, 1.63-, and 4.94-fold, respectively. So, it was apparent that imidacloprid-resistant N. lugens was negatively regulated by HNF4 (Cheng et al., 2021).
FTZ (Fushi Tarazu)
Upregulation of CYP6BG1 was responsible for chlorantraniliprole resistance in P. xylostella. The transcriptional factor FTZ-F1, an orphan nuclear receptor that binds to the FTZ gene, was found to regulate the transcriptional activity of CYP6BG1. Chlorantraniliprole potentiated the expression levels of FTZ-F1 and CYP6BG1 and was significantly higher in the resistant populations (Li X. et al., 2019).
CncC/Keap1 pathway
The Cap'n'collar isoform C/Kelch-like ECH associated protein 1 (CncC/Keap1) pathway, a “master regulator” of gene transcription coding for enzymes, was shown to be involved in response to xenobiotic and oxidative stress. CncC is an ortholog of mammalian NF-E2-Related Factor 2 (Nrf2), a transcription factor in the basic leucine zipper (bZIP) family (Suzuki and Yamamoto, 2015). CncC(Nrf2) is extensively expressed and rapidly destroyed by the action of Keap1 under normal physiological circumstances. Nonetheless, during oxidative stress, ROS (reactive oxygen species) modify the CncC (Nrf2)/Keap1 complex, preventing CncC (Nrf2) breakdown. CncC interacts with Muscle aponeurosis fibromatosis (Maf) to promote the expression of genes with Antioxidant Response Elements (AREs) motif in their upstream region, such as numerous P450s and GSTs coding genes, which increases cellular ROS levels.
Temporal expression profiles of B. dorsalis revealed that the transcription factor MafB and detoxification genes were strongly expressed in the fat body and that abamectin stimulated the expression of MafB, GSTz2, and CYP473A3 (Tang et al., 2019). Upstream sequence study of chlorpyrifos and cypermethrin-resistant Spodoptera exigua (Hubner) (Noctuidae: Lepidoptera) revealed that three GSTs (SeGSTo2, SeGSTe6, and SeGSTd3) have the same CncC/Maf binding site, while SeGSTo2 and SeGST6 share the AhR/ARNT binding site. In the presence of CncC and Maf proteins, luciferase activity driven by the GSTe6 promoter was raised, and the presence of AhR and ARNT also boosted the transcriptional activity of the GSTe6 promoter (Hu et al., 2019).
The SlituCncC gene of S. litura was shown to be more abundant in the Malpighian tubules, fat body, and midgut tissues of third and fourth instar larvae exposed to indoxacarb resistant strains than susceptible strains. When SlituCncC was knocked down in S. litura, 842 genes were downregulated, and 127 were upregulated. Six of these downregulated genes (CYP367A1, CYP367B1, CYP341B21, CYP340L2, SlituCXE1, and SlituABCH-1) were related with indoxacarb resistance in S. litura (CYP367A1, CYP367B1, CYP341B21, CYP340L2, SlituCXE1, and SlituABCH-1). The promoter regions of these six genes were predicted for the presence of CncC-Maf binding sites (Shi et al., 2021).
The CncC/Keap1 pathway was constitutively activated in two DDT-resistant Drosophila strains (RDDTR and 91R), together with over-expression of genes encoding possible DDT-detoxifying enzymes such as GSTD1, CYP6A2, and CYP6A8 (Misra et al., 2013). Among these detoxifying enzymes, miR-312-3p controlled the gene CYP6A8 (Seong et al., 2019). The imidacloprid-resistant N. lugens exhibits constitutive adipokinetic hormone (AKH) downregulation, which results in co-over-expression of CncC, Maf, and CYP6ER1 (Tang et al., 2020). The enzyme CYP6ER1, which was shown to be implicated in neonicotinoid, Nitenpyram resistant strains, was controlled by two new miRNAs, Novel_85 and Novel_191, which may be mediated through the CncC pathway (Mao et al., 2022).
MAPK/CREB pathway
The MAPK (Mitogen-Activated Protein Kinase) pathway is an evolutionarily conserved signaling system vital in many cellular functions in insects and other species. This system regulates growth, development, immunity, and responsiveness to environmental stimuli by transducing extracellular signals into intracellular responses. Extracellular ligands or signals, such as growth factors or cytokines, activate the route by attaching to their corresponding cell surface receptors. These receptors may be classified as receptor tyrosine kinases (RTKs) or G protein-coupled receptors (GPCRs). When ligands bind to the receptors, they alter conformation and activate intracellular signaling molecules. The renin-angiotensin system (RAS), a small GTPase that becomes activated by exchanging GDP for GTP, is one of the major molecules involved. Activated RAS then promotes the translocation of Rapidly Accelerated Fibrosarcoma (RAF), a serine/threonine protein kinase, to the cell membrane and induces conformational changes that lead to its activation. RAF then phosphorylates MEK (MAPK/Extracellular Signal-Regulated Kinase (ERK) Kinase), another serine/threonine kinase. MEK phosphorylates and activates ERK, also known as MAPK, the pathway’s terminal kinase. Active ERK enters the nucleus and phosphorylates different transcription factors, causing changes in gene expression. Depending on the environment, changes in gene expression generated by activated ERK result in a variety of biological responses. Cell proliferation and differentiation are examples of such reactions. Cell proliferation, differentiation, apoptosis, and immune response regulation are examples of such responses (Cruz et al., 2020). Recent research on whitefly shows that the MAPK/CREB signaling branch directly regulates Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) CYP6CM1 expression, which assists in imidacloprid detoxification (Yang et al., 2020).
MAPK pathway regulating Bt resistance in insects
Bacillus thuringiensis (Bt) is a Gram-positive, spore-forming bacteria capable of producing crystal proteins (Cry) that are poisonous to a broad range of insect species, mainly those of the orders Lepidoptera, Coleoptera, and Diptera. Bt also generates vegetative insecticidal protein (Vip) and secretory insecticidal protein (Sip), both of which are biodegradable and mainly target Coleoptera and Lepidoptera (Chakroun et al., 2016). Bt is highly particular in its activity and is extensively encouraged for pest management as an alternative to toxic chemical treatments. Nonetheless, insects have lately evolved resistance to Bt. Insect resistance to Cry toxins is thought to be caused by downregulation or mutation of the midgut proteinase and receptors, which results in decreased conversion of the protoxin to the active toxin and reduced toxin binding of Cry toxin to receptors (Ferre and Van Rie, 2002; Pigott and Ellar, 2007). Phenotypic association experiments and molecular expression analyses have revealed the required evidence for receptor-mediated Bt resistance developments. Currently, more than four types of functional receptors, including cadherin (Xu et al., 2005), aminopeptidase N (APN) (Tiewsiri and Wang, 2011), alkaline phosphatase (ALP) (Jurat-Fuentes and Adang, 2007), ATP binding cassettes (ABC) transporters (Atsumi et al., 2012; Xiao et al., 2014), and others, have been identified and validated in lepidopteran species related to the mode of action of Cry toxin action. The differential expression of the midgut membrane-bound ALP and ABC subfamily C (ABCC) genes have been implicated in high-level resistance to Cry1Ac in P. xylostella, and the connection between these genes is found to be regulated by MAPK cascades. The downregulation of ALP, ABCC2, and ABCC3 was strongly correlated with recessive Cry1Ac resistance but not with the upregulation of ABCC1. While inhibiting MAP4K4, a constitutively transcriptionally-activated MAPK upstream gene within the BtR-1 locus, led to a brief recovery of gene expression and restored the susceptibility in resistant larvae. However, silencing ABCC2 and ABCC3 in susceptible larvae reduced Cry1Ac susceptibility while having no effect on ALP expression (Guo et al., 2015).
MAPK cascade genes such as PxMAP4K4, PxRaf, Pxp38, and PxERK were upregulated in the midgut tissues of all resistant strains compared to the susceptible strain of P. xylostella. Increased phosphorylation of MAP3K7, Thousand and one amino acid (TAO) kinase, MAP2K6, ERK, and p38 was recorded in the resistant strain. In Bt resistant P. xylostella strain, upstream of these MAPKs, RAF is involved in ERK activation, TAO in JNK activation, and MAP3K7 in both p38 and JNK activation. MAP4K4 activates all three key MAPKs of p38, JNK, and ERK (Guo et al., 2021).
Using CRISPR/Cas9, the contribution of two paralogous ABC transporters, ABCC2 and ABCC3, and two aminopeptidases N, APN1 and APN3a to Bt Cry1Ac toxicity in P. xylostella was evaluated. A knockout strain containing deleted ABCC2 and ABCC3 genes exhibited 4482-fold resistance to Cry1A toxin. Similarly, knockout strains with deleted APN1 and APN3a genes exhibited 1425-fold resistance to Cry1Ac toxin, indicating their functional redundancy (Sun et al., 2022). Discovered that transcription factor PxGATAd activates membrane-bound PxmALP expression between the Cry1Ac susceptible and resistant strains by interacting with a non-canonical yet particular GATA-like cis-response element (CRE) present in the PxmALP promoter region to trigger the production of PxmALP directly. The ability of PxGATAd to regulate transcription was compromised by a six-nucleotide insertion mutation in this cis-acting area of the PxmALP promoter from the resistant strain. Additionally, PxGATAd silencing in susceptible larvae decreased PxmALP expression and sensitivity to Cry1Ac toxin. PxGATAd and PxmALP expression were both briefly restored when PxMAP4K4 expression was suppressed in resistant larvae, demonstrating that PxGATAd is a positive, responsive factor that is involved in the activation of PxmALP promoter and negatively controlled by the MAPK signaling pathway (Guo L. et al., 2022).
