With great interest, we have read the article by Clooney et al, which highlighted the regional effects on the heterogeneity of the gut microbiota among populations with inflammatory bowel disease (IBD).1 As a result, regional effects would largely limit the microbial-based diagnosis of diseases across regions. Although current machine learning methods based on microbial features have been applied to diagnosis of diseases such as IBD2 and type 2 diabetes,3 these methods are unable to mitigate the regional effects and meet the demand of microbial-based cross-regional diagnosis of diseases.
Here, we proposed a machine learning framework (online supplemental figure S1, accessible at: https://github.com/HUST-NingKang-Lab/EXPERT-Disease-GGMP), which integrated the neural network and transfer learning, to effectively reduce regional effects for microbial-based cross-regional diagnosis. Importantly, transfer learning can ‘borrow’ the mature knowledge about diseases from a source city to assist the disease diagnosis for a target city, especially when there is little knowledge about microbiota patterns in the target city.4
gutjnl-2022-328216supp001.pdf (2.4MB, pdf)
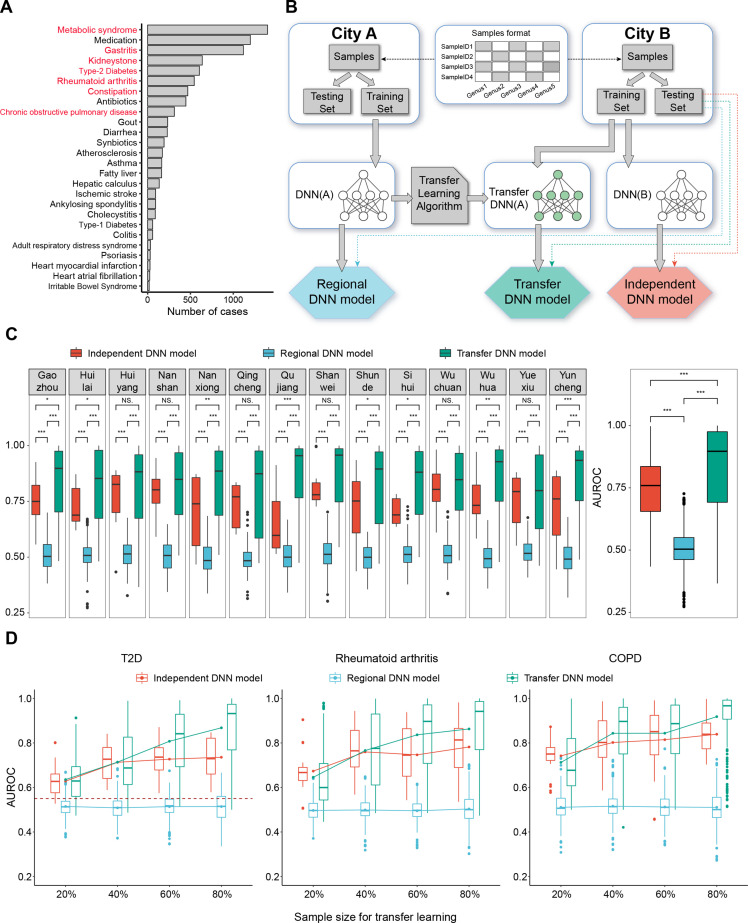
To assess the framework, we obtained genus-level taxonomy profiles from the Guangdong Gut Microbiome Project.5 These samples were collected from 14 cities and seven representative diseases were selected for assessment (figure 1A and online supplemental table S1). We randomly divided samples of each city into the training subset and the testing subset (80%:20% by default), then performed assessments for three models: (1) Independent disease neural network (DNN) model: ab initio training and testing the DNN model on the training subset and the testing subset of each city, respectively. (2) Regional DNN model: ab initio training the DNN model using the training subset of one city A (source city) and testing it on the testing subset of another city B (target city). (3) Transfer DNN model: ab initio training the DNN model using training subset of one city A, followed by applying transfer learning to a certain proportion (from 20% to 80%) of samples from city B, and then testing the transfer DNN model on the testing subset of city B (figure 1B and online supplemental figure S1).
Figure 1.
