Abstract
An outbreak of Enterobacter cloacae in the neonatal intensive care unit of a provincial hospital in Gauteng, South Africa, resulting in nine deaths was investigated. Macrorestriction analysis using pulsed-field gel electrophoresis revealed that three isolates of E. cloacae from blood cultures of patients, six from environmental sources, and one from the hands of a staff member belonged to the same genotypic cluster.
The molecular epidemiology of nosocomial outbreaks of Enterobacter cloacae has been well documented in recent years (1, 4, 6, 7). This is, however, not the case in developing countries, where nosocomial infections are a growing problem (9). The discriminatory power of phenotypic typing techniques is not adequate for the epidemiological analysis of Enterobacter outbreaks (1, 2, 5), thus increasing the difficulties of performing epidemiological studies in developing countries, where limited resources prevent the extensive use of costly molecular techniques.
In this study, an outbreak of necrotizing enterocolitis in the neonatal intensive care unit (ICU) of a provincial hospital in Gauteng, South Africa, which occurred during September and October 1996 was investigated. At the start of the investigation, nine deaths had already occurred. The organism implicated in the outbreak was E. cloacae. The primary purpose of the investigation was to elucidate the source of the organism and its possible mode of transmission so as to implement appropriate infection control measures to prevent further loss of lives. Macrorestriction analysis was used to determine the genetic relatedness of strains and to describe the molecular epidemiology of the outbreak.
A cursory investigation of the conditions and facilities available showed obvious breaches in infection control procedures. These were probably due to a shortage of hospital staff, equipment, and isolation facilities. Basic infection control practices, such as use of alcohol hand-rubs between instances of contact with patients, were not applied. Furthermore, predetermined aliquots of Vamin amino acid cocktail were drawn from a stock bottle into a syringe and injected as an additive to the infusates that were being administered. The stock solution was visibly turbid.
The following specimens were collected: (i) surface swabs from incubators, mattresses, and taps; (ii) samples of all in-use solutions and infusates; (iii) samples of sealed infusates and solutions from the same batch as those in use; (iv) samples of soaps and detergents; (v) hand impression plates from all staff members; (vi) rectal swabs from patients and staff. Columbia agar plates (Oxoid, Basingstoke, Hampshire, United Kingdom) with 5% horse blood were used for hand impression plates and for plating out surface swabs. All solutions were cultured with the BacT/Alert blood culture system (Organon Teknika Corporation, Durham, N.C.). Soaps and detergents were cultured in brain heart infusion broth (Oxoid).
A large variety of organisms was isolated, but attention was focused on isolates of E. cloacae (Table 1). E. cloacae was isolated from the in-use Vamin amino acid cocktail solution stock bottle. A culture of the contents of a sealed bottle from the same batch revealed no organisms, suggesting that the contamination was extrinsic. In view of the fact that E. cloacae was isolated from four in-use infusates, it was essential to exclude intrinsic contamination of solutions bearing the same batch number. These proved to be sterile, confirming that E. cloacae was introduced into the four bags. Three isolates of E. cloacae were found on hand plates from members of the nursing staff. A single isolate of E. cloacae was obtained from a rectal swab taken from a patient.
TABLE 1.
