Abstract
A total of 148 isolates from 55 bacteremic patients were examined by pulsed-field gel electrophoresis. Genetically different nonblood strains were isolated from 13.9% of patients with bacteremia caused by gram-positive cocci and 42.1% with Pseudomonas aeruginosa bacteremia, indicating that antibiograms of a single nonblood P. aeruginosa isolate are not always informative for treatment of bacteremia.
Bacteremia arises from preexisting local infections such as pneumonia, bile tract infection, and wounds in some patients, while in immunocompromised patients, it frequently arises as a result of invasion by endogenous microflora (1, 9, 15). Moreover, there are bacteremias with no identifiable source, such as occult bacteremia of childhood. For patients with leukemia, surveillance culture of stool or throat samples is clinically important since primary bacteremia frequently occurs during chemotherapy through colonization of the intestinal tract or throat (8, 15). In patients with primary lung cancer, infections, including endogenous bacteremia, also occur frequently during chemotherapy (5, 7). If strains isolated from nonblood clinical specimens prior to acquisition of a positive blood culture are identical to blood isolates, antibiograms and other information should be useful in the prophylaxis and treatment of bacteremia. However, it remains to be clarified whether blood isolates are genetically identical to isolates from nonblood specimens (6, 11). Nor is it clear whether there are any differences between blood and other isolates among bacterial species. To address these points, we examined genetic relationships between clinical blood isolates and nonblood isolates of the same species of bacteria by pulsed-field gel electrophoresis (PFGE).
The study was carried out in Nagasaki University Hospital, an 829-bed hospital in Nagasaki, Japan. Freeze-dried stocked clinical strains isolated between January 1992 and December 1996 were used. Clinical isolates were identified by Vitek Gram-Positive and -Negative Identification cards (bioMerieux-Vitek, Inc., Hazelwood, Mo.). Staphylococci were also identified simultaneously with an ID 32 STAPH kit (bioMerieux Sa, Marcy l’Etoile, France), and streptococci and enterococci were identified with a rapid ID 32 STREP kit (bioMerieux). Patients with episodes of positive blood culture associated with isolation of the same bacterial species from other clinical specimens within a month before positive blood culture was obtained were consecutively enrolled in this study. A septic episode of bacteremia was defined as described previously (2, 13), i.e., as follows: (i) the first positive culture or (ii) a new positive blood culture occurring 48 h after the preceding positive culture. The numbers of cases and isolates tested for each bacterial species are listed in Table 1. In total, 148 isolates, including methicillin-susceptible Staphylococcus aureus (MSSA), methicillin-resistant S. aureus (MRSA), coagulase-negative staphylococci (CoNS), Streptococcus pneumoniae, enterococci, and Pseudomonas aeruginosa, from 55 patients were examined.
TABLE 1.
Number of bacteremic patients and related isolates used for the PFGE study
| Organism(s) | No. of bacteremic patients | No. of related isolates testeda |
|---|---|---|
| MSSA | 4 | 13 |
| MRSA | 18 | 57 |
| CoNS | 11 | 27 |
| S. pneumoniae | 1 | 2 |
| Enterococci | 2 | 4 |
| P. aeruginosa | 19 | 45 |
| Total | 55 | 148 |
Both blood and nonblood isolates from patients with bacteremia were included.
Chromosomal DNA was prepared from each strain and digested with restriction enzymes by using GenePath group reagent kits (Bio-Rad Laboratories, Hercules, Calif.) except for S. pneumoniae isolates. SmaI was used for staphylococci and enterococci, while SpeI was used for P. aeruginosa in accordance with the manufacturer’s instructions. The samples were electrophoresed with the Gene Navigator system (Pharmacia LKB Biotechnology, Uppsala, Sweden) at 170 V for 24 h, with pulse times ranging from 5 to 60 s for staphylococci and enterococci, and at 180 V for 22 h, with pulse times ranging from 10 to 45 s for P. aeruginosa (10). Genomic DNA of S. pneumoniae was prepared, digested with SmaI, and processed for PFGE as described previously (4, 14). Electrophoresis was carried out at 200 V for 20 h, with a pulse time of 9.5 s. A lambda DNA ladder (Bio-Rad) was used for each PFGE as a molecular size marker. Thereafter, gels were stained with ethidium bromide before being photographed under UV transillumination. The criteria reported by F. C. Tenover et al. (12) were applied to the interpretation of the DNA restriction patterns produced by PFGE. Briefly, each strain was classified as indistinguishable, closely related, possibly related, or different if the number(s) of fragment differences compared with a reference strain was 0, 1 to 3, 4 to 6, or ≥7, respectively. If the strains were indistinguishable, closely related, or probably related on the basis of DNA restriction patterns, they were considered derivatives from a common ancestor.
