Abstract
Recent studies have shown that ultrafast enzymatic digestion of proteins can be achieved in microdroplet within 250 μs. Further investigation of peptides resulting from microdroplet digestion (MD) would be necessary to evaluate it as an alternative to the conventional bulk digestion for bottom-up and biotherapeutic protein characterization. Herein we examined and compared protein tryptic digestion in both MD and bulk solution. In the case of MD of β-lactoglobulin B, the preservation of long peptides was observed due to short digestion time. In addition, MD is applicable to digest both high- and low-abundance proteins in mixture. In the case of digesting NIST 8671 mAb antibody containing a low level of commonly encountered host cell protein (HCP) PLBL2 (mAb:PLBL2= 100:1 by weight), MD produced lower levels of digestion-induced chemical modifications of asparagine/glutamine deamidation, compared with overnight digestion. No significant difference between MD and bulk digestion was observed in terms of trypsin digestion specificity based on examination of semi- and unspecific-cleaved peptides. Our study suggests that MD, a fast digestion approach, could be adopted for bottom-up proteomics research and for peptide mapping of mAbs to characterize site-specific deamidation and glycosylation, for the purpose of development of biopharmaceuticals.
Keywords: enzymatic digestion, microdroplet reaction, antibody, deamidation
Graphic abstract
Introduction
Numerous studies in the past decade have shown that reaction rates can be dramatically accelerated if reactions are conducted in microdroplet rather than in bulk solution.1–4 The reaction acceleration has mainly been attributed to the special reaction environment at the air-liquid surface of microdroplet where partial solvation of the reagents, the presence of a strong electric filed, enhanced concentration of solute, rapid desolvation, and surface excess charge accumulation could be the influencing factors to speed up the reaction.5–10 This “catalytic” feature allows microdroplet chemistry to have impactful applications including rapid chemical derivatization,11–14 reaction kinetic studies,15 high-throughput reaction screening,16 nanomaterial preparation,17–19 and the rapid, green and sustainable synthesis or degradation.4, 20–26 However, most of these reactions involved small organic molecules, large molecule protein reactions in microdroplets are rare, except enzymatic digestion of proteins.20–22 The microdroplet digestion of proteins is very fast and completes in sub-millisecond time scale (e.g., within 250 μs).20–22
Protein digestion is an important step for bottom-up protemoics research, as large proteins can be cleaved into small peptides that can be easily analyzed with mass spectrometry (MS). Tratidional digestion of proteins is time-consuming and often needs overnight incubation of proteins and enzymes like trypsin. To accelerate enzymatic protein digestion, a variety of novel methods have been reported, including increasing the digestion temperature, adding organic solvents, applying microwave energy, using high intensity focused ultrasound or employing microchip reactor.27–35 Microdroplet digestion appears to be an ultrafast approach in this regard. However, detailed investigation of microdroplet protein digestion has not been done, including the examination of peptide mis-cleavage, cleavage specificity and chemical modification involved in digestion.
Monoclonal antibodies (mAbs) and related products are the fastest growing class of human therapeutics in the pharmaceutical industry,36 and many mAb products have been approved for treatment of different pathologies including cancers, and immunological disorders.36–40 Most therapeutic mAbs are a class of recombinant proteins that are susceptible to a variety of enzymatic or chemical modifications, also called post-translational modifications (PTMs), during cell culture, purification, formulation and long-term storage.41–44 Common PTMs previously identified include glycosylation, deamidation, isomerization, and oxidation, incomplete disulfide bond formation, glycation, N-terminal glutamine cyclization, and C-terminal lysine processing.41, 43, 45–47 Deamidation is one of the most common chemical modifications observed during the degradation processes of mAbs, which may result in changes in physiochemical properties, such as hydrophobicity, charge, secondary or/and tertiary structure, and may lower the thermodynamic or kinetic barrier to unfold.44 This may affect the stability, binding affinity, half-life and efficacy of mAb products, potentially causing immunogenicity48 Therefore, characterization and control of such modifications is important for the quality control of antibody drugs throughout the product life cycle. Currently, peptide mapping, a bottom-up strategy, via reversed-phase liquid chromatography coupled with mass spectrometry (LC-MS) is considered as a gold standard for monitoring and characterization of site-specific modifications including deamidation that may arise during production, processing or storage.49 However, to generate a peptide map, the therapeutic protein must first be digested into its constituent peptides through enzymatic reaction in which the sample needs to be exposed to various buffers, even elevated temperatures for up to 24 h to reach the maximum digestion efficiency. Unfortunately, the conditions that are amenable for effective digestion are often similar to conditions that promote unwanted modifications. For example, it has been reported that asparagine deamidation is promoted by conditions such as elevated temperatures or high pH and are further exacerbated by exposure to these extreme conditions for extended incubation time.50–52 Thus, it is often challenging to find a balance between maximum digestion efficiency and while minimizing artificial modifications. Considering the ultrafast speed of microdroplet protein digestion,20 we reason that chemical modifications on antibody peptides could be reduced if microdroplet digestion is used to replace traditional digestion protocols.