Isolates of E. cloacae from environmental specimens, hand impression plates from ward staff, rectal swabs from staff and patients, and blood cultures of patients in a neonatal ICU of a Gauteng hospital
| Isolate | Source or strain | API profile | Antibiotic resistance patterna | Typeb |
|---|---|---|---|---|
| 1 | In-use neolyte infusion, patient 1 | 3305573 | AMC, AMP, KZ, FOX | A |
| 2 | In-use infusate, patient 1 | 3305573 | AMC, AMP, KZ, FOX | A |
| 3 | Blood culture, patient 1 | 3305573 | AMC, AMP, KZ, FOX | A |
| 4 | In-use infusate, patient 2 | 3305573 | AMC, AMP, KZ, FOX | A |
| 5 | Blood culture, patient 2 | 3305573 | AMC, AMP, KZ, FOX | A |
| 6 | Nebulizer water, patient 2 | 3305573 | AMC, AMP, KZ, FOX | A |
| 7 | In-use infusate, patient 3 | 3305573 | AMC, AMP, KZ, FOX | A |
| 8 | Blood culture, patient 4 | 3305573 | AMC, AMP, KZ, FOX | A |
| 9 | Rectal swab, patient 5 | 3305773 | AMC, AMP, KZ, FOX | B |
| 10 | Nursing assistant hand plate | 3305573 | AMC, AMP, KZ, FOX | C |
| 11 | Nursing Sister hand plate | 3305573 | AMC, AMP, KZ, FOX | A |
| 12 | Nursing assistant hand plate | 3305573 | AMC, AMP, KZ, FOX | D |
| 13 | In-use Vamin amino acid cocktail solution | 3305573 | AMC, AMP, KZ, FOX | A |
| 14 | ATCC 13047 | 3305773 | AMC, CXM, AMP, KZ, FOX | E |
| 15 | ATCC 35591 | 3305773 | AMC, AMP, KZ, FOX | F |
| 16 | ATCC 29005 | 3305773 | AMC, AMP, KZ, FOX | G |
| 17 | ATCC 35930 | 3305573 | AMC | H |
| 18 | ATCC 29941 | 3305773 | AMC, AMP, KZ, FOX | I |
Antibiotic abbreviations and content per disk: AMC, amoxicillin-clavulanic acid (30 μg); AMP, ampicillin (10 μg); CXM, cefuroxime (30 μg); KZ, cefazolin (30 μg); CAZ, ceftazidime (30 μg); PRL, piperacillin (100 μg); TZP, piperacillin-tazobactam (110 μg); FOX, cefoxitin (30 μg); TOB, tobramycin (10 μg); AK, amikacin (30 μg); CN, gentamicin (10 μg); W, trimethoprim (5 μg); IPM, imipenem (10 μg); MEM, meropenem (10 μg); FEP, cefepime (30 μg); CIP, ciprofloxacin (5 μg); OFX, ofloxacin (5 μg).
Types were assigned after examination of restriction fragment length polymorphism patterns.
E. cloacae was isolated from three blood cultures, six environmental samples, three hand impression plates, and one rectal swab by conventional microbiological identification techniques and the API system (bioMérieux S.A., Marcy l’Etoile, France). Five American Type Culture Collection (ATCC) strains of E. cloacae were used as controls. Table 1 shows a list of isolates and strains tested and the sources from which they were obtained. Antibiotic susceptibilities of the isolates were determined by the Kirby-Bauer disk diffusion technique in accordance with National Committee for Clinical Laboratory Standards standards; resistance patterns are shown in Table 1.
DNA for pulsed-field gel electrophoresis (PFGE) analysis was isolated within agarose blocks to prevent shearing of DNA as described previously (3). Restriction digests using XbaI and NotI (Amersham, Little Chalfont, United Kingdom) (8) were conducted within agarose blocks for 5 h at 37°C in 2 volumes of reaction buffer (provided by the manufacturer) containing a final enzyme concentration of 0.5 U/μl. PFGE was carried out immediately on a Chef DRII system (Bio-Rad, Hercules, Calif.) in GTBE running buffer (45 mM Tris-borate, pH 8.0; 1 mM EDTA; 0.1 M glycine). XbaI digests were run for 20 h at 6 V/cm with pulse times ramped linearly from 5 to 50 s. NotI digests were run for 16 h at 6 V/cm with pulse times ramped linearly from 0.5 to 5 s.
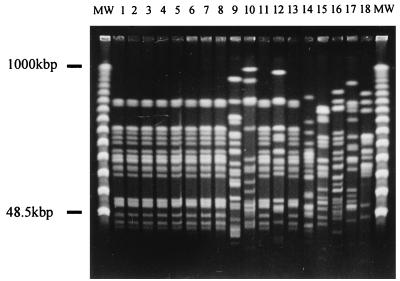
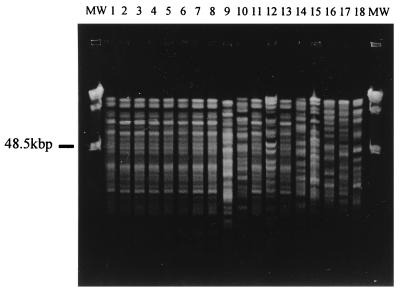
PFGE results for the 13 isolates collected during the investigation and the 5 ATCC reference strains (Fig. 1 and Fig. 2) show convincing genetic relatedness between the organisms isolated from the blood of patients and those isolated from other sources (except isolates 9, 10, and 12). Each isolate was assigned a genotype (A through I) based on the patterns obtained by PFGE analysis (Table 1). Antibiograms and API profiles of the isolates were considerably less discriminatory than molecular techniques. This is consistent with the findings of other investigators (4, 10). It is unclear whether contamination of the nebulizer bottle of patient 2 played a role in the infection of the patient. The results, however, do indicate prevalence of this particular strain of E. cloacae within the neonatal ICU.