Data distribution, assessment workflow and framework evaluation. (A) data distribution. The number of samples of different diseases. The seven diseases marked in red were assessed, including metabolic syndrome, gastritis, kidney stones, T2D, rheumatoid arthritis, constipation and COPD. (B) The workflow of assessment. The genera abundance profiles of samples from each city were randomly divided into the training subset (80%) and the testing subset (20%). Three assessment workflows for each model were marked by three different colours. The testing subset of city B was used to test all the three models. (C) Framework evaluation: comparison of the AUROC of three models. Boxplots in the left panel show the AUROC of the three models for diagnosing seven diseases using samples in each of city, and the right panel shows these values collectively. *, p<0.05; **, p<0.01; ***, p<0.005; Mann-Whitney-Wilcoxon test. (D) The relationship between sample size and the performance of three models. Boxplots show AUROC of three models for diagnosing three diseases (COPD, rheumatoid arthritis and T2D). The lines show the change in average AUROC of three models with sample size increasing. The dashed line shows the average AUROC of cross-regional diagnosis of T2D using random forest model.5 For all the boxplots, boxes represent the IQR between the first and third quartiles and the line inside represents the median. Whiskers denote the lowest and highest values within the 1.5×IQR from the first and third quartiles, respectively. AUROC, area under the receiver operating characteristic; COPD, chronic obstructive pulmonary disease; T2D, type 2 diabetes.
We found that the regional DNN model across cities presented a low average accuracy of 0.506 compared with the independent DNN model with an average accuracy of 0.743 (pWilcox=2.22×10−16; figure 1C and online supplemental figure S2). It suggested that regional factors largely limited the cross-regional diagnosis, as also indicated in previous studies.5 However, the transfer DNN model profoundly increased prediction accuracy across cities with an average accuracy of 0.829 (pWilcox=2.22×10−16, compared with the independent DNN model; figure 1C and online supplemental figure S2). Intriguingly, once the proportion of samples used in the target city exceeded 50% for transfer learning, the transfer DNN model could even present higher prediction accuracy than that of the independent DNN model (figure 1D). Furthermore, the transfer DNN models also had good performance when we have applied this approach on two intercontinental cohorts (online supplemental figures S3 and S4).
Moreover, our machine learning framework is advantageous in identification of region-specific microbes, as well as microbes shared across all regions. We used the ‘leave-one-feature-out’ method to discover certain microbes which were strongly affected by regions, such as Enterobacteriaceae and Clostridium, while others were less affected by regions, such as Parabacteroides and Faecalibacterium (online supplemental table S2). We speculated that the region-specific microbes may contribute to the effectiveness of the transfer DNN model in the cross-regional diagnosis of diseases.
Collectively, our study demonstrates that transfer learning can realise microbial-based cross-regional diagnosis of diseases with high accuracy and robustness, by using knowledge about microbial features across regions. This study provides a new venue to exceed the regional limitation, and facilitate microbial-based cross-regional diagnosis of diseases in clinical trials by artificial intelligence techniques.
Data accession: metagenomic sequencing samples are available in the European Bioinformatics Institute (EBI) database of European Molecular Biology Laboratory (EBI accession number PRJEB18535) at https://www.ebi.ac.uk/ena/browser/view/PRJEB18535.
Footnotes
Contributors: NW and KN designed the study, conceived and proposed the idea. NW performed the experiments and analysed the data. NW visualised the data. NW, MC and KN contributed to editing and proofreading the manuscript. All authors read and approved the final manuscript.
Funding: This work was partially supported by the National Natural Science Foundation of China (Grant Nos. 32071465, 31871334, and 31671374), and the National Key R&D Programme of China (Grant No. 2018YFC0910502).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1. Clooney AG, Eckenberger J, Laserna-Mendieta E, et al. Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut 2021;70:499–510. 10.1136/gutjnl-2020-321106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weng F, Meng Y, Lu F, et al. Differentiation of intestinal tuberculosis and crohn's disease through an explainable machine learning method. Sci Rep 2022;12:1714. 10.1038/s41598-022-05571-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gou W, Ling C-W, He Y, et al. Interpretable machine learning framework reveals robust gut microbiome features associated with type 2 diabetes. Diabetes Care 2021;44:358–66. 10.2337/dc20-1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sun Q, Liu Y, Chua T. Meta-transfer learning for few-shot learning. 2019 IEEE Conf Comput Vis Pattern Recognit, 2019:403–12. [Google Scholar]
- 5. He Y, Wu W, Zheng H-M, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med 2018;24:1532–5. 10.1038/s41591-018-0164-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2022-328216supp001.pdf (2.4MB, pdf)