Abstract
A PCR test based on insertion sequence IS1081 was developed to detect Mycobacterium tuberculosis complex organisms in the peripheral blood. The method was applied to blood samples from immunocompetent individuals with localized pulmonary tuberculosis. Seven of 16 (43.75%) blood samples were found to be positive for the circulating DNA copies of M. tuberculosis complex.
Tuberculosis is a persistent problem in the developing world and the biggest cause of mortality. The advent of AIDS has made the disease a major public health problem which has recently been exacerbated by increasing numbers of high-risk patients. A rapid and timely diagnosis of tuberculosis is thus essential to combat this disease. The tests based on PCR have shown promise for the detection of mycobacteria in clinical samples (2, 5, 12). However, several different PCR systems that have been described for the diagnosis of tuberculosis have produced widely differing results with regard to the sensitivity of the assay with different types of clinical samples (7, 8, 11). Peripheral blood appears to be the clinical material of choice for PCR, especially in cases of disseminated and extrapulmonary forms of the disease (6, 10). Earlier studies with blood-based PCR assays in humans suggested that PCR with peripheral blood mononuclear cells for the diagnosis of tuberculosis may be useful only in those who are substantially immunocompromised (3, 6, 9, 10), due either to AIDS or to conditions such as alcohol abuse, renal disorders, diabetes mellitus, etc. It appears that more data are required to determine the effectiveness of the blood-based PCR assay for the diagnosis of tuberculosis, especially in immunocompetent patients (1). In this study, we prospectively used the PCR technique for the detection of Mycobacterium tuberculosis complex in the peripheral blood of immunocompetent, HIV-negative individuals with localized pulmonary lesions of tuberculosis.
Blood samples from 16 patients reporting at the District TB Hospital, Karnal, India, with localized pulmonary tuberculosis were collected. All 16 patients were HIV negative with no history of any immunosuppressive condition, such as renal transplantation, diabetes mellitus, alcoholism, radiotherapy, and cancer. These patients were not hospitalized and had been receiving antitubercular therapy beginning 2 weeks after a confirmatory diagnosis based on acid-fast-bacillus-positive sputum smears and chest X-ray findings. Ten blood samples were collected from healthy individuals of the same age group as that of the tuberculosis-positive individuals. Five milliliters of peripheral blood was collected in EDTA-anticoagulated tubes from each of the patients. Leukocytes were pelleted after the lysis of erythrocytes by a hypotonic erythrocyte lysis buffer under chilled conditions (1). These leukocytes were resuspended in 1 ml of Tris-EDTA buffer (10 mM Tris, 1 mM EDTA [pH 8.0]), and a 400-μl aliquot of this was processed for PCR. The leukocyte samples were subjected to supercooling in a liquid nitrogen container for about 15 min and then thawed abruptly in a boiling water bath for 10 min. This was followed by lysis with lysozyme, digestion with proteinase K, and extraction with hexadecyltrimethylammonium bromide and sodium chloride essentially as described previously (13, 14). DNA was precipitated with 0.6 volume of isopropanol in the aqueous phase and separated by chloroform treatment and extraction. The DNA was then dissolved in 50 μl of Tris-EDTA buffer containing RNase A (Sigma) at the rate of 20 μg/ml. A 10-μl aliquot of total DNA was used for a 25-μl PCR mixture.