FIG. 1.
Restriction fragment length polymorphism analysis of E. cloacae DNA using XbaI separated on PFGE. Lanes 1 through 18 represent isolates 1 through 18. MW, molecular weight markers (PFGE marker 1, λ ladder; Boehringer Mannheim, GmbH, Mannheim, Germany).
FIG. 2.
E. cloacae DNA of isolates 1 through 18 (lanes 1 through 18) digested with NotI and separated by PFGE. MW, molecular weight markers (PFGE marker 1, λ ladder; Boehringer Mannheim).
Two of the isolates of E. cloacae from hand impression plates of staff members were not of the same strain as those from the blood cultures. However, the presence of these and other pathogenic organisms on the hands of staff members who are in constant contact with patients is worrying. According to the charts of patients who died, Vamin amino acid cocktail solution was administered to patients who were already seriously ill. Because the solution was contaminated with the same strain as that found in the blood cultures of patients, the condition of the patients was probably aggravated, resulting in death. Thus, the administration of Vamin amino acid cocktail seems to have played a significant role in the development of necrotizing enterocolitis and the subsequent high death rate of infants.
The outbreak was controlled by retrieving all solutions suspected of contamination; reviewing with ward staff all infection control procedures, with particular emphasis on handwashing; and discouraging the use of stock solutions. A total breakdown of infection control procedures evidently had led to an outbreak of catastrophic dimensions. By providing comprehensive feedback from the investigation and using the molecular epidemiology of the outbreak as an educational tool, we were able to underscore the importance of maintaining basic infection control procedures at all times in the hospital in spite of a shortage of staff, funding, and equipment. If such measures are not implemented, the direct and indirect consequences of nosocomial infection are immeasurable.
REFERENCES
- 1.Bingen E, Denamur E, Lambert-Zechovsky N, Brahimi N, El Lakamy M, Elion J. Rapid genotyping shows the absence of cross-contamination in Enterobacter cloacae nosocomial infections. J Hosp Infect. 1992;21:95–101. doi: 10.1016/0195-6701(92)90028-k. [DOI] [PubMed] [Google Scholar]
- 2.De Gheldre Y, Maes N, Rost F, De Ryck R, Clevenbergh P, Vincent J L, Struelens M J. Molecular epidemiology of an outbreak of multidrug-resistant Enterobacter aerogenes infections and in vivo emergence of imipenem resistance. J Clin Microbiol. 1997;35:152–160. doi: 10.1128/jcm.35.1.152-160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finney M. Pulsed-field gel electrophoresis. In: Janssen K, editor. Current protocols in molecular biology. 1st ed. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 2.5.9–2.5.17. [Google Scholar]
- 4.Garaizar J, Kaufmann M E, Pitt T L. Comparison of ribotyping with conventional methods for the type identification of Enterobacter cloacae. J Clin Microbiol. 1991;29:1303–1307. doi: 10.1128/jcm.29.7.1303-1307.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaston M A. Enterobacter: an emerging nosocomial pathogen. J Hosp Infect. 1988;11:197–208. doi: 10.1016/0195-6701(88)90098-9. [DOI] [PubMed] [Google Scholar]
- 6.Graser Y, Meyer W, Halle E, Presber W, Schonian G. Optimisation of a PCR-based assay for fingerprinting microorganisms. Med Microbiol Lett. 1993;2:379–385. [Google Scholar]
- 7.Grattard F, Pozzetto B, Berthelot P, Rayet I, Ros A, Lauras B, Gaudin O G. Arbitrarily primed PCR, ribotyping, and plasmid pattern analysis applied to investigation of a nosocomial outbreak due to Enterobacter cloacae in a neonatal intensive care unit. J Clin Microbiol. 1994;32:596–602. doi: 10.1128/jcm.32.3.596-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haertl R, Bandlow G. Epidemiological fingerprinting of Enterobacter cloacae by small-fragment restriction endonuclease analysis and pulsed-field gel electrophoresis of genomic restriction fragments. J Clin Microbiol. 1993;31:128–133. doi: 10.1128/jcm.31.1.128-133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponce-de-Leon, S. 1991. The needs of developing countries and the resources required. J. Hosp. Infect. 18(Suppl. A):376–381. [DOI] [PubMed]
- 10.Van Belkum A. Molecular typing techniques for epidemiological studies in medical microbiology. Med Microbiol Lett. 1996;5:271–283. [Google Scholar]