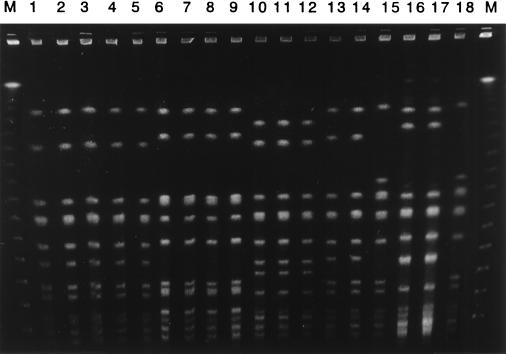
Figure 1 shows PFGE patterns of MRSA strains from patients with bacteremic episodes. All PFGE profiles of nonblood isolates were identical to those of blood isolates for patients A through D. Although an isolate from the throat was identical to a blood isolate from patient E, sputum and pus isolates, which were identical, showed distinguishable PFGE patterns. Table 2 shows the genetic relationship of MRSA isolated from nonblood specimens to blood isolates of MRSA. In total, 37 of 39 (94.9%) nonblood isolates of MRSA were genetically related to blood isolates; these consisted of 33 indistinguishable and four closely related isolates.
FIG. 1.
PFGE profiles of MRSA isolated from blood and nonblood specimens. Genomic DNA of MRSA strains was digested with restriction enzyme SmaI. Samples were ordered by set of blood isolate and nonblood isolate(s) from each patient. Lanes 1, 6, 10, 13, and 15 are blood isolates from various patients. Lanes 2 to 5 are isolates from sputum, the tip of a central venous catheter, pus, and feces from patient A, respectively. Lanes 7 to 9 are isolates from sputum and nasal and throat swabs from patient B, respectively. Lanes 11 and 12 are isolates from the tip of a central venous catheter and sputum from patient C, respectively. Lane 14 is an isolate from pus from patient D. Lanes 16 to 18 are isolates from pus, sputum, and a throat swab from patient E, respectively. M, molecular size marker.
TABLE 2.
Genetic relationships of MRSA strains isolated from nonblood specimens and blood
| PFGE interpretation | No. of nonblood isolates from:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Sputum | Throat swab | Nasal swab | Pus | Urine | Feces | Tip of catheter | Other | |
| Indistinguishable | 9 | 4 | 3 | 5 | 2 | 2 | 5 | 3 |
| Closely related | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| Possibly related | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Different | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Total | 11 | 4 | 4 | 5 | 2 | 3 | 7 | 3 |
For the patients with MSSA bacteremia, all nonblood isolates, including the strains isolated from sputum (one strain), a throat swab (two strains), a nasal swab (one strain), pus (two strains), ascites fluid (one strain), and the tip of a central venous catheter (two strains), were genetically indistinguishable from blood isolates. For the patients with CoNS bacteremia, 14 of 16 (87.5%) nonblood isolates, including the strains isolated from sputum (2 strains), a throat swab (6 strains), a nasal swab (2 strains), ascites fluid (1 strain), the tip of an intravenous catheter (1 strain), and feces (2 strains), were genetically related to blood isolates, while 2 strains, 1 from throat swab and 1 from the tip of an intravenous catheter, were different from the blood isolates from the respective patients. The 14 genetically related isolates consisted of 10 indistinguishable, 2 closely related (isolates from a throat swab), and 2 possibly related (isolates from a throat swab and ascites fluid) strains. An isolate of S. pneumoniae from cerebrospinal fluid was genetically identical to a blood isolate from the same patient. A strain of Enterococcus faecium isolated from sputum was identical to the blood isolate from the same patient, while a strain of Enterococcus faecalis isolated from the urine of another patient was different from a blood isolate from that patient.
