Abstract
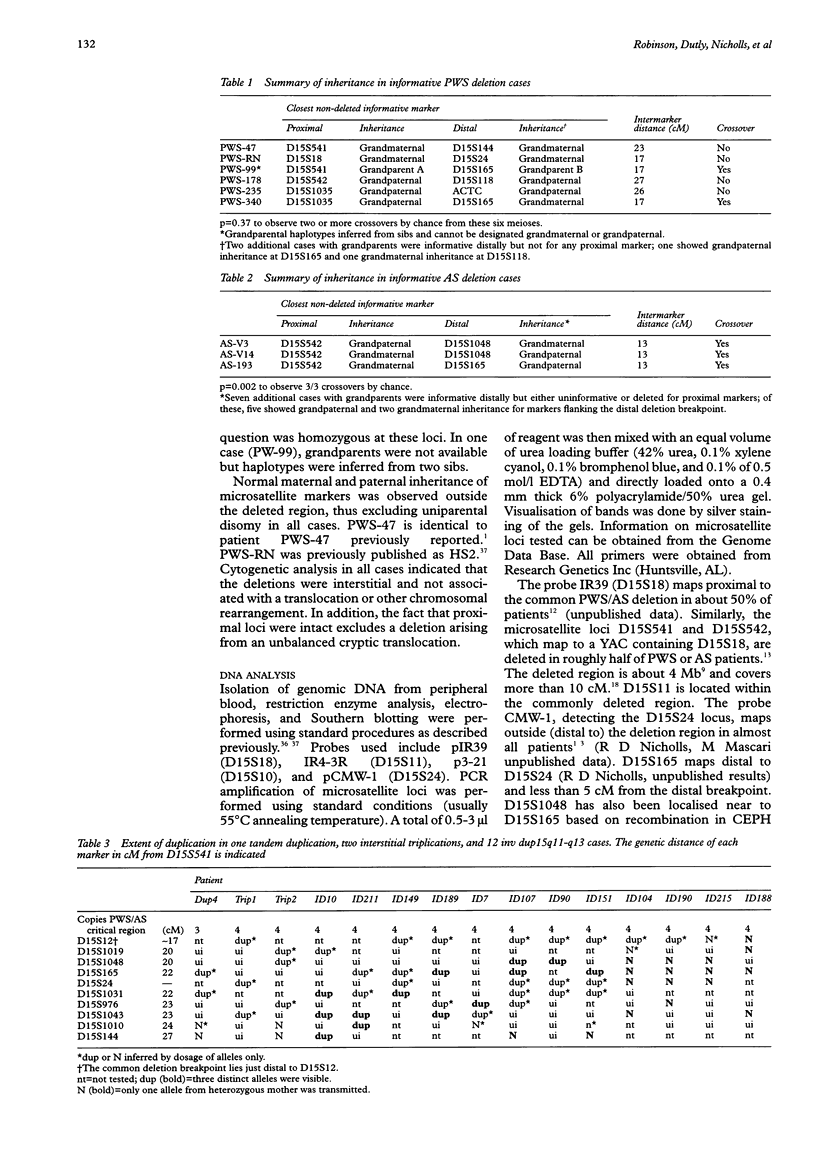
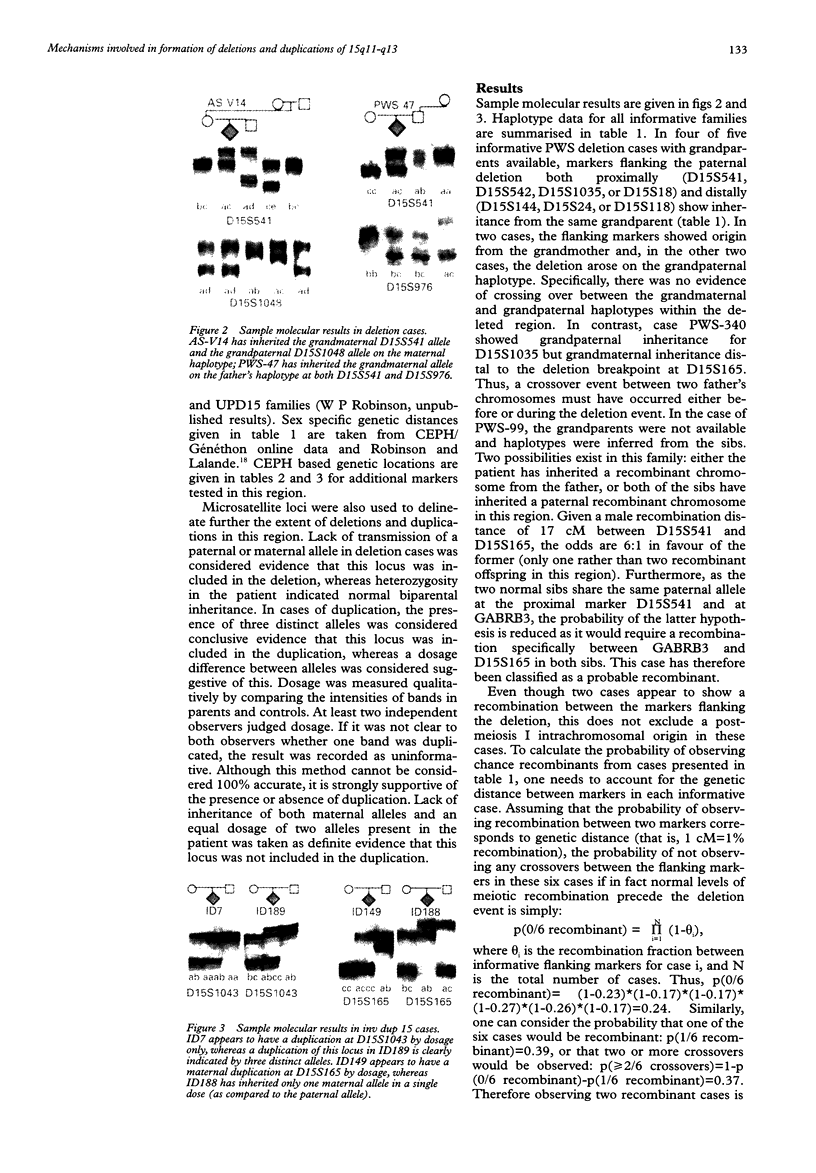
Haplotype analysis was undertaken in 20 cases of 15q11-q13 deletion associated with Prader-Willi syndrome (PWS) or Angelman syndrome (AS) to determine if these deletions arose through unequal meiotic crossing over between homologous chromosomes. Of these, six cases of PWS and three of AS were informative for markers on both sides of the deletion. For four of six cases of paternal 15q11-q13 deletion (PWS), markers on both sides of the deletion breakpoints were inferred to be of the same grandparental origin, implying an intrachromosomal origin of the deletion. Although the remaining two PWS cases showed evidence of crossing over between markers flanking the deletion, this was not more frequent than expected by chance given the genetic distance between proximal and distal markers. It is therefore possible that all PWS deletions were intrachromosomal in origin with the deletion event occurring after normal meiosis I recombination. Alternatively, both sister chromatid and homologous chromosome unequal exchange during meiosis may contribute to these deletions. In contrast, all three cases of maternal 15q11-q13 deletion (AS) were associated with crossing over between flanking markers, which suggests significantly more recombination than expected by chance (p = 0.002). Therefore, there appears to be more than one mechanism which may lead to PWS/AS deletions or the resolution of recombination intermediates may differ depending on the parental origin of the deletion. Furthermore, 13 of 15 cases of 15q11-q13 duplication, triplication, or inversion duplication had a distal duplication breakpoint which differed from the common distal deletion breakpoint. The presence of at least four distal breakpoint sites in duplications indicates that the mechanisms of rearrangement may be complex and multiple repeat sequences may be involved.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeliovich D., Dagan J., Werner M., Lerer I., Shapira Y., Meiner V. Simultaneous formation of inv dup(15) and dup(15q) in a girl with developmental delay: origin of the abnormal chromosomes. Eur J Hum Genet. 1995;3(1):49–55. doi: 10.1159/000472273. [DOI] [PubMed] [Google Scholar]
- Antonarakis S. E., Avramopoulos D., Blouin J. L., Talbot C. C., Jr, Schinzel A. A. Mitotic errors in somatic cells cause trisomy 21 in about 4.5% of cases and are not associated with advanced maternal age. Nat Genet. 1993 Feb;3(2):146–150. doi: 10.1038/ng0293-146. [DOI] [PubMed] [Google Scholar]
- Blair I. P., Nash J., Gordon M. J., Nicholson G. A. Prevalence and origin of de novo duplications in Charcot-Marie-Tooth disease type 1A: first report of a de novo duplication with a maternal origin. Am J Hum Genet. 1996 Mar;58(3):472–476. [PMC free article] [PubMed] [Google Scholar]