Interesting to document is that P. xylostella has evolved a mechanism that makes it resistant to Bt toxins without impairing fitness. Fushi Tarazu Factor 1 (FTZ-F1), a MAPK-modulated transcription factor, downregulates Bt Cry1Ac toxin receptors while up-regulating non-receptor paralogs of MAPK cascades that regulate downstream transcription factors via the p38, Jun N-terminal kinase (JNK), or ERK pathways (Yang et al., 2013). Phosphorylated FTZ-F1 activates non-receptor genes through the motif “TAMAGTC,” whereas unphosphorylated FTZ-F1 initiates receptor genes via the binding site “YCAAGGYCR.” The activated MAPK cascade raises the amount of phosphorylation of FTZ-F1, conferring P. xylostella resistance to Cry1Ac toxin without impairing growth (Guo Z. et al., 2022). Host adaptation to the primary Bt virulence factors in P. xylostella was due to insertion of short interspersed nuclear element (SINE), SE2 in the promoter of MAP4K4 gene that enhances the effect of transcription factor forkhead box O (FOXO) to induce MAPK pathway, which in turn potentiates host defense mechanism against the pathogen (Guo et al., 2023).
GPCR/cAMP/PKA pathway
G protein-coupled receptors (GPCRs) are a large family of transmembrane receptors. It is a seven-transmembrane protein containing an extracellular domain for ligand binding and signal transduction into the cells regulating various cellular processes through an intracellular domain linked to a heterotrimeric G protein consisting of alpha (Gα), beta (Gβ), and gamma (Gγ) subunits. When GPCR interacts with its ligand, it induces cytosolic G-protein exchanges of guanosine diphosphate (GDP)-bound G protein for guanosine triphosphate (GTP). The G proteins are now separated into Gα and βγ-subunits (Gβγ). The Gα subunit triggers adenylate cyclase (Ac) to form the secondary messenger cAMP (cyclic adenosine monophosphate) from ATP, which then activates protein kinase A (PKA). PKA, in turn, induces a phosphorylation cascade that activates the cAMP-response element binding (CREB) transcription factor that finally regulates the expression of target genes (Hilger et al., 2018).
Caffeine induction of CYP6A2 and CYP6A8 in D. melanogaster was due to increased levels of cAMP due to the suppression of caffeine by cAMP-hydrolyzing cAMP phosphodiesterase (PDE) and downregulation of the AP-1 transcription factor Jun (Bhaskara et al., 2008). GPCR-related genes regulated the P450 gene expression in Culex quinquefasciatus (Diptera: Culicidae) for the first time where, suppressing GPCR with RNAi reduced permethrin resistance due to the decreased expression of related genes (CYP6AA7, CYP9M10, CYP9J34, and CYP9J40) in the high-resistance C. quinquefasciatus strain (Li T. et al., 2014).
The relative expression levels of the 94 GPCR genes in M. domestica among the near-isogenic imidacloprid resistance resistant strain (N-IRS), the susceptible strain (CSS) and another strain generated from field populations with imidacloprid resistance (IRS) were compared. It was found that compared to CSS strain, five GPCR genes were elevated in the N-IRS strain, and eight GPCR genes were upregulated in the IRS strains. Among the over-expressed GPCRs, LOC101899380 and LOC101895664 were heterologously expressed in D. melanogaster, and they were found upregulating CYP6G1, CYP6A2, CYP6A8, and CYP12D1 genes. Moreover, the transgenic D. melanogaster created with the GPCR gene LOC101899380 affected the expression of all four P450 genes, but LOC101895664 only affected CYP6G1 and CYP6A2, indicating that the latter is less involved in imidacloprid resistance via control of CYP6A8 and CYP12D1 expression (Ma et al., 2020).
The intermediary effectors engaged in insecticide resistance functions between GPCR020021 and the four target P450 genes, including 1 Gαs (Gαs006458), two AC (AC007240 and AC004739), and two PKAs (PKA000798 and PKA018257), were identified using a variety of functional genomics approaches. These include transgenic expression of GPCR020021 in D. melanogaster and heterologous expression of GPCR020021 and its downstream Gαs, AC, and PKA in Sf9 cells (Li and Liu, 2019) as well as in vivo RNAi knockdown of GPCR020021, Gαs006458, AC007240, AC004739, and the four target P450 genes in permethrin-susceptible (Li and Liu, 2018) and -resistant strains (Li et al., 2015; Li and Liu, 2017).
Epitranscriptomic regulation of insecticide resistance
Epitranscriptome refers to different dynamic and reversible chemical modifications affecting RNA transcripts (coding and non-coding RNAs), which is the post-transcriptional regulation of gene expression. So, this structure and functions of dynamic RNA modifications during the developmental process and environmental stress and their effects on gene expression have emerged as a new branch of functional genomics known as “epi-transcriptomics.”
The dynamic and reversible RNA base modifications are catalyzed by enzymes like methyltransferases (writers) and removed by demethylases (erasers). Readers are the modification-specific binding proteins that interpret these modifications. It is similar to epigenetic DNA modification (Kumar et al., 2018). Many of the mRNA base modifications involve attachment of a methyl (CH3) group at a particular position either on the base [e.g., N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-methylcytidine (m5C), 3-methylcytidine (m3C), N7-methylguanosine (m7G), and 1-methylguanosine (m1G)], ribose sugar (e.g., 2-O-methyladenosine), or on both base and sugar [e.g., N6,2′-O-dimethyl adenosine (m6Am)] (Dominissini et al., 2016; Molinie et al., 2016).
m6A RNA is among eukaryotic mRNA’s most abundant chemical modifications. In D. melanogaster, the m6A pathway was involved in neuronal functions and sex determination (Lence et al., 2016). The transcriptome-wide profiling of m6A in the silkworm Bombyx mori has been used to identify its role in regulating gene expression, chromosome alignment and segregation, and nucleopolyhedrovirus (BmNPV) infection (Li B. et al., 2019).
CYP4C64 has a crucial role in resistance to the neonicotinoid thiamethoxam and regulation of the gene by the m6A pathway in whitefly B. tabaci (Yang et al., 2021). CYP4C64 was strongly over-expressed in the thiamethoxam-resistant strains compared with the susceptible strains. A polymorphism (T to A at position-206 bp) was observed in the 5′ UTR of CYP4C64 at a greater frequency than 3′ UTR and CDS in the thiamethoxam-resistant strains compared with the susceptible strain. The T-206A transversion was predicted to have a sequence (CGACA) that resembles an N6 -methyladenine (m6A) sequence. The m6A modification on target RNAs, namely, methyltransferase-like 3 (METTL3) and 14 (METTL14), were confirmed in Western blot studies. METTL3 was over-expressed in resistant strains compared with the susceptible strain. These results help us understand the epi-transcriptomic regulation of the xenobiotic response in insects and the role of m6A in developing insecticide resistance (Figure 5C).
Future perspectives
For pest management or study, effective delivery mechanisms are required for introducing agomirs and antagomirs into insects. Feeding, injection, topical treatment, transgenic approaches, viral vectors, and nanoparticle-mediated delivery are all typical ways researchers introduce agomirs and antagomirs. Because insects are more receptive to external stimuli and may display more excellent feeding rates during this stage, the larval/nymphal stage is commonly selected for RNAi investigations linked to insecticide resistance (Jain et al., 2020). The incorporation of agomirs or antagomirs into the larval diet or by injection can result in efficient absorption and targeted gene silencing. For example, in the agomir and antagomir studies for insecticide resistance, fourth-instar nymphs of N. lugens (Li et al., 2022), third-instar larvae of P. xylostella (Zhu et al., 2020), and 1-day post-emergence female mosquitoes (Sun et al., 2019) were often employed.
Recently, pest control using miRNA has been achieved through a process known as trans-kingdom RNA interference (TK-RNAi). It involves transfer of miRNAs through the diet, across kingdoms, to recipient organisms where they influence their biological activity. Escherichia coli has been engineered to express precursors for artificial miRNAs (amiRNAs) with insect targets for pest management by TK-RNAi via bacterial-mediated precursor miRNA expression. A reduction in oogenesis, significant mortality, and developmental abnormalities was seen in H. armigera larvae fed with E. coli-expressing a precursor for an RNA that targets EcR (Yogindran and Rajam, 2016). To control pests, another TK-RNAi method uses an insect or plant precursor backbone that expresses target amiRNAs or insect-specific miRNAs in transgenic plants. In transgenic tobacco plants, Nicotiana tabacum and Nicotiana benthamiana, expressing amiRNAs targeting H. armigera acetylcholinesterase, AChE 1 and AChE 2, respectively. Continuous feeding of first instar larvae with these plants led to increased mortality, developmental abnormalities, and delayed growth rates (Saini et al., 2018). Nymphs of the B. tabaci species exhibited aberrant egg hatching and poor development when they were raised on transgenic N. tabacum plants that expressed three separate amiRNAs (amiRNASxl, amiRNAAChE, and amiRNAOrc), which each target the sex lethal protein (Sxl), AChE, and orcokinin (Orc), respectively (Zubair et al., 2020). However, the stability of miRNA is a major concern to be considered. RNA chemical changes, such as ribose 20 hydroxyl group modification, unlocked or locked nucleic acids, and phosphorothioate backbone modification, have been researched to solve difficulties with miRNA stability. In comparison to a genetic modification strategy, these possible answers will be more practical with a spray technique.
Conclusion
This review covered intricate cellular processes at the gene level in insecticide resistance responses as a survival mechanism in insects exposed to frequent insecticide applications in the agriculture and public health protection fronts. Recent outcomes on insecticide resistance research explored in high-throughput molecular methods demonstrate that resistance to insecticides has been linked to transcriptional activities of miRNAs and lncRNAs, a better understanding of these events that can be used in developing insecticide resistance management tactics. Substantial evidence has emerged on miRNAs regulating genes involved in detoxifying insecticides and modifying their target sites, the most common pesticide resistance strategies reported in insects.
By studying the miRNA profiles of insecticide-resistant insects, researchers can identify particular miRNAs that are upregulated or downregulated in response to insecticide exposure. Using these newly discovered miRNAs as potential therapeutic targets can reduce resistant populations and their expansions. Techniques for altering the expression or activity of these miRNAs can be developed to restore insecticide sensitivity or increase the efficacy of insecticide-based control strategies. Artificial mimics or inhibitors of specific miRNAs, for example, might modify their expression levels, potentially reversing or reducing pesticide resistance.