In this study, we evaluated the performance of microdroplet protein digestion including digestion efficiency, sequence coverage, peptide cleavage specificity and deamidation resulted from digestion. Both model protein β-lactoglobulin B and antibody mAb were tested. In general, in comparison to bulk solution digestion, long peptides tend to be well preserved in microdroplet digest probably due to the much shorter digestion time and room temperature used. Equally importantly, deamidation is reduced. Our results suggest that microdroplet-assisted enzymatic reaction could serve as an alternative to traditional digestion method commonly used in bottom-up proteomic studies as well as for the biotherapeutic protein characterization.
Experimental section
Chemicals
Monoclonal antibody reference material 8671 (NIST 8671 mAb) was obtained from the National Institute of Standards and Technology (NIST, Gaithersburg, MD). Phospholipase B-like 2 protein (PLBL2, CHO-S–Purified) was purchased from Immunology Consultants Laboratory (Portland, OR). β-Lactoglobulin B from bovine milk, trypsin (sequencing grade), urea (electrophoresis grade), ammonium bicarbonate (bioultra grade), dithiothreitol (DTT, bioultra grade), and iodoacetamide (IAM, bioultra grade), were all sourced from Sigma-Aldrich (St. Louis, MO). Peptide gp 100 (25-33) human (KVPRNQDWL, HPLC grade) was purchased from AnaSpec (Fremont, CA). Tuftsin (TKPR, HPLC grade) were obtained from Genscript Biotech (Piscataway, NJ). Acetonitrile (ACN, HPLC grade), acetic acid (LCMS grade) and formic acid (FA, LCMS grade) were purchased from Fisher Scientific (Fair Lawn, NJ). A Millipore Direct-Q5 purification system (Burlington, MA) was used to obtain purified water for sample preparation.
Microdroplet and bulk digestions of β-lactoglobulin B
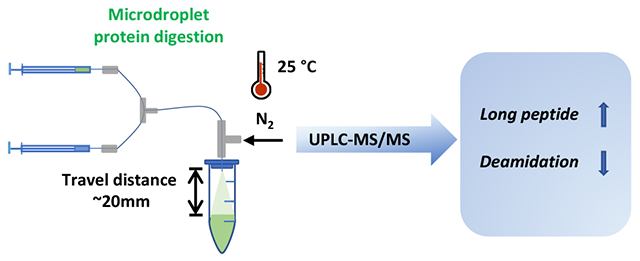
400 μg β-lactoglobulin B from bovine milk was dissolved in 50 mM ammonium bicarbonate (ABC, NH4HCO3, pH 8.0) containing 8 M urea. The denatured protein solution (1 μg/μL) was reduced with 20 mM DTT at 37 °C for 1 h and alkylated with 40 mM IAM at room temperature in the dark for 30 min. A second aliquot of DTT (20 mM) was added again to quench extra amount of IAM. The alkylated protein sample was loaded into Amicon Ultra-0.5, 3 kDa filters (Millipore Sigma), then centrifuged at 14,000 g for buffer exchange to remove small molecule reagents like urea, DTT and IAM. The concentrated protein solution was diluted with 100 mM ABC buffer to 1 μg/μL for digestion. As shown in Scheme 1, for microdroplet digestion of β-lactoglobulin B, 200 μL of 1 μg/μL protein in ABC buffer was added in one syringe and 200 μL of 0.05 μg/μL trypsin in 0.1% acetic acid was loaded in the other syringe. The injection flow rate of both syringes was 5 μL/min. The protein and enzyme were mixed via a T-mixer and then delivered to a home-made sonic spray ionization (SSI)53 emitter for generating microdroplets of the reaction mixture. The connection capillary between T-mixer and emitter was 8 cm long with 98 μm ID (the dead volume: 0.6 μL). A 1.5 mL Eppendorf tube containing a quenching solvent of 50 μL of H2O/ACN/FA (50:50:1, v/v) was used to collect the sprayed microdroplets (the distance between the emitter tip and the quenching solution in the vial was about 2 cm). The SSI emitter was composed of an inner fused-silica capillary (98 μm ID, 200 μm OD) and a coaxial outer fused-silica capillary (250 μm ID and 355 μm OD) and N2 gas of 120 psi was used as the spray sheath gas to assist formation of microdroplets (no high voltage was applied). The coaxial capillary outlets were flush to reach the optimum spray geometry with smallest droplet sizes previously measured.54 Based on the previous report21 of microdroplet digestion employing an emitter with the same capillary dimensions, the same nebulizing gas pressure and the same spray distance as used in this work, the microdroplet digestion time of this experiment (i.e., the flight time of the sprayed microdroplets between the sprayer and the quenching solution) was estimated to be 250 μs (the same as previously reported21). The sprayed microdroplet was collected for 5 min for subsequent nanoESI-MS analysis.