- Buiting K., Greger V., Brownstein B. H., Mohr R. M., Voiculescu I., Winterpacht A., Zabel B., Horsthemke B. A putative gene family in 15q11-13 and 16p11.2: possible implications for Prader-Willi and Angelman syndromes. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5457–5461. doi: 10.1073/pnas.89.12.5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buiting K., Saitoh S., Gross S., Dittrich B., Schwartz S., Nicholls R. D., Horsthemke B. Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet. 1995 Apr;9(4):395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- Butler M. G. Prader-Willi syndrome: current understanding of cause and diagnosis. Am J Med Genet. 1990 Mar;35(3):319–332. doi: 10.1002/ajmg.1320350306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy S. B., Thuline H. C., Holm V. A. Deletion of chromosome 15 (q11-13) in a Prader-Labhart-Willi syndrome clinic population. Am J Med Genet. 1984 Feb;17(2):485–495. doi: 10.1002/ajmg.1320170211. [DOI] [PubMed] [Google Scholar]
- Chance P. F., Abbas N., Lensch M. W., Pentao L., Roa B. B., Patel P. I., Lupski J. R. Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet. 1994 Feb;3(2):223–228. doi: 10.1093/hmg/3.2.223. [DOI] [PubMed] [Google Scholar]
- Cheng S. D., Spinner N. B., Zackai E. H., Knoll J. H. Cytogenetic and molecular characterization of inverted duplicated chromosomes 15 from 11 patients. Am J Hum Genet. 1994 Oct;55(4):753–759. [PMC free article] [PubMed] [Google Scholar]
- Christian S. L., Robinson W. P., Huang B., Mutirangura A., Line M. R., Nakao M., Surti U., Chakravarti A., Ledbetter D. H. Molecular characterization of two proximal deletion breakpoint regions in both Prader-Willi and Angelman syndrome patients. Am J Hum Genet. 1995 Jul;57(1):40–48. [PMC free article] [PubMed] [Google Scholar]
- Clayton-Smith J., Pembrey M. E. Angelman syndrome. J Med Genet. 1992 Jun;29(6):412–415. doi: 10.1136/jmg.29.6.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton-Smith J., Webb T., Cheng X. J., Pembrey M. E., Malcolm S. Duplication of chromosome 15 in the region 15q11-13 in a patient with developmental delay and ataxia with similarities to Angelman syndrome. J Med Genet. 1993 Jun;30(6):529–531. doi: 10.1136/jmg.30.6.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutly F., Schinzel A. Unequal interchromosomal rearrangements may result in elastin gene deletions causing the Williams-Beuren syndrome. Hum Mol Genet. 1996 Dec;5(12):1893–1898. doi: 10.1093/hmg/5.12.1893. [DOI] [PubMed] [Google Scholar]