In the same way, lncRNAs have been linked to insecticide resistance affecting gene expression and biological processes by engaging with DNA, RNA, or proteins. lncRNAs are expressed differently in insecticide-resistant pests than in susceptible ones. These lncRNAs can regulate genes involved in insecticide detoxification, target site changes, or other resistance mechanisms. Understanding the roles and activities of these lncRNAs can give insights into the underlying molecular processes of insecticide resistance. It may lead to the development of resistance-fighting techniques against insects. Various technologies, such as RNA interference (RNAi) or gene editing techniques, can manipulate the expression or activity-specific lncRNAs. It may disrupt or regulate the regulatory networks involved in insecticide resistance by targeting resistance-associated lncRNAs, resulting in enhanced control strategies.
In conclusion, both miRNAs and lncRNAs can help manage insecticide resistance by providing intervention targets and a better knowledge of resistance mechanisms. However, it is essential to emphasize that RNA-based pest control is still in its early stages, and more study is needed to properly investigate and harness the potential of miRNAs and lncRNAs in countering insecticide resistance.
m6A is a well-known and thoroughly researched RNA modification cellular process in organisms. The m6A modification modulates RNA metabolism, such as mRNA stability, splicing, and translation. A recent study has shown that m6A mutations have a role in insecticide resistance in some insect species. Changes in the m6A landscape have been detected in resistant insect populations, indicating a potential function in modifying resistance-related gene expression.
Researchers may gain insight into the regulatory processes driving resistance development by examining the epi-transcriptomic alterations associated with insecticide resistance. Understanding how particular RNA changes impact the expression and function of resistance genes might give helpful knowledge for creating resistance-fighting tactics. Furthermore, altering RNA changes might be a focused way to manage insecticide resistance. It may modulate the expression of resistance-related genes and restore insecticide sensitivity by regulating the enzymes responsible for adding or deleting particular RNA modifications.
However, it is essential to highlight that the epitranscriptomics and insecticide resistance area is still in its early phases. More study is needed to understand the exact epi-transcriptomic changes associated with resistance and their functional implications. Furthermore, finding tools and procedures for precise manipulation of RNA changes in insects is a problem that must be overcome. Continued study in this area will help us understand the epitranscriptomic control of pesticide resistance and may lead to novel techniques for managing resistant pest populations.
Researchers can uncover crucial molecular targets and propose creative techniques to manage resistance by unraveling the regulatory processes linked with insecticide resistance. Combining insecticides with synergists that inhibit detoxifying enzymes, modifying essential regulatory proteins or pathways via RNA interference or gene editing technologies, and developing novel insecticides that target specific resistance mechanisms are all examples of such methods. It is critical to note that resistance regulatory mechanisms vary and develop between pest species and populations. Continuous research and observations are essential to stay tuned in and gather more profound information on the insecticide resistance phenomena and build sustainable management approaches.
Acknowledgments
All the authors acknowledge the Department of Agricultural Entomology, Tamil Nadu Agricultural University, Coimbatore, India for providing necessary support and facilities. The first author acknowledges the Department of Science and Technology, Ministry of Science and Technology, Government of India, New Delhi, India for DST INSPIRE FELLOWSHIP/2021/IF210049.
Funding Statement
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Author contributions
CM: Conceptualization, Resources, Supervision, Writing–original draft, Writing–review and editing. MM: Conceptualization, Supervision, Writing–original draft, Writing–review and editing. SP: Writing–original draft, Writing–review and editing. KN: Writing–review and editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2023.1257859/full#supplementary-material
Glossary
| ABC | ATP-Binding cassettes |
| ABCG | ATP-Binding cassettes group |
| ACC | Acetyl-CoA carboxylase |
| AChE | Acetylcholine esterase |
| AhR | Aryl hydrocarbon Receptor |
| AKH | Adipokinetic hormone |
| ARE | Antioxidant Response Element |
| ARNT | Aryl Hydrocarbon Receptor Nuclear Translocator |
| BPH | Brown planthopper |
| bZIP | Basic leucine zipper |
| CarE | Carboxyl esterase |
| CCE | Carboxyl choline esterase |
| CDS | Coding sequence |
| CncC/Keap1 | Cap'n'collar isoform C/Kelch-like ECH associated protein 1 |
| CNV | Copy Number Variation |
| CYP | Cytochrome P450 |
| CyR | Cyflumetofen-resistant strain of T. cinnabarinus |
| DBD | DNA binding domain |
| DDT | Dichloro-diphenyl-trichloroethane |
| DHR96 | Drosophila Hormone receptor-like in 96 (HR96) |
| EcR | Ecdysone receptor |
| ERK | Extracellular Signal-Regulated Kinase |
| EST | Esterase |
| FTZ | Fushi Tarazu |
| GPCR | G protein-coupled receptor |
| GST | Glutathione S Transferases |
| HNF | Hepatocyte nuclear factor |
| HR96 | Hormone receptor-like in 96 |
| IR | Insecticide resistance |
| LBD | Ligand binding domain |
| Maf | Muscle aponeurosis fibromatosis |
| MAPK | Mitogen-activated phosphate kinase pathway |
| METTL | Methyltransferase-like |
| MFO | Mixed function oxidase |
| miR | Mature microRNA |
| NGS | Next-generation sequencing |
| NR | Nuclear receptor |
| Nrf2 | NF-E2-Related Factor 2 |
| PxEcR-B | Plutella xylostella Ecdysone receptor isoform B |
| PxJHE | Plutella xylostella juvenile hormone esterase |
| PxRyR | Plutella xylostella ryanodine receptor |
| RAS | Renin-angiotensin system |
| RAF | Rapidly Accelerated Fibrosarcoma |
| RISC | RNA induced silencing complex |
| RTK | Receptor tyrosine kinases |
| RNA | Ribo Nucleic acid |
| RNAi | RNA interference |
| ROS | Reactive oxygen species |
| RXR | Retinoid-X-receptor |
| RyR | Ryanodine receptor |
| SBPH | Small backed planthopper |
| SXR/PXR | Steroid and xenobiotic receptor/Pregnane X Receptor |
| TAO | Thousand and one amino acid |
| USP | Ultraspiracle protein |
| UTR | Untranslated region |
| VGSC | Voltage-gated sodium channel |
| XRE | Xenobiotic Response Element |
References
- APRD (2023). Arthropod pesticide resistance database Michigan state university . https://www.pesticideresistance.org/ (Accessed May 5, 2023). [Google Scholar]
- Atsumi S., Miyamoto K., Yamamoto K., Narukawa J., Kawai S., Sezutsu H., et al. (2012). Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori . Proc. Natl. Acad. Sci. U.S.A. 109, E1591–E1598. 10.1073/pnas.1120698109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banazeer A., Afzal M. B., Hassan S., Ijaz M., Shad S. A., Serrão J. E. (2021). Status of insecticide resistance in Plutella xylostella (Linnaeus) (Lepidoptera: plutellidae) from 1997 to 2019: cross-resistance, genetics, biological costs, underlying mechanisms, and implications for management. Phytoparasitica 50, 465–485. 10.1007/s12600-021-00959-z [DOI] [Google Scholar]
- Bao Y. Y., Zhang C. X. (2019). Recent advances in molecular biology research of a rice pest, the brown planthopper. J. Integr. Agric. 18, 716–728. 10.1016/S2095-3119(17)61888-4 [DOI] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behura S. K. (2007). Insect microRNAs: structure, function and evolution. Insect biochem. Mol. Biol. 37, 3–9. 10.1016/j.ibmb.2006.10.006 [DOI] [PubMed] [Google Scholar]
- Berenbaum M. R., Johnson R. M. (2015). Xenobiotic detoxification pathways in honey bees. Curr. Opin. Insect Sci. 10, 51–58. 10.1016/j.cois.2015.03.005 [DOI] [PubMed] [Google Scholar]
- Bhaskara S., Chandrasekharan M. B., Ganguly R. (2008). Caffeine induction of Cyp6a2 and Cyp6a8 genes of Drosophila melanogaster is modulated by cAMP and D-JUN protein levels. Gene 415, 49–59. 10.1016/j.gene.2008.02.017 [DOI] [PubMed] [Google Scholar]
- Blackman R. L., Eastop V. F. (2000). Aphids on the world's crops: An identification and information guide. Chichester, United Kingdom: John Wiley and Sons Ltd. [Google Scholar]
- Bo H., Miaomiao R., Jianfeng F., Sufang H., Xia W., Elzaki M. E. A., et al. (2020). Xenobiotic transcription factors CncC and maf regulate expression of CYP321A16 and CYP332A1 that mediate chlorpyrifos resistance in Spodoptera exigua . J. Hazard. Mat. 398, 122971. 10.1016/j.jhazmat.2020.122971 [DOI] [PubMed] [Google Scholar]
- Boaventura D., Bolzan A., Padovez F. E., Okuma D. M., Omoto C., Nauen R. (2020). Detection of a ryanodine receptor target‐site mutation in diamide insecticide resistant fall armyworm, Spodoptera frugiperda . Pest Manag. Sci. 76, 47–54. 10.1002/ps.5505 [DOI] [PubMed] [Google Scholar]
- Bracht J., Hunter S., Eachus R., Weeks P., Pasquinelli A. E. (2004). Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA 10, 1586–1594. 10.1261/rna.7122604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D. R., Stark A., Russell R. B., Cohen S. M. (2003). Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36. 10.1016/s0092-8674(03)00231-9 [DOI] [PubMed] [Google Scholar]