PCR protocols were thoroughly standardized with respect to sensitivity and specificity by an extensive series of spiking studies with pure genomic DNA and cells of M. tuberculosis for seeding into cattle and goat blood. Primers (BW-6 [5′ CGA CAC CGA GCA GCT TCT GGC TG 3′] and BW-7 [5′ GTC GGC ACC ACG CTG GCT AGT G 3′]) aimed at the 306-bp region of the multicopy insertion sequence IS1081 (14) were used to amplify mycobacterial DNA in the blood leukocyte samples. The amplification parameters included an initial denaturation at 94°C for 5 min followed by 35 cycles each of denaturation at 94°C for 1 min, annealing at 68°C for 1.5 min, and extension at 72°C for 2 min. The extension step in the 35th cycle was held for 10 min before the samples were shifted to 4°C for storage. The PCR products were identified by agarose gel electrophoresis and combined ethidium bromide staining followed by Southern blot hybridization to an α-32P-labeled 306-bp PCR product from the genomic DNA of M. tuberculosis H37Ra with the BW6 and BW7 primers directed against the IS1081 sequence. Positive and negative controls with or without the genomic DNA of M. tuberculosis complex were included in each run. Of a total of 16 blood samples, 7 (43.75%) were found to be clearly positive for the presence of circulating DNA copies of M. tuberculosis complex in blood. PCR product or signal could not be detected on PCR and probe assays performed with blood donated by healthy individuals with no history of tuberculosis. The results were confirmed by retesting the samples by PCR assay. In all the samples, PCR products were clearly visible on the gel without any confusion (Fig. 1). Our results in this regard are contrary to those of Schluger et al. (10), Kolk et al. (6), Rolfs et al. (9), and Folgueira et al. (3), who recorded a much lower rate of positive PCR with blood samples. We do not believe that the higher rate of positive PCR results in our study were due to PCR-generated contamination, since we retested the samples and routinely included positive and negative controls during extractions, PCR mixture preparation, and electrophoresis. Separate lab spaces were dedicated to the processes of DNA extraction, PCR mixture preparation, and cycling to rule out the problem of artifactual contamination. Comparison of our data with those of all the earlier reports indicates that we used a fairly sensitive assay which could detect less than 10 copies of M. tuberculosis DNA per 5 ml of EDTA-anticoagulated blood. The test also could detect other species of the M. tuberculosis complex, particularly Mycobacterium bovis, and hence could not be taken as a species-specific assay for M. tuberculosis.
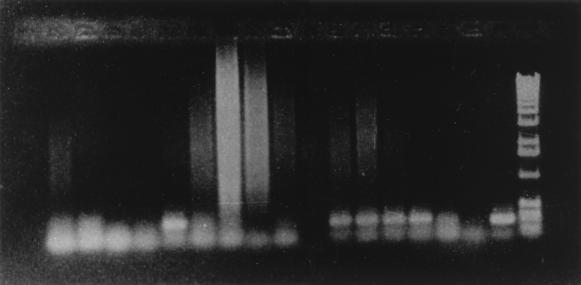
FIG. 1.
Screening of human blood samples from immunocompetent tuberculosis patients by PCR with BW6 and BW7 primers. From left to right, the first 16 lanes contain PCR products from blood samples; the rightmost 2 lanes show PCR product from M. bovis DNA (positive control) and a 1-kb DNA ladder (marker), respectively.
All the patients in our study had been undergoing antitubercular therapy for approximately 2 weeks. This could be a reason for the low sensitivity (<50%) of the assay. However, the PCR results conflicted with earlier findings which suggested that CD4 cells play a critical role in the containment of hematogenous dissemination, and thus blood-based PCR and culture methods were of less significance in case of HIV-negative patients and the HIV-positive individuals having mean CD4 counts of more than 200 cells/μl (3). The observation of M. tuberculosis DNA in the blood of patients included in this study underlines the facts that the incidence of hematogenous dissemination is underestimated in HIV-negative patients and that it is worthwhile to depend on the blood-based PCR assays based on multicopy target sequences for rapid diagnosis of tuberculosis by using blood as a convenient clinical specimen. In our opinion, the PCR technique evaluated in this study could be a good candidate for use in routine diagnosis of tuberculosis from blood samples, once standardization is accomplished. This, however, will require a thorough investigation regarding the sensitivity of the assay with a large number of patient populations. Furthermore, modifications in the DNA extraction process may yield better sensitivity. Multiplex PCR with multicopy sequences (like IS1081) along with highly specific target regions such as those based on 16S/23S ribosomal RNA sequences (4) may eliminate the need for Southern hybridization with 32P, making the assay cost effective for field purposes.
In conclusion, this study has demonstrated the reliability of the blood-based PCR assay, which was more sensitive than the previous PCR protocols for the diagnosis of tuberculosis in immunocompetent patients. These findings support the hypothesis that the escape of tubercle bacilli from alveolar spaces to the bloodstream may be more frequent in case of immunocompetent patients than previously thought.
Acknowledgments
We are thankful to Kuldeep Singh, Incharge, TB Hospital, Karnal, for providing patient data and blood samples for this study and to the Director of NDRI, Karnal, for providing necessary facilities.