- Haber J. E., Leung W. Y., Borts R. H., Lichten M. The frequency of meiotic recombination in yeast is independent of the number and position of homologous donor sequences: implications for chromosome pairing. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1120–1124. doi: 10.1073/pnas.88.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley R. S., Arbel T. Yeast genetics and the fall of the classical view of meiosis. Cell. 1993 Feb 12;72(3):301–303. doi: 10.1016/0092-8674(93)90108-3. [DOI] [PubMed] [Google Scholar]
- Hawley R. S., Frazier J. A., Rasooly R. Separation anxiety: the etiology of nondisjunction in flies and people. Hum Mol Genet. 1994 Sep;3(9):1521–1528. doi: 10.1093/hmg/3.9.1521. [DOI] [PubMed] [Google Scholar]
- Knoll J. H., Nicholls R. D., Magenis R. E., Glatt K., Graham J. M., Jr, Kaplan L., Lalande M. Angelman syndrome: three molecular classes identified with chromosome 15q11q13-specific DNA markers. Am J Hum Genet. 1990 Jul;47(1):149–154. [PMC free article] [PubMed] [Google Scholar]
- Knoll J. H., Nicholls R. D., Magenis R. E., Graham J. M., Jr, Lalande M., Latt S. A. Angelman and Prader-Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am J Med Genet. 1989 Feb;32(2):285–290. doi: 10.1002/ajmg.1320320235. [DOI] [PubMed] [Google Scholar]
- Kuwano A., Mutirangura A., Dittrich B., Buiting K., Horsthemke B., Saitoh S., Niikawa N., Ledbetter S. A., Greenberg F., Chinault A. C. Molecular dissection of the Prader-Willi/Angelman syndrome region (15q11-13) by YAC cloning and FISH analysis. Hum Mol Genet. 1992 Sep;1(6):417–425. doi: 10.1093/hmg/1.6.417. [DOI] [PubMed] [Google Scholar]
- LaSalle J. M., Lalande M. Homologous association of oppositely imprinted chromosomal domains. Science. 1996 May 3;272(5262):725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- LeGuern E., Gouider R., Ravisé N., Lopes J., Tardieu S., Gugenheim M., Abbas N., Bouche P., Agid Y., Brice A. A de novo case of hereditary neuropathy with liability to pressure palsies (HNPP) of maternal origin: a new mechanism for deletion in 17p11.2? Hum Mol Genet. 1996 Jan;5(1):103–106. doi: 10.1093/hmg/5.1.103. [DOI] [PubMed] [Google Scholar]
- Leana-Cox J., Jenkins L., Palmer C. G., Plattner R., Sheppard L., Flejter W. L., Zackowski J., Tsien F., Schwartz S. Molecular cytogenetic analysis of inv dup(15) chromosomes, using probes specific for the Prader-Willi/Angelman syndrome region: clinical implications. Am J Hum Genet. 1994 May;54(5):748–756. [PMC free article] [PubMed] [Google Scholar]
- Mascari M. J., Gottlieb W., Rogan P. K., Butler M. G., Waller D. A., Armour J. A., Jeffreys A. J., Ladda R. L., Nicholls R. D. The frequency of uniparental disomy in Prader-Willi syndrome. Implications for molecular diagnosis. N Engl J Med. 1992 Jun 11;326(24):1599–1607. doi: 10.1056/NEJM199206113262404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowery-Rushton P. A., Hanchett J. M., Zipf W. B., Rogan P. K., Surti U. Identification of mosaicism in Prader-Willi syndrome using fluorescent in situ hybridization. Am J Med Genet. 1996 Dec 30;66(4):403–412. doi: 10.1002/(SICI)1096-8628(19961230)66:4<403::AID-AJMG4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Nicholls R. D., Fischel-Ghodsian N., Higgs D. R. Recombination at the human alpha-globin gene cluster: sequence features and topological constraints. Cell. 1987 May 8;49(3):369–378. doi: 10.1016/0092-8674(87)90289-3. [DOI] [PubMed] [Google Scholar]