- Burgin J., Ahamed A., Cummins C., Devraj R., Gueye K., Gupta D., et al. (2023). The European nucleotide archive in 2022. Nucleic Acids Res. 51, D121–D125. 10.1093/nar/gkac1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho R. A., Omoto C., Field L. M., Williamson M. S., Bass C. (2013). Investigating the molecular mechanisms of organophosphate and pyrethroid resistance in the fall armyworm Spodoptera frugiperda . PLoS One 17, e62268. 10.1371/journal.pone.0062268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayirlioglu P., Kadow I. G., Zhan X., Okamura K., Suh G. S., Gunning D., et al. (2008). Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science 319, 1256–1260. 10.1126/science.1149483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakroun M., Banyuls N., Bel Y., Escriche B., Ferré J. (2016). Bacterial vegetative insecticidal proteins (Vip) from entomopathogenic bacteria. Microbiol. Mol. Biol. Rev. 80, 329–350. 10.1128/MMBR.00060-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W., Lei Z., Chongyu L., Kongming W., Yutao X. (2019). Research progress of resistance mechanism and management techniques of fall armyworm Spodoptera frugiperda to insecticides and Bt Crops. Plant Dis. pests. 10, 10–17. 10.19579/j.cnki.plant-d.p.2019.04.004 [DOI] [Google Scholar]
- Cheesman M. J., Traylor M. J., Hilton M. E., Richards K. E., Taylor M. C., Daborn P. J., et al. (2013). Soluble and membrane-bound Drosophila melanogaster CYP6G1 expressed in Escherichia coli: purification, activity, and binding properties toward multiple pesticides. Insect biochem. Mol. Biol. 43, 455–465. 10.1016/j.ibmb.2013.02.003 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Li Y., Li W., Song Y., Zeng R., Lu K. (2021). Inhibition of hepatocyte nuclear factor 4 confers imidacloprid resistance in Nilaparvata lugens via the activation of cytochrome P450 and UDP-glycosyltransferase genes. Chemosphere 263, 128269. 10.1016/j.chemosphere.2020.128269 [DOI] [PubMed] [Google Scholar]
- Choi I. K., Hyun S. (2012). Conserved microRNA miR-8 in fat body regulates innate immune homeostasis in Drosophila. Dev. Comp. Immunol. 37, 50–54. 10.1016/j.dci.2011.12.008 [DOI] [PubMed] [Google Scholar]
- Choudhary C., Sharma S., Meghwanshi K. K., Patel S., Mehta P., Shukla N., et al. (2021). Long non-coding RNAs in insects. Animals 11, 1118. 10.3390/ani11041118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz J., Martín D., Franch-Marro X. (2020). Egfr signaling is a major regulator of ecdysone biosynthesis in the Drosophila prothoracic gland. Curr. Biol. 30, 1547–1554. 10.1016/j.cub.2020.01.092 [DOI] [PubMed] [Google Scholar]
- Daborn P., Boundy S., Yen J., Pittendrigh B., Ffrench-Constant R. (2001). DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Mol. Gen. Genom. 266, 556–563. 10.1007/s004380100531 [DOI] [PubMed] [Google Scholar]
- Denison M. S., Fisher J. M., Whitlock J. P. (1988). Inducible, receptor-dependent protein-DNA interactions at a dioxin-responsive transcriptional enhancer. Proc. Natl. Acad. Sci. U. S. A. 85, 2528–2532. 10.1073/pnas.85.8.2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., Peer E., Kol N., Ben-Haim M. S., et al. (2016). The dynamic N 1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446. 10.1038/nature16998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Ge F., Zhu S., Parajulee M. N. (2004). Effect of cotton cultivar on development and reproduction of Aphis gossypii (Homoptera: aphididae) and its predator Propylaea japonica (Coleoptera: coccinellidae). J. Econ. Entomol. 97 (4), 1278–1283. 10.1093/jee/97.4.1278 [DOI] [PubMed] [Google Scholar]
- Eggleton P. (2020). The state of the world's insects. Annu. Rev. Environ. Resour. 45, 61–82. 10.1146/annurev-environ-012420-050035 [DOI] [Google Scholar]
- Etebari K., Afrad M. H., Tang B., Silva R., Furlong M. J., Asgari S. (2018). Involvement of microRNA miR‐2b‐3p in regulation of metabolic resistance to insecticides in Plutella xylostella . Insect Mol. Biol. 27, 478–491. 10.1111/imb.12387 [DOI] [PubMed] [Google Scholar]
- Etebari K., Furlong M. J., Asgari S. (2015). Genome wide discovery of long intergenic non-coding RNAs in Diamondback moth (Plutella xylostella) and their expression in insecticide resistant strains. Sci. Rep. 5, 14642–14644. 10.1038/srep14642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahmy N. T., Osman A., Badr M. S., Morcos N., Diclaro J. W., Abd-ElSamie E. M. (2020). Deciphering pyrethroid resistance in Cx. pipiens (L): implications of cytochrome P450; expression profiling and regulatory microRNA. Mol. Cell Probes. 52, 101579. 10.1016/j.mcp.2020.101579 [DOI] [PubMed] [Google Scholar]
- Feng K., Liu J., Wei P., Ou S., Wen X., Shen G., et al. (2020). lincRNA_Tc13743 2-miR-133-5p-TcGSTm02 regulation pathway mediates cyflumetofen resistance in Tetranychus cinnabarinus . Insect biochem. Mol. Biol. 123, 103413. 10.1016/j.ibmb.2020.103413 [DOI] [PubMed] [Google Scholar]
- Feng X., Li M., Liu N. (2018). Carboxylesterase genes in pyrethroid resistant house flies, Musca domestica . Insect bio. Chem. Mol. Biol. 92, 30–39. 10.1016/j.ibmb.2017.11.007 [DOI] [PubMed] [Google Scholar]
- Ferre J., Van Rie J. (2002). Biochemistry and genetics of insect resistance to Bacillus thuringiensis . Annu. Rev. Entomol. 47, 501–533. 10.1146/annurev.ento.47.091201.145234 [DOI] [PubMed] [Google Scholar]
- Feyereisen R., Dermauw W., Van Leeuwen T. (2015). Genotype to phenotype, the molecular and physiological dimensions of resistance in arthropods. Pestic. Biochem. Physiol. 121, 61–77. 10.1016/j.pestbp.2015.01.004 [DOI] [PubMed] [Google Scholar]
- Foster S. P., Denholm I., Rison J. L., Portillo H. E., Margaritopoulis J., Slater R. (2012). Susceptibility of standard clones and European field populations of the green peach aphid, Myzus persicae, and the cotton aphid, Aphis gossypii (Hemiptera: aphididae), to the novel anthranilic diamide insecticide cyantraniliprole. Pest Manag. Sci. 68, 629–633. 10.1002/ps.2306 [DOI] [PubMed] [Google Scholar]
- Fromm B., Domanska D., Høye E., Ovchinnikov V., Kang W., Aparicio-Puerta E., et al. (2020). MirGeneDB 2.0: the metazoan microRNA complement. Nucleic Acids Res. 48, D132-D141–D141. 10.1093/nar/gkz885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong M. J., Wright D. J., Dosdall L. M. (2013). Diamondback moth ecology and management: problems, progress, and prospects. Annu. Rev. Entomol. 58, 517–541. 10.1146/annurev-ento-120811-153605 [DOI] [PubMed] [Google Scholar]
- Gaddelapati S. C., Kalsi M., Roy A., Palli S. R. (2018). Cap'n'collar C regulates genes responsible for imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata . Int. J. Mol. Sci. 99, 54–62. 10.1016/j.ibmb.2018.05.006 [DOI] [PubMed] [Google Scholar]
- Ganehiarachchi M., Knodel J. J. (2008). Diamondback moth in canola: biology and integrated pest management. NDSU Ext. Serv., E-1346. [Google Scholar]
- Ge W., Chen Y. W., Weng R., Lim S. F., Buescher M., Zhang R., et al. (2012). Overlapping functions of microRNAs in control of apoptosis during Drosophila embryogenesis. Cell death. 19, 839–846. 10.1038/cdd.2011.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain P., Staels B., Dacquet C., Spedding M., Laudet V. (2006). Overview of nomenclature of nuclear receptors. Pharmacol. Rev. 58, 685–704. 10.1124/pr.58.4.2 [DOI] [PubMed] [Google Scholar]
- Gimenez S., Abdelgaffar H., Goff G. L., Hilliou F., Blanco C. A., Hanniger S., et al. (2020). Adaptation by copy number variation increases insecticide resistance in the fall armyworm. Commun. Biol. 3, 664. 10.1038/s42003-020-01382-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo M., Hilliou F., Fricaux T., Audant P., Feyereisen R., Le Goff G. (2015). Cytochrome P450s from the fall armyworm (Spodoptera frugiperda): responses to plant allelochemicals and pesticides. Insect Mol. Biol. 24, 115–128. 10.1111/imb.12140 [DOI] [PubMed] [Google Scholar]
- Grbic M., Van Leeuwen T., Clark R. M., Rombauts S., Rouzé P., Grbić V., et al. (2011). The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479, 487–492. 10.1038/nature10640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H. K., Van Dongen S., Enright A. J. (2007). miRBase: tools for microRNA genomics. Nucleic Acids Res. 36, D154–D158. 10.1093/nar/gkm952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo K., Yang P., Chen J., Lu H., Cui F. (2017). Transcriptomic responses of three aphid species to chemical insecticide stress. Sci. China Life Sci. 60, 931–934. 10.1007/s11427-017-9104-5 [DOI] [PubMed] [Google Scholar]