Scheme 1.

Workflows of microdroplet and bulk digestions in this work
During the microdroplet digestion experiment, as a control experiment to assess the contribution of bulk digestion inside the short connection capillary after mixing of protein and trypsin via the Tee, the outlet end of the connection capillary was directly immersed in the quenching buffer of H2O/ACN/FA (50:50:1, v/v) contained in a vial (setup shown in Scheme S1, Supporting Information). 50 μL of 1 μg/μL protein in ABC buffer was loaded in one syringe. Then 50 μL 0.05 μg/μL trypsin in 0.1% acetic acid was loaded in the other syringe. The flow rate of both syringes was 5 μL/min. The protein and enzyme were mixed via a Tee and collected for 5 min for nanoESI-MS analysis.
For bulk digestion, an equal amount of protein and enzyme was mixed (25 μL of 1 μg/μL β-lactoglobulin B and 25 μL of 0.05 μg/μL trypsin) and incubated at 37 °C overnight (14 h). FA was added to 1% (v/v) to quench the digestion. The collected digests from both microdroplet and bulk digestions were vacuum dried and reconstituted with 60 μL H2O/ACN/FA (50:50:1, v/v). 15 μL peptide TKPR (355.9 μM) was added as the internal standard for nanoESI-MS analysis. The injection flow rate was 2 μL/min, and +3.5 kV was applied for nanoESI ionization. The temperature of the MS inlet was 250 °C. A Thermo Scientific Q Exactive mass spectrometer (San Jose, CA) was used with resolution set to 140,000 for the MS analysis.
Microdroplet digestion of a mixture of mAb and PLBL2
1 mg NIST 8671 mAb was spiked with 10 μg PLBL2 to mimic an antibody sample containing an impurity protein. The sample was then diluted to 1 mg/mL mAb and 0.01 mg/mL PLBL2 with 50 mM ABC buffer (pH 8.0) containing 8 M urea. The denatured protein mixture was reduced with 20 mM DTT at 37 °C for 1 h and alkylated with 40 mM IAM at room temperature in the dark for 30 min. A second aliquot of DTT (20 mM) was added again to quench excess amount of IAM. The alkylated protein sample was loaded into Amicon Ultra-0.5, 10 kDa filters (Millipore Sigma), then centrifuged at 14,000 g for buffer exchange. The concentrated protein solution was diluted with 100 mM ABC to 250 μL. The final concentration of mAb and PLBL2 were 4 μg/μL and 0.04 μg/μL, respectively. For microdroplet digestion, digestion was performed in triplicates using the setup as described in Scheme 1. 200 μL of mAb (4 μg/μL) and PLBL2 (0.04 μg/μL) was loaded in one syringe and 200 μL of trypsin (0.1 μg/μL) in 0.1% acetic acid was loaded in the other syringe. The flow rate of both syringes was set at 5 μL/min. A 1.5 mL tube containing a quenching solvent of 50 μL of H2O/ACN/FA (50:50:1, v/v) was used to collect the digest for 10 min. Nitrogen sheath gas was applied at 120 psi to assist the spray. For bulk solution digestion, 50 μL of 4 μg/μL mAb and 0.04 μg/μL PLBL2 was mixed with 50 μL of 0.1 μg/μL trypsin and then incubated at 37 °C for overnight (14 h). FA was added to 1% (v/v) to quench the digestion. The collected digests from both microdroplet and bulk digestions were vacuum dried and reconstituted with 60 μL H2O/ACN/FA (95:5:0.1, v/v) followed by centrifuge at 14000 g for 5 min. The supernatant was transferred for LC/MS analysis.