- Nicholls R. D., Knoll J. H., Glatt K., Hersh J. H., Brewster T. D., Graham J. M., Jr, Wurster-Hill D., Wharton R., Latt S. A. Restriction fragment length polymorphisms within proximal 15q and their use in molecular cytogenetics and the Prader-Willi syndrome. Am J Med Genet. 1989 May;33(1):66–77. doi: 10.1002/ajmg.1320330109. [DOI] [PubMed] [Google Scholar]
- Nicholls R. D. New insights reveal complex mechanisms involved in genomic imprinting. Am J Hum Genet. 1994 May;54(5):733–740. [PMC free article] [PubMed] [Google Scholar]
- Nicholls R. D. Recombination model for generation of a submicroscopic deletion in familial Angelman syndrome. Hum Mol Genet. 1994 Jan;3(1):9–11. doi: 10.1093/hmg/3.1.9. [DOI] [PubMed] [Google Scholar]
- Palau F., Löfgren A., De Jonghe P., Bort S., Nelis E., Sevilla T., Martin J. J., Vilchez J., Prieto F., Van Broeckhoven C. Origin of the de novo duplication in Charcot-Marie-Tooth disease type 1A: unequal nonsister chromatid exchange during spermatogenesis. Hum Mol Genet. 1993 Dec;2(12):2031–2035. doi: 10.1093/hmg/2.12.2031. [DOI] [PubMed] [Google Scholar]
- Pellegrino J. E., Schnur R. E., Kline R., Zackai E. H., Spinner N. B. Mosaic loss of 15q11q13 in a patient with hypomelanosis of Ito: is there a role for the P gene? Hum Genet. 1995 Oct;96(4):485–489. doi: 10.1007/BF00191813. [DOI] [PubMed] [Google Scholar]
- Pentao L., Wise C. A., Chinault A. C., Patel P. I., Lupski J. R. Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet. 1992 Dec;2(4):292–300. doi: 10.1038/ng1292-292. [DOI] [PubMed] [Google Scholar]
- Reiter L. T., Murakami T., Koeuth T., Pentao L., Muzny D. M., Gibbs R. A., Lupski J. R. A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet. 1996 Mar;12(3):288–297. doi: 10.1038/ng0396-288. [DOI] [PubMed] [Google Scholar]
- Robinson W. P., Bernasconi F., Basaran S., Yüksel-Apak M., Neri G., Serville F., Balicek P., Haluza R., Farah L. M., Lüleci G. A somatic origin of homologous Robertsonian translocations and isochromosomes. Am J Hum Genet. 1994 Feb;54(2):290–302. [PMC free article] [PubMed] [Google Scholar]
- Robinson W. P., Bernasconi F., Mutirangura A., Ledbetter D. H., Langlois S., Malcolm S., Morris M. A., Schinzel A. A. Nondisjunction of chromosome 15: origin and recombination. Am J Hum Genet. 1993 Sep;53(3):740–751. [PMC free article] [PubMed] [Google Scholar]