- Guo L., Cheng Z., Qin J., Sun D., Wang S., Wu Q., et al. (2022a). MAPK-mediated transcription factor GATAd contributes to Cry1Ac resistance in diamondback moth by reducing PxmALP expression. PLoS Genet. 18, e1010037. 10.1371/journal.pgen.1010037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Guo L., Bai Y., Kang S., Sun D., Qin J., et al. (2023). Retrotransposon-mediated evolutionary rewiring of a pathogen response orchestrates a resistance phenotype in an insect host. Proc. Natl. Acad. Sci. U.S.A. 120, e2300439120. 10.1073/pnas.2300439120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Guo L., Qin J., Ye F., Sun D., Wu Q., et al. (2022b). A single transcription factor facilitates an insect host combating Bacillus thuringiensis infection while maintaining fitness. Nat. Commun. 13, 6024. 10.1038/s41467-022-33706-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Kang S., Chen D., Wu Q., Wang S., Xie W., et al. (2015). MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. PLoS Genet. 11, e1005124. 10.1371/journal.pgen.1005124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Kang S., Wu Q., Wang S., Crickmore N., Zhou X., et al. (2021). The regulation landscape of MAPK signaling cascade for thwarting Bacillus thuringiensis infection in an insect host. PLoS Pathog. 17, e1009917. 10.1371/journal.ppat.1009917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Qin J., Zhou X., Zhang Y. (2018). Insect transcription factors: A landscape of their structures and biological functions in Drosophila and beyond. Int. J. Mol. Sci. 19, 3691. 10.3390/ijms19113691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. E. (2002). Aryl hydrocarbon receptors: diversity and evolution. Chem. Biol. Interact. 141, 131–160. 10.1016/S0009-2797(02)00070-4 [DOI] [PubMed] [Google Scholar]
- Heckel D. G. (2021). Perspectives on gene copy number variation and pesticide resistance. Pest Manag. Sci. 78, 12–18. 10.1002/ps.6631 [DOI] [PubMed] [Google Scholar]
- Hereward J. P., Cai X., Matias A. M., Walter G. H., Xu C., Wang Y. (2020). Migration dynamics of an important rice pest: the brown planthopper (Nilaparvata lugens) across Asia-Insights from population genomics. Evol. Appl. 13, 2449–2459. 10.1111/eva.13047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilger D., Masureel M., Kobilka B. K. (2018). Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol. 25, 4–12. 10.1038/s41594-017-0011-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V., Bushati N., Cohen S. M. (2010). Drosophila microRNAs 263a/b confer robustness during development by protecting nascent sense organs from apoptosis. PLoS Biol. 8, e1000396. 10.1371/journal.pbio.1000396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Huang H., Hu S., Ren M., Wei Q., Tian X., et al. (2021). Changes in both trans-and cis-regulatory elements mediate insecticide resistance in a lepidopteron pest, Spodoptera exigua . PLoS Genet. 17, e1009403. 10.1371/journal.pgen.1009403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Huang H., Wei Q., Ren M., Mburu D. K., Tian X., et al. (2019). Transcription factors CncC/Maf and AhR/ARNT coordinately regulate the expression of multiple GSTs conferring resistance to chlorpyrifos and cypermethrin in Spodoptera exigua . Pest Manag. Sci. 75, 2009–2019. 10.1002/ps.5316 [DOI] [PubMed] [Google Scholar]
- Huang Y., Qin J., Wu P., Zheng J., Qiu L. (2022). Comparison of cytochrome P450 CYP332A1 gene in resistant and susceptible strains of the cotton bollworm, Helicoverpa armigera (Lepidoptera: noctuidae). Appl. Entomol. Zool. 57, 171–181. 10.1007/s13355-022-00777-7 [DOI] [Google Scholar]
- Huang Y., Zheng J., Wu P., Zhang Y., Qiu L. (2023). A comparative study of transcriptional regulation mechanism of cytochrome P450 CYP6B7 between resistant and susceptible strains of Helicoverpa armigera . J. Agric. Food Chem. 71, 9314–9323. 10.1021/acs.jafc.3c01593 [DOI] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan J., Pasquinelli A. E., Bálint É., Tuschl T., Zamore P. D. (2001). A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838. 10.1126/science.1062961 [DOI] [PubMed] [Google Scholar]
- Hyun S., Lee J. H., Jin H., Nam J., Namkoong B., Lee G., et al. (2009). Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 139, 1096–1108. 10.1016/j.cell.2009.11.020 [DOI] [PubMed] [Google Scholar]
- Ingham V. A., Pignatelli P., Moore J. D., Wagstaff S., Ranson H. (2017). The transcription factor Maf-S regulates metabolic resistance to insecticides in the malaria vector Anopheles gambiae . BMC genom 18, 669–711. 10.1186/s12864-017-4086-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IRAC (2023). Insecticide resistance action committee (IRAC) . https://irac-online.org/mode-of-action/ (Accessed March 6, 2023). [Google Scholar]
- Jain R. G., Robinson K. E., Fletcher S. J., Mitter N. (2020). RNAi-based functional genomics in Hemiptera. Insects 11, 557. 10.3390/insects11090557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F. L., Chen Y. J., Chen J., Wang D. D., Dai G. H. (2011). Biological activity of extracts from 8 species of plants against Tetranychus cinnabarinus . Chin. Agric. Sci. Bull. 27, 286–291. [Google Scholar]
- Jover R., Bort R., Gómez‐Lechón M. J., Castell J. V. (2001). Cytochrome P450 regulation by hepatocyte nuclear factor 4 in human hepatocytes: A study using adenovirus‐mediated antisense targeting. Hepato 33, 668–675. 10.1053/jhep.2001.22176 [DOI] [PubMed] [Google Scholar]
- Jurat-Fuentes J. L., Adang M. J. (2007). A proteomic approach to study Cry1Ac binding proteins and their alterations in resistant Heliothis virescens larvae. J. Invertebr. Pathol. 95, 187–191. 10.1016/j.jip.2007.01.008 [DOI] [PubMed] [Google Scholar]
- Kalsi M., Palli S. R. (2017). Transcription factor cap n collar C regulates multiple cytochrome P450 genes conferring adaptation to potato plant allelochemicals and resistance to imidacloprid in Leptinotarsa decemlineata (Say). Int. J. Mol. Sci. 83, 1–12. 10.1016/j.ibmb.2017.02.002 [DOI] [PubMed] [Google Scholar]
- Karres J. S., Hilgers V., Carrera I., Treisman J., Cohen S. M. (2007). The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell 131, 136–145. 10.1016/j.cell.2007.09.020 [DOI] [PubMed] [Google Scholar]
- Katz K., Shutov O., Lapoint R., Kimelman M., Brister J. R., O’Sullivan C. (2022). The sequence read archive: A decade more of explosive growth. Nucleic Acids Res. 50, D387–D390. 10.1093/nar/gkab1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy C. J., Tierney K. B. (2012). “Xenobiotic protection/resistance mechanisms in organisms,” in Laws EA environmental toxicology: Selected entries from the encyclopedia of sustainability science and technology (New York: Springer; ), 689–721. 10.1007/978-1-4614-5764-0_23 [DOI] [Google Scholar]
- Kim I. Y., Choi B., Park W. R., Kim Y. J., Kim B. E., Mun S., et al. (2021). Nuclear receptor HR96 up‐regulates cytochrome P450 for insecticide detoxification in Tribolium castaneum . Pest Manag. Sci. 78, 230–239. 10.1002/ps.6626 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Moreau J. A., Zina J. M., Mazgaeen L., Yoon K. S., Pittendrigh B. R., et al. (2018). Identification and interaction of multiple genes resulting in DDT resistance in the 91-R strain of Drosophila melanogaster by RNAi approaches. Pestic. Biochem. Physiol. 151, 90–99. 10.1016/j.pestbp.2018.03.003 [DOI] [PubMed] [Google Scholar]
- King-Jones K., Horner M. A., Lam G., Thummel C. S. (2006). The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 4, 37–48. 10.1016/j.cmet.2006.06.006 [DOI] [PubMed] [Google Scholar]
- Kirkland L. S., Chirgwin E., Ward S. E., Congdon B. S., Rooyen A., Umina P. A. (2023). P450‐mediated resistance in Myzus persicae (Sulzer) (Hemiptera: aphididae) reduces the efficacy of neonicotinoid seed treatments in Brassica napus. Pest Manag. Sci. 79, 1851–1859. 10.1002/ps.7362 [DOI] [PubMed] [Google Scholar]
- Kozomara A., Birgaoanu M., Griffiths-Jones S. (2019). miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162. 10.1093/nar/gky1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Chinnusamy V., Mohapatra T. (2018). Epigenetics of modified DNA bases: 5-methylcytosine and beyond. Front. Genet. 9, 640. 10.3389/fgene.2018.00640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahm G. P., Cordova D., Barry J. D. (2009). New and selective ryanodine receptor activators for insect control. Bioorg. Med. Chem. 17, 4127–4133. 10.1016/j.bmc.2009.01.018 [DOI] [PubMed] [Google Scholar]
- Lai E. C., Tomancak P., Williams R. W., Rubin G. M. (2003). Computational identification of Drosophila microRNA genes. Genome Biol. 4, R42–R20. 10.1186/gb-2003-4-7-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin A., Marygold S. J., Antonazzo G., Attrill H., Dos Santos G., Garapati P. V., et al. (2021). FlyBase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 49, D899–D907. 10.1093/nar/gkaa1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie R. D., Mitchell R. D., Deguenon J. M., Ponnusamy L., Reisig D., Pozo-Valdivia A. D., et al. (2022). Characterization of long non-coding RNAs in the bollworm, Helicoverpa zea, and their possible role in Cry1Ac-resistance. Insects 13, 12. 10.3390/insects13010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff G., Hilliou F. (2017). Resistance evolution in Drosophila: the case of CYP6G1. Pest Manag. Sci. 73, 493–499. 10.1002/ps.4470 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. 10.1016/0092-8674(93)90529-y [DOI] [PubMed] [Google Scholar]
- Legeai F., Rizk G., Walsh T., Edwards O., Gordon K., Lavenier D., et al. (2010). Bioinformatic prediction, deep sequencing of microRNAs and expression analysis during phenotypic plasticity in the pea aphid, Acyrthosiphon pisum . BMC Genomics 11, 281–289. 10.1186/1471-2164-11-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B., Hoffmann J. (2007). The host defense of Drosophila melanogaster . Annu. Rev. Immunol. 25, 697–743. 10.1146/annurev.immunol.25.022106.141615 [DOI] [PubMed] [Google Scholar]