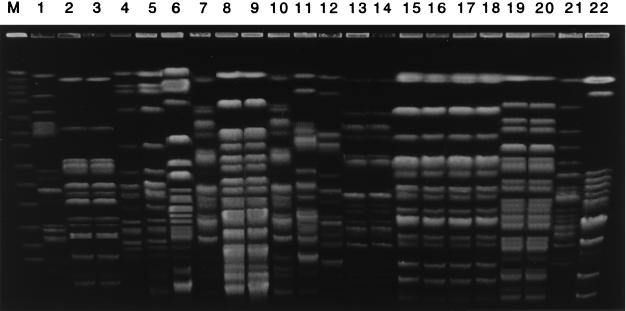
Figure 2 shows PFGE patterns of P. aeruginosa strains from some of the patients with bacteremic episodes. In contrast to those of MRSA, PFGE profiles of nonblood isolates of P. aeruginosa were frequently different from those of blood isolates. Although the genotypes of nonblood isolates were identical to those of blood isolates for patients J through M, genetically distinct nonblood P. aeruginosa strains were isolated from other patients. Although the genotypes of nasal and throat isolates were the same for patient F, they differed from that of the blood isolate. A fecal isolate was genetically identical to a blood isolate from patient G, while a different P. aeruginosa organism was isolated from the patient’s sputum. Although an isolate from pus was identical to a blood isolate for patient H, sputum and throat isolates, showing the same genotype, were distinguishable. Cross transmission between patients K and L was suggested since the PFGE profiles of all isolates were genetically identical between the two. Table 3 shows the results of genetic analysis of P. aeruginosa isolated from nonblood specimens and of blood isolates. The PFGE profiles of two isolates from the tip of a central venous catheter differed from those of blood isolates. In total, 16 of 27 (59.3%) nonblood isolates of P. aeruginosa were genetically related to blood isolates, including 15 indistinguishable and 1 closely related isolates. Additionally, more than two strains with different morphology or hemolysis activity were isolated from some samples (one blood, one throat swab, and two sputa); these showed identical PFGE patterns. Genetically different nonblood isolates were rare for patients with bacteremia caused by gram-positive cocci (5 of 36 patients; 13.9%), while genetically distinct nonblood strains were isolated from 8 of 19 patients (42.1%) with P. aeruginosa bacteremia.
FIG. 2.
PFGE profiles of P. aeruginosa isolated from blood and nonblood specimens. Genomic DNA of P. aeruginosa strains was digested with restriction enzyme SpeI. Samples were ordered by set of blood isolate and nonblood isolate(s) from each patient. Lanes 1, 4, 7, 11, 13, 15, 17, 19, and 21 are blood isolates from various patients. Lanes 2 and 3 are isolates from nasal and throat swabs from patient F, respectively. Lanes 5 and 6 are isolates from feces and sputum from patient G, respectively. Lanes 8 to 10 are isolates from sputum, a throat swab, and pus from patient H, respectively. Lanes 12, 14, 16, 18, 20, and 22 are isolates from the tip of a central venous catheter from patient I, a throat swab from patient J, sputum from patient K, pleural effusion from patient L, urine from patient M, and feces from patient N, respectively. M, molecular size marker.
TABLE 3.
Genetic relationships of P. aeruginosa strains isolated from nonblood specimens and blood
| PFGE interpretation | No. of nonblood isolates from:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Sputum | Throat swab | Nasal swab | Pus | Urine | Feces | Tip of catheter | Other | |
| Indistinguishable | 3 | 4 | 1 | 3 | 2 | 1 | 0 | 1 |
| Closely related | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Possibly related | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Different | 3 | 3 | 1 | 0 | 0 | 2 | 2 | 0 |
| Total | 6 | 7 | 2 | 4 | 2 | 3 | 2 | 1 |
Most nonblood isolates of gram-positive cocci, especially staphylococci, were genetically related to blood isolates for each patient. Therefore, antibiograms, the presence of resistance genes or specific virulence factors (e.g., toxic shock syndrome toxin 1 or enterotoxins), and other information concerning nonblood isolates may help in the treatment of patients who have contracted bacteremia or other infections.
In contrast, nonblood isolates of P. aeruginosa frequently differed in genotype from blood isolates. Moreover, three different genotypes of P. aeruginosa were isolated from some patients. Since P. aeruginosa widely exists both in the hospital environment and in nature (e.g., in water, soil, and vegetables), it is possible that patients and healthy individuals are colonized by several genetically different P. aeruginosa strains.
Of interest, for one patient with CoNS bacteremia and for two patients with P. aeruginosa bacteremia, the PFGE profiles of isolates from the tips of central venous catheters were different from those of blood isolates. These findings suggest that two or more genetically distinct members of the same species of bacteria can invade the bloodstream of a patient.
Although surveillance cultures are performed prior to chemotherapy or organ transplantation in compromised hosts (3, 8, 15), our results suggest that a single nonblood isolate of P. aeruginosa is not always informative. The findings of this study suggest that several specimens from different sites should be cultured and that antimicrobial susceptibility testing of the isolates should be performed.