- Robinson W. P., Lalande M. Sex-specific meiotic recombination in the Prader--Willi/Angelman syndrome imprinted region. Hum Mol Genet. 1995 May;4(5):801–806. doi: 10.1093/hmg/4.5.801. [DOI] [PubMed] [Google Scholar]
- Robinson W. P., Spiegel R., Schinzel A. A. Deletion breakpoints associated with the Prader-Willi and Angelman syndromes (15q11-q13) are not sites of high homologous recombination. Hum Genet. 1993 Mar;91(2):181–184. doi: 10.1007/BF00222722. [DOI] [PubMed] [Google Scholar]
- Saitoh S., Buiting K., Rogan P. K., Buxton J. L., Driscoll D. J., Arnemann J., König R., Malcolm S., Horsthemke B., Nicholls R. D. Minimal definition of the imprinting center and fixation of chromosome 15q11-q13 epigenotype by imprinting mutations. Proc Natl Acad Sci U S A. 1996 Jul 23;93(15):7811–7815. doi: 10.1073/pnas.93.15.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S., Harada N., Jinno Y., Hashimoto K., Imaizumi K., Kuroki Y., Fukushima Y., Sugimoto T., Renedo M., Wagstaff J. Molecular and clinical study of 61 Angelman syndrome patients. Am J Med Genet. 1994 Aug 15;52(2):158–163. doi: 10.1002/ajmg.1320520207. [DOI] [PubMed] [Google Scholar]
- Schinzel A. A., Brecevic L., Bernasconi F., Binkert F., Berthet F., Wuilloud A., Robinson W. P. Intrachromosomal triplication of 15q11-q13. J Med Genet. 1994 Oct;31(10):798–803. doi: 10.1136/jmg.31.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantravahi U., Nicholls R. D., Stroh H., Ringer S., Neve R. L., Kaplan L., Wharton R., Wurster-Hill D., Graham J. M., Jr, Cantú E. S. Quantitative calibration and use of DNA probes for investigating chromosome abnormalities in the Prader-Willi syndrome. Am J Med Genet. 1989 May;33(1):78–87. doi: 10.1002/ajmg.1320330110. [DOI] [PubMed] [Google Scholar]
- Turleau C., Taillard F., Doussau de Bazignan M., Delépine N., Desbois J. C., de Grouchy J. Hypomelanosis of Ito (incontinentia pigmenti achromians) and mosaicism for a microdeletion of 15q1. Hum Genet. 1986 Oct;74(2):185–187. doi: 10.1007/BF00282090. [DOI] [PubMed] [Google Scholar]
- Urbán Z., Helms C., Fekete G., Csiszár K., Bonnet D., Munnich A., Donis-Keller H., Boyd C. D. 7q11.23 deletions in Williams syndrome arise as a consequence of unequal meiotic crossover. Am J Hum Genet. 1996 Oct;59(4):958–962. [PMC free article] [PubMed] [Google Scholar]
- Van Dyke D. L., Babu V. R., Weiss L. Parental age, and how extra isochromosomes (secondary trisomy) arise. Clin Genet. 1987 Jul;32(1):75–79. doi: 10.1111/j.1399-0004.1987.tb03328.x. [DOI] [PubMed] [Google Scholar]
- Zackowski J. L., Nicholls R. D., Gray B. A., Bent-Williams A., Gottlieb W., Harris P. J., Waters M. F., Driscoll D. J., Zori R. T., Williams C. A. Cytogenetic and molecular analysis in Angelman syndrome. Am J Med Genet. 1993 Apr 1;46(1):7–11. doi: 10.1002/ajmg.1320460104. [DOI] [PubMed] [Google Scholar]