- Lence T., Akhtar J., Bayer M., Schmid K., Spindler L., Ho C. H., et al. (2016). m6A modulates neuronal functions and sex determination in Drosophila. Nature 540, 242–247. 10.1038/nature20568 [DOI] [PubMed] [Google Scholar]
- Li B., Wang X., Li Z., Lu C., Zhang Q., Chang L., et al. (2019c). Transcriptome‐wide analysis of N6‐methyladenosine uncovers its regulatory role in gene expression in the lepidopteran Bombyx mori . Insect Mol. Biol. 28, 703–715. 10.1111/imb.12584 [DOI] [PubMed] [Google Scholar]
- Li F., Liu X. N., Zhu Y., Ma J., Liu N., Yang J. H. (2014a). Identification of the 2-tridecanone responsive region in the promoter of cytochrome P450 CYP6B6 of the cotton bollworm, Helicoverpa armigera (Lepidoptera: noctuidae). Bull. Entomol. Res. 104, 801–808. 10.1017/S0007485314000698 [DOI] [PubMed] [Google Scholar]
- Li S., Hussain F., Unnithan G. C., Dong S., UlAbdin Z., Gu S., et al. (2019a). A long non-coding RNA regulates cadherin transcription and susceptibility to Bt toxin Cry1Ac in pink bollworm, Pectinophora gossypiella . Pestic. Biochem. Physiol. 158, 54–60. 10.1016/j.pestbp.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Li T., Cao C. W., Yang T., Zhang L., He L., Xi Z. Y., et al. (2015). A G-protein-coupled receptor regulation pathway in cytochrome P450-mediated permethrin-resistance in mosquitoes, Culex quinquefasciatus . Culex Quinquefasciatus. Sci. Rep. 5, 17772–17813. 10.1038/srep17772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Liu L., Zhang L., Liu N. (2014b). Role of G-protein-coupled receptor-related genes in insecticide resistance of the mosquito, Culex quinquefasciatus . Sci. Rep. 4, 6474. 10.1038/srep06474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Liu N. N. (2017). Regulation of P450-mediated permethrin resistance in Culex quinquefasciatus by the GPCR/Gas/AC/cAMP/PKA signaling cascade. Biochem. Biophys. Rep. 12, 12–19. 10.1016/j.bbrep.2017.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Liu N. N. (2019). Role of the G-protein-coupled receptor signaling pathway in insecticide resistance. Int. J. Mol. Sci. 20, 4300. 10.3390/ijms20174300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Liu N. N. (2018). The function of G-protein-coupled receptor regulatory cascade in southern house mosquitoes (Diptera: culicidae). J. Med. Entomol. 55, 862–870. 10.1093/jme/tjy022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Cassidy J. J., Reinke C. A., Fischboeck S., Carthew R. W. (2009). A microRNA imparts robustness against environmental fluctuation during development. Cell 137, 273–282. 10.1016/j.cell.2009.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Deng Z., Chen X. (2021b). Regulation of insect P450s in response to phytochemicals. Curr. Opin. Insect. Sci. 43, 108–116. 10.1016/j.cois.2020.12.003 [DOI] [PubMed] [Google Scholar]
- Li X., Hu S., Yin H., Zhang H., Zhou D., Sun Y., et al. (2021a). MiR-4448 is involved in deltamethrin resistance by targeting CYP4H31 in Culex pipiens pallens . Parasit. Vectors 14, 159–163. 10.1186/s13071-021-04665-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ren X., Liu Y., Smagghe G., Liang P., Gao X. (2020). MiR-189942 regulates fufenozide susceptibility by modulating ecdysone receptor isoform B in Plutella xylostella (L). Pestic. Biochem. Physiol. 163, 235–240. 10.1016/j.pestbp.2019.11.021 [DOI] [PubMed] [Google Scholar]
- Li X., Shan C., Li F., Liang P., Smagghe G., Gao X. (2019b). Transcription factor FTZ-F1 and cis acting elements mediate expression of CYP6BG1, conferring resistance to chlorantraniliprole in Plutella xylostella . Pest Manag. Sci. 75, 1172–1180. 10.1002/ps.5279 [DOI] [PubMed] [Google Scholar]
- Li Y., Padgett R. W. (2012). Bantam is required for optic lobe development and glial cell proliferation. PLoS One 7, e32910. 10.1371/journal.pone.0032910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Mao K., Jin R., Cai T., Qin Y., Zhang Y., et al. (2022). miRNA novel_268 targeting NlABCG3 is involved in nitenpyram and clothianidin resistance in Nilaparvata lugens. Int. J. Biol. Macromol. 217, 615–623. 10.1016/j.ijbiomac.2022.07.096 [DOI] [PubMed] [Google Scholar]
- Liao X., Xu P. F., Gong P. P., Wan H., Li J. H. (2021). Current susceptibilities of brown planthopper Nilaparvata lugens to triflumezopyrim and other frequently used insecticides in China. Insect Sci. 28, 115–126. 10.1111/1744-7917.12764 [DOI] [PubMed] [Google Scholar]
- Liu B., Tang M., Chen H. (2022). Activation of the ROS/CncC signaling pathway regulates cytochrome P450 CYP4BQ1 responsible for (+)-α-Pinene tolerance in Dendroctonus armandi . Int. J. Mol. Sci. 23, 11578. 10.3390/ijms231911578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Tian M., Guo Q., Ma L., Zhou D., Shen B., et al. (2016). MiR-932 regulates pyrethroid resistance in Culex pipiens pallens (Diptera: culicidae). J. Med. Entomol. 53, 1205–1210. 10.1093/jme/tjw083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Hsu Y. H. (2007). The role of comparative pathology in the investigation of zoonoses. Pestic. Shenyang 46, 127–133. 10.1016/S1016-3190(10)60004-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Peng W., Li Z., Li W., Li L., Pan J., et al. (2012b). Next‐generation small RNA sequencing for microRNAs profiling in Apis mellifera: comparison between nurses and foragers. Insect Mol. Biol. 21, 297–303. 10.1111/j.1365-2583.2012.01135.x [DOI] [PubMed] [Google Scholar]
- Liu N., Landreh M., Cao K., Abe M., Hendriks G. J., Kennerdell J. R., et al. (2012a). The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila . Nature 482, 519–523. 10.1038/nature10810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang H. Y., Ning Y. B., Qiao K., Wang K. Y. (2015). Resistance selection and characterization of chlorantraniliprole resistance in Plutella xylostella (Lepidoptera: plutellidae). J. Econ. Entomol. 108, 1978–1985. 10.1093/jee/tov098 [DOI] [PubMed] [Google Scholar]
- Lu K., Cheng Y., Li W., Li Y., Zeng R., Song Y. (2020). Activation of CncC pathway by ROS burst regulates cytochrome P450 CYP6AB12 responsible for λ-cyhalothrin tolerance in Spodoptera litura . J. Hazard. Mat. 387, 121698. 10.1016/j.jhazmat.2019.121698 [DOI] [PubMed] [Google Scholar]
- Lu K., Li Y., Cheng Y., Li W., Song Y., Zeng R., et al. (2021). Activation of the NR2E nuclear receptor HR83 leads to metabolic detoxification-mediated chlorpyrifos resistance in Nilaparvata lugens . Pestic. Biochem. Physiol. 173, 104800. 10.1016/j.pestbp.2021.104800 [DOI] [PubMed] [Google Scholar]
- Ma K., Li F., Tang Q., Liang P., Liu Y., Zhang B., et al. (2019). CYP4CJ1-mediated gossypol and tannic acid tolerance in Aphis gossypii Glover. Chemosphere 219, 961–970. 10.1016/j.chemosphere.2018.12.025 [DOI] [PubMed] [Google Scholar]
- Ma Z., Zhang Y., You C. M., Zeng X. P., Gao X. W. (2020). The role of G protein-coupled receptor-related genes in cytochrome P450- mediated resistance of the house fly, Musca domestica (Diptera: muscidae) to imidacloprid. Insect. Mol. Biol. 29, 92–103. 10.1111/imb.12615 [DOI] [PubMed] [Google Scholar]
- Manyilizu W. B. (2019). “Pesticides, anthropogenic activities, history and the health of our environment: lessons from africa,” in Pesticides-use and misuse and their impact in the environment. Editors Larramendy, M. L., Soloneski S. (London: IntechOpen; ), 111–120. [Google Scholar]
- Mao K., Jin R., Ren Z., Zhang J., Li Z., He S., et al. (2022). miRNAs targeting CYP6ER1 and CarE1 are involved in nitenpyram resistance in Nilaparvata lugens . Insect Sci. 29, 177–187. 10.1111/1744-7917.12910 [DOI] [PubMed] [Google Scholar]
- Markov G. V., Laudet V. (2011). Origin and evolution of the ligand-binding ability of nuclear receptors. Mol. Cell Endocrinol. 334, 21–30. 10.1016/j.mce.2010.10.017 [DOI] [PubMed] [Google Scholar]
- Mei Y., Jing D., Tang S., Chen X., Chen H., Duanmu H., et al. (2022). InsectBase 2.0: A comprehensive gene resource for insects. Nucleic Acids Res. 50, D1040–D1045. 10.1093/nar/gkab1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melander A. L. (1914). Can insects become resistant to sprays? J. Eco. Entomol. 7, 167–173. 10.1093/jee/7.2.167 [DOI] [Google Scholar]
- Meng L. W., Yuan G. R., Chen M. L., Dou W., Jing T. X., Zheng L. S., et al. (2021). Genome-wide identification of long non-coding RNAs (lncRNAs) associated with malathion resistance in Bactrocera dorsalis . Pest Manag. Sci. 77, 2292–2301. 10.1002/ps.6256 [DOI] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E., Mattick J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159. 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- Milesi P., Claret J. L., Unal S., Weill M., Labbe P. (2022). Evolutionary trade-offs associated with copy number variations in resistance alleles in Culex pipiens mosquitoes. Parasit. Vectors 15, 484. 10.1186/s13071-022-05599-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra J. R., Lam G., Thummel C. S. (2013). Constitutive activation of the Nrf2/Keap1 pathway in insecticide-resistant strains of Drosophila. Insect biochem. Mol. Biol. 43, 1116–1124. 10.1016/j.ibmb.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K., Okada T. N., Siomi H., Siomi M. C. (2009). Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA 15, 1282–1291. 10.1261/rna.1541209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinie B., Wang J., Lim K. S., Hillebrand R., Lu Z. X., Van Wittenberghe N., et al. (2016). m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods. 13, 692–698. 10.1038/nmeth.3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyes C. L., Vontas J., Martins A. J., Ng L. C., Koou S. Y., Dusfour I., et al. (2017). Contemporary status of insecticide resistance in the major Aedes vectors of arboviruses infecting humans. PLoS Negl. Trop. Dis. 11, e0005625. 10.1371/journal.pntd.0005625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K., Tanaka Y., Nakano T., Adachi T., Tanaka H., Kaminuma T., et al. (2006). Nuclear receptor-mediated transcriptional regulation in Phase I, II, and III xenobiotic metabolizing systems. Drug Metab. Pharmacokinet. 21, 437–457. 10.2133/dmpk.21.437 [DOI] [PubMed] [Google Scholar]