REFERENCES
- 1.Body G P. Epidemiological studies of Pseudomonas species in patients with leukemia. Am J Med Sci. 1970;260:82–89. doi: 10.1097/00000441-197008000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Hirakata Y, Furuya N, Iwata M, Kashitani F, Ishikawa M, Yumoto S, Yasui K, Endoh H, Marui A, Kaku M, Tateda K, Yamaguchi K. Assessment of clinical significance of positive blood cultures of relatively low-virulence isolates. J Med Microbiol. 1996;44:195–198. doi: 10.1099/00222615-44-3-195. [DOI] [PubMed] [Google Scholar]
- 3.Hirakata Y, Katoh T, Tsukagoshi M, Hayashi M, Sugiyama Y, Kitamura S. Bacterial colonization of the upper respiratory tract of patients with primary lung cancer and nonmalignant lung disease. Chemotherapy (Basel) 1997;43:400–405. doi: 10.1159/000239598. [DOI] [PubMed] [Google Scholar]
- 4.Lefevre J C, Faucon G, Sicard A M, Gasc A M. DNA fingerprinting of Streptococcus pneumoniae strains by pulsed-field gel electrophoresis. J Clin Microbiol. 1993;31:2724–2728. doi: 10.1128/jcm.31.10.2724-2728.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oshita F, Tamura T, Okamoto H, Miya T, Kojima A, Ohe Y, Sasaki Y, Eguchi K, Schinkai T, Saijo N. The frequency and management of infectious episodes and sepsis in small cell lung cancer patients receiving intensive chemotherapy with granulocyte-colony stimulating factor. Jpn J Clin Oncol. 1991;21:353–359. [PubMed] [Google Scholar]
- 6.Pfaller M A, Wendt C, Hollis R J, Wenzel R P, Fritschel S J, Neubauer J J, Herwaldt L A. Comparative evaluation of an automated ribotyping system versus pulsed-field gel electrophoresis for epidemiological typing of clinical isolates of Escherichia coli and Pseudomonas aeruginosa from patients with recurrent gram-negative bacteremia. Diagn Microbiol Infect Dis. 1996;25:1–8. doi: 10.1016/0732-8893(96)00082-x. [DOI] [PubMed] [Google Scholar]
- 7.Radford J A, Ryder W D, Dodwell D, Anderson H, Thatcher N. Predicting septic complications of chemotherapy: an analysis of 382 patients treated for small cell lung cancer without dose reduction after major sepsis. Eur J Cancer. 1993;29A:81–86. doi: 10.1016/0959-8049(93)90581-y. [DOI] [PubMed] [Google Scholar]
- 8.Richet H M, Andremont A, Tancrede C, Pico J L, Jarvis W R. Risk factors for candidemia in patients with acute lymphocytic leukemia. Rev Infect Dis. 1991;13:211–215. doi: 10.1093/clinids/13.2.211. [DOI] [PubMed] [Google Scholar]
- 9.Schimpf S C, Young V M, Greene W H, Vermeulen G D, Moody M R, Wiernik P H. Origin of infection in acute nonlymphocytic leukemia: significance of hospital acquisition of potential pathogens. Ann Intern Med. 1972;77:707–714. doi: 10.7326/0003-4819-77-5-707. [DOI] [PubMed] [Google Scholar]
- 10.Senda K, Arakawa Y, Nakashima K, Ito H, Ichiyama S, Shimokata K, Kato N, Ohta M. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob Agents Chemother. 1996;40:349–353. doi: 10.1128/aac.40.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan T Q, Musser J M, Shulman R J, Mason E O, Jr, Mahoney D H, Jr, Kaplan S L. Molecular epidemiology of coagulase-negative Staphylococcus blood isolates from neonates with persistent bacteremia and children with central venous catheter infections. J Infect Dis. 1994;169:1393–1397. doi: 10.1093/infdis/169.6.1393. [DOI] [PubMed] [Google Scholar]
- 12.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinstein M P, Reller L B, Murphy J R, Lichtenstein K A. The clinical significance of positive blood culture: a comprehensive analysis of 500 episodes of bacteremia and fungemia in adults. I. Laboratory and epidemiologic observation. Rev Infect Dis. 1983;5:35–53. doi: 10.1093/clinids/5.1.35. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida R, Hirakata Y, Kaku M, Takemura H, Tanaka H, Tomono K, Koga H, Kohno S, Kamihira S. Genetic relationship of penicillin resistant Streptococcus pneumoniae serotype 19B strains in Japan. Epidemiol Infect. 1997;118:105–110. doi: 10.1017/s0950268896007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young L S, Stevens P, Kaijser B. Gram-negative pathogens in septicaemic infections. Scand J Infect Dis. 1982;31:78–94. [PubMed] [Google Scholar]