REFERENCES
- 1.Ahmed N. Rapid diagnosis of tuberculosis in dairy cattle by polymerase chain reaction and gene probes. M.Sc. thesis. Karnal, India: National Dairy Research Institute; 1997. [Google Scholar]
- 2.Brisson-Noel A, Gicquel B, Lecossier D, Levy-Frebault V, Nassif X, Hance A J. Rapid diagnosis of tuberculosis by amplification of mycobacterial DNA in clinical samples. Lancet. 1992;2:1069–1071. doi: 10.1016/s0140-6736(89)91082-9. [DOI] [PubMed] [Google Scholar]
- 3.Folgueira L, Delgado R, Palenque E, Aguado J M, Noriega A R. Rapid diagnosis of Mycobacterium tuberculosis bacteremia by PCR. J Clin Microbiol. 1996;34:512–515. doi: 10.1128/jcm.34.3.512-515.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glennon M, Smith T, Cormican M, Noone D, Barry T, Maher M, Dawson M, Gilmartin J J, Gannon F. The ribosomal intergenic spacer region: a target for PCR-based diagnosis of tuberculosis. Tubercle Lung Dis. 1994;75:353–360. doi: 10.1016/0962-8479(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 5.Khandekar P, Amin A, Reddy P, Banwalikar J, Shivannaure C, Katoch V. Evaluation of PCR-based test for the detection of Mycobacterium tuberculosis in coded sputum specimens. Indian J Med Res. 1994;100:167–171. [PubMed] [Google Scholar]
- 6.Kolk A H J, Kox L F F, Kuijper S, Richter C. Detection of Mycobacterium tuberculosis in peripheral blood. Lancet. 1994;344:694. doi: 10.1016/s0140-6736(94)92135-0. [DOI] [PubMed] [Google Scholar]
- 7.Kox L F F, Rhienthong D, Miranda A M, Udomsantisuk N, Ellis K, van Leeuwen J, van Heusden S, Kuijper S, Kolk A H J. A more reliable PCR for detection of Mycobacterium tuberculosis in clinical samples. J Clin Microbiol. 1994;32:672–678. doi: 10.1128/jcm.32.3.672-678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noordhoek G T, van Embden J D A, Kolk A H J. Questionable reliability of the polymerase chain reaction in the detection of Mycobacterium tuberculosis. N Engl J Med. 1993;329:2036. doi: 10.1056/NEJM199312303292713. [DOI] [PubMed] [Google Scholar]
- 9.Rolfs A, Beige J, Finckh U, Köhler B, Schaberg T, Lokies J, Lode H. Amplification of Mycobacterium tuberculosis from peripheral blood. J Clin Microbiol. 1995;33:3312–3314. doi: 10.1128/jcm.33.12.3312-3314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schluger N W, Condos R, Lewis S, Rom W N. Amplification of DNA of Mycobacterium tuberculosis from peripheral blood of patients with pulmonary tuberculosis. Lancet. 1994;344:232–233. doi: 10.1016/s0140-6736(94)92999-8. [DOI] [PubMed] [Google Scholar]
- 11.Seth P, Ahuja G, Vijaya Bhanu N, Behari M, Bhowmik S, Dar L, Chakraborty M. Evaluation of polymerase chain reaction for rapid diagnosis of clinically suspected tuberculous meningitis. Tubercle Lung Dis. 1996;77:353–357. doi: 10.1016/s0962-8479(96)90101-x. [DOI] [PubMed] [Google Scholar]
- 12.Sritharan V, Barker R H., Jr A simple method for diagnosing M. tuberculosis infection in clinical samples using PCR. Mol Cell Probes. 1991;5:385–395. doi: 10.1016/s0890-8508(06)80011-3. [DOI] [PubMed] [Google Scholar]
- 13.van Soolingen D, de Haas P E W, Hermans P W M, van Embden J D A. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 1994;235:196–205. doi: 10.1016/0076-6879(94)35141-4. [DOI] [PubMed] [Google Scholar]
- 14.Wards B J, Collins D, de Lisle G. Detection of Mycobacterium bovis in tissues by polymerase chain reaction. Vet Microbiol. 1995;43:227–240. doi: 10.1016/0378-1135(94)00096-f. [DOI] [PubMed] [Google Scholar]