- Nauen R., Reckmann U., Thomzik J., Thielert W. (2008). Biological profile of spirotetramat (Movento®)–a new two-way systemic (ambimobile) insecticide against sucking pest species. Bayer. Crop. 61, 245–278. [Google Scholar]
- Oppold A. M., Müller R. (2017). “Epigenetics: A hidden target of insecticides,” in Insect epigenetics, advances in insect physiology. Editor Verlinden H. (Cambridge: Academic Press; ), 313–324. [Google Scholar]
- Pan Y., Peng T., Xu P., Zeng X., Tian F., Song J., et al. (2019). Transcription factors AhR/ARNT regulate the expression of CYP6CY3 and CYP6CY4 switch conferring nicotine adaptation. Int. J. Mol. Sci. 20, 4521. 10.3390/ijms20184521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhu E., Gao X., Nauen R., Xi J., Peng T., et al. (2017). Novel mutations and expression changes of acetyl coenzyme A carboxylase are associated with spirotetramat resistance in Aphis gossypii Glover. Insect Mol. Biol. 26, 383–391. 10.1111/imb.12300 [DOI] [PubMed] [Google Scholar]
- Pasquinelli A. E., Reinhart B. J., Slack F., Martindale M. Q., Kuroda M. I., Maller B., et al. (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89. 10.1038/35040556 [DOI] [PubMed] [Google Scholar]
- Peng T., Chen X., Pan Y., Zheng Z., Wei X., Xi J., et al. (2017). Transcription factor aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator is involved in regulation of the xenobiotic tolerance‐related cytochrome P450 CYP6DA2 in Aphis gossypii Glover. Insect Mol. Biol. 26, 485–495. 10.1111/imb.12311 [DOI] [PubMed] [Google Scholar]
- Peng T., Liu X., Tian F., Xu H., Yang F., Chen X., et al. (2022). Functional investigation of lncRNAs and target cytochrome P450 genes related to spirotetramat resistance in Aphis gossypii Glover. Pest Manag. Sci. 78, 1982–1991. 10.1002/ps.6818 [DOI] [PubMed] [Google Scholar]
- Peng T., Pan Y., Gao X., Xi J., Zhang L., Ma K., et al. (2016). Reduced abundance of the CYP6CY3-targeting let-7 and miR-100 miRNAs accounts for host adaptation of Myzus persicae nicotianae . Insect biochem. Mol. Biol. 75, 89–97. 10.1016/j.ibmb.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Peng T., Pan Y., Tian F., Xu H., Yang F., Chen X., et al. (2021). Identification and the potential roles of long non-coding RNAs in regulating acetyl-CoA carboxylase ACC transcription in spirotetramat-resistant Aphis gossypii . Pestic. Biochem. Physiol. 179, 104972. 10.1016/j.pestbp.2021.104972 [DOI] [PubMed] [Google Scholar]
- Pigott C. R., Ellar D. J. (2007). Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol. Mol. Biol. Rev. 71, 255–281. 10.1128/mmbr.00034-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J., Du Y., Yu J., Guo J. (2019). MicroRNAs as potential biomarkers of insecticide exposure: A review. Chem. Res. Toxicol. 32, 2169–2181. 10.1021/acs.chemrestox.9b00236 [DOI] [PubMed] [Google Scholar]
- Qin J., Ye F., Xu L., Zhou X., Crickmore N., Zhou X., et al. (2021). A cis-acting mutation in the Px ABCG1 promoter is associated with Cry1Ac resistance in Plutella xylostella (L). Int. J. Mol. Sci. 22, 6106. 10.3390/ijms22116106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J. J., Chang H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- Ranson H., N’guessan R., Lines J., Moiroux N., Nkuni Z., Corbel V. (2011). Pyrethroid resistance in african anopheline mosquitoes: what are the implications for malaria control? Trends Parasitol. 27, 91–98. 10.1016/j.pt.2010.08.004 [DOI] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature 403, 901–906. 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Riveron J. M., Yunta C., Ibrahim S. S., Djouaka R., Irving H., Menze B. D., et al. (2014). A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 15, R27–R20. 10.1186/gb-2014-15-2-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini R. P., Raman V., Dhandapani G., Malhotra E. V., Sreevathsa R., Kumar P. A., et al. (2018). Silencing of HaAce1 gene by host-delivered artificial microRNA disrupts growth and development of Helicoverpa armigera. PLoS One 13, e0194150. 10.1371/journal.pone.0194150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. M., Good R. T., Appleton B., Sherrard J., Raymant G. C., Bogwitz M. R., et al. (2010). Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6, e1000998. 10.1371/journal.pgen.1000998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong K. M., Coates B. S., Pittendrigh B. R. (2019). Impacts of sub-lethal DDT exposures on microRNA and putative target transcript expression in DDT resistant and susceptible Drosophila melanogaster strains. Front. Genet. 10, 45. 10.3389/fgene.2019.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Li W. L., Zeng H. X., Shi Y., Liao X. L. (2022). Systematic identification and functional analysis of long noncoding RNAs involved in indoxacarb resistance in Spodoptera litura . Insect Sci. 29, 1721–1736. 10.1111/1744-7917.13015 [DOI] [PubMed] [Google Scholar]
- Shi L., Shi Y., Liu M. F., Zhang Y., Liao X. L. (2021). Transcription factor CncC potentially regulates the expression of multiple detoxification genes that mediate indoxacarb resistance in Spodoptera litura . Insect Sci. 28, 1426–1438. 10.1111/1744-7917.12860 [DOI] [PubMed] [Google Scholar]
- Shi L., Wang M., Zhang Y., Shen G., Di H., Wang Y., et al. (2017). The expression of P450 genes mediating fenpropathrin resistance is regulated by CncC and Maf in Tetranychus cinnabarinus (Boisduval). Comp. Biochem. Physiol. Part - C. Toxicol. 198, 28–36. 10.1016/j.cbpc.2017.05.002 [DOI] [PubMed] [Google Scholar]
- Siddiqui J. A., Fan R., Naz H., Bamisile B. S., Hafeez M., Ghani M. I., et al. (2023). Insights into insecticide-resistance mechanisms in invasive species: challenges and control strategies. Front. Physiol. 13, 1112278. 10.3389/fphys.2022.1112278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. Y. (2014). The toxicology and biochemistry of insecticides. Boca Raton: CRC Press, 249–251. [Google Scholar]
- Sogawa K., Nakano R., Kobayashi A., Kikuchi Y., Ohe N., Matsushita N., et al. (1995). Possible function of Ah receptor nuclear translocator (Arnt) homodimer in transcriptional regulation. Proc. Natl. Acad. Sci. U. S. A. 92, 1936–1940. 10.1073/pnas.92.6.1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon K. R., Giddings J. M., Maund S. J. (2001). Probabilistic risk assessment of cotton pyrethroids: I. Distributional analyses of laboratory aquatic toxicity data. Environ. Toxicol. Chem. 20, 652–659. [DOI] [PubMed] [Google Scholar]
- Sparks T. C., Nauen R. (2015). Irac: mode of action classification and insecticide resistance management. Pestic. Biochem. Physiol. 121, 122–128. 10.1016/j.pestbp.2014.11.014 [DOI] [PubMed] [Google Scholar]
- Sparks T. C., Wessels F. J., Lorsbach B. A., Nugent B. M., Watson G. B. (2019). The new age of insecticide discovery-the crop protection industry and the impact of natural products. Pestic. Biochem. Physiol. 161, 12–22. 10.1016/j.pestbp.2019.09.002 [DOI] [PubMed] [Google Scholar]
- Sun D., Zhu L., Guo L., Wang S., Wu Q., Crickmore N., et al. (2022). A versatile contribution of both aminopeptidases N and ABC transporters to Bt Cry1Ac toxicity in the diamondback moth. BMC Biol. 20, 33. 10.1186/s12915-022-01226-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. H., Xu N., Xu Y., Zhou D., Sun Y., Wang W. J., et al. (2019). A novel miRNA, miR-13664, targets CpCYP314A1 to regulate deltamethrin resistance in Culex pipiens pallens . Parasitol. Res. 146, 197–205. 10.1017/S0031182018001002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Yamamoto M. (2015). Molecular basis of the keap1–nrf2 system. Free Radic. Biol. Med. 8, 93–100. 10.1016/j.freeradbiomed.2015.06.006 [DOI] [PubMed] [Google Scholar]
- Tang B., Cheng Y., Li Y., Li W., Ma Y., Zhou Q., et al. (2020). Adipokinetic hormone regulates cytochrome P450-mediated imidacloprid resistance in the brown planthopper, Nilaparvata lugens . Chemosphere 259, 127490. 10.1016/j.chemosphere.2020.127490 [DOI] [PubMed] [Google Scholar]
- Tang G. H., Xiong Y., Liu Y., Song Z. H., Yang Y., Shen G. M., et al. (2019). The Transcription factor mafB regulates the susceptibility of Bactrocera dorsalis to abamectin via GSTz2. Front. Physiol. 10, 1068. 10.3389/fphys.2019.01068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiewsiri K., Wang P. (2011). Differential alteration of two aminopeptidases N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc. Nat.l Acad. Sci. U.S.A. 34, 14037–14042. 10.1073/pnas.1102555108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troczka B., Zimmer C. T., Elias J., Schorn C., Bass C., Davies T. E., et al. (2012). Resistance to diamide insecticides in diamondback moth, Plutella xylostella (Lepidoptera: plutellidae) is associated with a mutation in the membrane-spanning domain of the ryanodine receptor. Insect bio. Chem. Mol. Biol. 42, 873–880. 10.1016/j.ibmb.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Truscott M., Islam A. B., López-Bigas N., Frolov M. V. (2011). mir-11 limits the proapoptotic function of its host gene, dE2f1. Genes Dev. 25, 1820–1834. 10.1101/gad.16947411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C. P., Akgül B. (2005). Drosophila glutathione S‐transferases. Meth. Enzymol. 401, 204–226. 10.1016/S0076-6879(05)01013-X [DOI] [PubMed] [Google Scholar]
- Van Leeuwen T., Dermauw W., Mavridis K., Vontas J. (2020). Significance and interpretation of molecular diagnostics for insecticide resistance management of agricultural pests. Curr. Opin. Insect Sci. 39, 69–76. 10.1016/j.cois.2020.03.006 [DOI] [PubMed] [Google Scholar]
- Varghese J., Cohen S. M. (2007). microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 21, 2277–2282. 10.1101/gad.439807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese J., Lim S. F., Cohen S. M. (2010). Drosophila miR-14 regulates insulin production and metabolism through its target, sugarbabe. Genes Dev. 24, 2748–2753. 10.1101/gad.1995910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velarde R. A., Robinson G. E., Fahrbach S. E. (2006). Nuclear receptors of the honey bee: annotation and expression in the adult brain. Insect Mol. Biol. 15, 583–595. 10.1111/j.1365-2583.2006.00679.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jin R., Liu C., Gao Y., Deng X., Wan H., et al. (2021). Functional characterization of the transcription factors AhR and ARNT in Nilaparvata lugens . Pestic. Biochem. Physiol. 176, 104875. 10.1016/j.pestbp.2021.104875 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhao S., Shi L., Xu Z., He L. (2014). Resistance selection and biochemical mechanism of resistance against cyflumetofen in Tetranychus cinnabarinus (Boisduval). Pestic. Biochem. Physiol. 111, 24–30. 10.1016/j.pestbp.2014.04.004 [DOI] [PubMed] [Google Scholar]
- Weetman D., Djogbenou L. S., Lucas E. (2018). Copy number variation (CNV) and insecticide resistance in mosquitoes: evolving knowledge or an evolving problem? Curr. Opin. Insect Sci. 27, 82–88. 10.1016/j.cois.2018.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Zheng C., Peng T., Pan Y., Xi J., Chen X., et al. (2016). miR-276 and miR-3016-modulated expression of acetyl-CoA carboxylase accounts for spirotetramat resistance in Aphis gossypii Glover. Insect biochem. Mol. Biol. 79, 57–65. 10.1016/j.ibmb.2016.10.011 [DOI] [PubMed] [Google Scholar]
- World Health Organisation (2012). World malaria report. Switzerland: WHO Press. [Google Scholar]
- World Health Organization (2016). Test procedures for insecticide resistance monitoring in malaria vector mosquitoes . https://apps.who.int/iris/bitstream/handle/10665/250677/9789241511575-eng.pdf (Accessed February 25, 2023). [Google Scholar]
- Xiao Y. T., Zhang T., Liu C. X., Heckel D. G., Li X. C., Tabashnik B. E., et al. (2014). Mis-splicing of the ABCC2 gene linked with Bt toxin resistance in Helicoverpa armigera . Sci. Rep. 4, 6184. 10.1038/srep06184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Tan A., Palli S. R. (2010). The function of nuclear receptors in regulation of female reproduction and embryogenesis in the red flour beetle, Tribolium castaneum . Tribolium Castaneum. J. Insect Physiol. 56, 1471–1480. 10.1016/j.jinsphys.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Vernooy S. Y., Guo M., Hay B. A. (2003). The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13, 790–795. 10.1016/s0960-9822(03)00250-1 [DOI] [PubMed] [Google Scholar]
- Xu X. J., Yu L. Y., Wu Y. D. (2005). Disruption of a cadherin gene associated with resistance to Cry1Ac δ-endotoxin of Bacillus thuringiensis in Helicoverpa armigera . Appl. Environ. Microbiol. 71, 948–954. 10.1128/aem.71.2.948-954.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Chen S., Xu X., Lin G., Lin S., Bai J., et al. (2022). Novel-miR-310 mediated response mechanism to Cry1Ac protoxin in Plutella xylostella (L). Int. J. Biol. Macromol. 219, 587–596. 10.1016/j.ijbiomac.2022.08.017 [DOI] [PubMed] [Google Scholar]
- Yang J., Chen S., Xu X., Lin S., Wu J., Lin G., et al. (2023). Novel miR-108 and miR-234 target juvenile hormone esterase to regulate the response of Plutella xylostella to Cry1Ac protoxin. Ecotoxicol. Environ. Saf. 254, 114761. 10.1016/j.ecoenv.2023.114761 [DOI] [PubMed] [Google Scholar]
- Yang S. H., Sharrocks A. D., Whitmarsh A. J. (2013). MAP kinase signalling cascades and transcriptional regulation. Gene 513, 1–13. 10.1016/j.gene.2012.10.033 [DOI] [PubMed] [Google Scholar]
- Yang X., Deng S., Wei X., Yang J., Zhao Q., Yin C., et al. (2020). MAPK-directed activation of the whitefly transcription factor CREB leads to P450-mediated imidacloprid resistance. Proc. Natl. Acad. Sci. U.S.A. 117, 10246–10253. 10.1073/pnas.1913603117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Wei X., Yang J., Du T., Yin C., Fu B., et al. (2021). Epitranscriptomic regulation of insecticide resistance. Sci. Adv. 7, eabe5903. 10.1126/sciadv.abe5903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogindran S., Rajam M. V. (2016). Artificial miRNA-mediated silencing of ecdysone receptor (EcR) affects larval development and oogenesis in Helicoverpa armigera . Insect biochem. Mol. Biol. 77, 21–30. 10.1016/j.ibmb.2016.07.009 [DOI] [PubMed] [Google Scholar]
- Zhang B. Z., Zhang M. Y., Li Y. S., Hu G. L., Fan X. Z., Guo T. X., et al. (2022b). MicroRNA-263b confers imidacloprid resistance in Sitobion miscanthi (Takahashi) by regulating the expression of the nAChRβ1 subunit. Pestic. Biochem. Physiol. 187, 105218. 10.1016/j.pestbp.2022.105218 [DOI] [PubMed] [Google Scholar]
- Zhang C., Liu P., Sun L., Cao C. (2023a). Integration of miRNA and mRNA expression profiles in Asian spongy moth Lymantria dispar in response to cyantraniliprole. Pestic. Biochem. Physiol. 191, 105364. 10.1016/j.pestbp.2023.105364 [DOI] [PubMed] [Google Scholar]
- Zhang C., Wang X., Tai S., Qi L., Yu X., Dai W. (2023c). Transcription factor CncC potentially regulates cytochrome P450 CYP321A1-mediated flavone tolerance in Helicoverpa armigera . Pestic. Biochem. Physiol. 191, 105360. 10.1016/j.pestbp.2023.105360 [DOI] [PubMed] [Google Scholar]
- Zhang H., Lin X., Yang B., Zhang L., Liu Z. (2023b). Two point mutations in CYP4CE1 promoter contributed to the differential regulation of CYP4CE1 expression by FoxO between susceptible and nitenpyram resistant Nilaparvata lugens . Res. Sq. 10.21203/rs.3.rs-2799058/v1 [DOI] [PubMed] [Google Scholar]
- Zhang M. Y., Zhang P., Su X., Guo T. X., Zhou J. L., Zhang B. Z., et al. (2022a). MicroRNA-190-5p confers chlorantraniliprole resistance by regulating CYP6K2 in Spodoptera frugiperda (Smith). Pestic. Biochem. Physiol. 184, 105133. 10.1016/j.pestbp.2022.105133 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Dou W., Taning C. N., Smagghe G., Wang J. J. (2021). Regulatory roles of microRNAs in insect pests: prospective targets for insect pest control. Curr. Opin. Biotechnol. 70, 158–166. 10.1016/j.copbio.2021.05.002 [DOI] [PubMed] [Google Scholar]
- Zhang X., Jie D., Liu J., Zhang J., Zhang T., Zhang J., et al. (2019). Aryl hydrocarbon receptor regulates the expression of LmGSTd7 and is associated with chlorpyrifos susceptibility in Locusta migratoria . Pest Manag. Sci. 75, 2916–2924. 10.1002/ps.5600 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng K., Hu J., Shi L., Wei P., Xu Z., et al. (2018). A microRNA‐1 gene, tci‐miR‐1‐3p, is involved in cyflumetofen resistance by targeting a glutathione S‐transferase gene, TCGSTM4, in Tetranychus cinnabarinus . Insect Mol. Biol. 27 (3), 352–364. 10.1111/imb.12375 [DOI] [PubMed] [Google Scholar]
- Zhao Y. X., Huang J. M., Ni H., Guo D., Yang F. X., Wang X., et al. (2020). Susceptibility of fall armyworm, Spodoptera frugiperda (JE Smith), to eight insecticides in China, with special reference to lambda-cyhalothrin. Pestic. Biochem. Physiol. 168, 104623. 10.1016/j.pestbp.2020.104623 [DOI] [PubMed] [Google Scholar]
- Zhou Q. Z., Zhang B., Yu Q. Y., Zhang Z. (2016). BmncRNAdb: A comprehensive database of non-coding RNAs in the silkworm, Bombyx mori . BMC Bioinforma. 17, 370. 10.1186/s12859-016-1251-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Li L., Wei R., Liang P., Gao X. (2021). Regulation of GSTu1-mediated insecticide resistance in Plutella xylostella by miRNA and lncRNA. PLoS Genet. 17, e1009888. 10.1371/journal.pgen.1009888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Sun X., Nie X., Liang P., Gao X. (2020). MicroRNA-998–3p contributes to Cry1Ac-resistance by targeting ABCC2 in lepidopteran insects. Insect bio. Chem. Mol. Biol. 117, 103283. 10.1016/j.ibmb.2019.103283 [DOI] [PubMed] [Google Scholar]
- Zhu B., Xu M., Shi H., Gao X., Liang P. (2017). Genome-wide identification of lncRNAs associated with chlorantraniliprole resistance in diamondback moth Plutella xylostella (L). BMC Genom 18, 380–411. 10.1186/s12864-017-3748-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubair M., Khan M. Z., Rauf I., Raza A., Shah A. H., Hassan I., et al. (2020). Artificial microRNA (amiRNA)-mediated resistance against whitefly (Bemisia tabaci) targeting three genes. Crop Prot. 137, 105308. 10.1016/j.cropro.2020.105308 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.