UPLC-MS/MS Analysis of Tryptic Digests
The prepared tryptic peptides were analyzed using UPLC-MS/MS. Samples were directly separated using a Waters Acquity UPLC equipped with a BEH Cl8 column (2.1 × 150 mm, 1.7 μm particle size; Milford, MA, U.S.A) at an injection volume of 5 μL. Separation was performed with mobile phase A consisting of 0.1% formic acid in water and mobile phase B consisting of 0.1% formic acid in acetonitrile. The mobile phase flow rate was kept at 200 μL/min. The gradient conditions were as follows: 0-65 min, increased from 5% to 35% B; 65-75 min, linear ramp from 35% to 40% B; 75-80 min, climbed to 90% B and remained at 90% for 5 min before returning to 5% B. Total run time per sample was 90 min. Mass spectrometric analysis was performed on the Thermo Scientific Q Exactive mass spectrometer (San Jose, CA). Data-dependent MS/MS was performed as follows: the first event was the survey positive mass scan (m/z range of 250–1800) followed by 20 HCD events (30% NCE) on the 20 most abundant ions detected from the first event. Ions were generated using a sheath gas flow rate of 35, an auxiliary gas flow rate of 10, a spray voltage of 4.0 kV, a capillary temperature of 300 °C, an auxiliary gas temperature of 100 °C and an S-Lens RF level of 50. Resolution was set at 70 000 (AGC target of 3E6) and 17 500 (AGC target of 5E4) for survey scans and MS/MS events, respectively. The maximum ion injection time was 100 ms for survey scan, and 100 ms for the other scans. The dynamic exclusion duration of 8 s was used with a single repeat count.
Data analysis
Raw files were searched using MaxQuant software (version 2.0.3.1) equipped with the Andromeda search engine (Max Planck Institute, Martinsreed, Germany). The first search peptide mass tolerance was set to 20 ppm, and the main search peptide mass tolerance was 4.5 ppm. Trypsin was set as the digestion enzyme with a maximum of two miss-cleavages. Carbamidomethylation was set as a fixed modification, while oxidation (on methionine M), acetylation (protein N-term), and deamidation (occurring to either asparagine N or glutamine Q) were set as variable modifications. Spectra were searched against the sequence of NIST 8671 mAb light chain and heavy chain plus the sequence of PLBL2. The “match between runs” feature was used with a matching time window of 0.7 min and an alignment time window of 20 min. Target decoy analysis was performed by searching a reverse database with a PSM FDR of 1% and protein FDR of 5%. Label-free quantification (LFQ) was performed using the MaxLFQ algorithm in MaxQuant software (MaxPlanck) according to default LFQ parameters with minimum ratio count set as 2, normalization type as classic, minimum number of neighbors equal to 3 and average number of neighbors equal to 6. A separate database searching was performed with digest mode set to semispecific and unspecific, respectively. The MaxQuant output tables were used for further analysis and visualization of data.
Results and Discussion
To evaluate the performance of microdroplet protein digestion, we started with trypsin digestion of a single protein first and β-lactoglobulin B was chosen as the test sample. We used an online mixing setup as shown in Scheme 1 for microdroplet digestion, for the reason that the online mixing would minimize the contribution of protein digestion in bulk phase before performing microdroplet digestion. As shown in the microdroplet digestion setup (Scheme 1), the residential time of protein and trypsin in the connection capillary before spray was estimated to be 4 s, based on the capillary dead volume of 0.6 μL and the mixed solution flow rate of 10 μL/min. In the control experiment (Scheme S1 and Figure S1, Supporting Information), online mixing of β-lactoglobulin B and trypsin for 5 s led to little digestion of the protein and the acquired MS spectrum (Figure S1–b, Supporting Information) showed intact protein ions (+10 ~ +21 ions) along with two peptide ions at m/z 419 and m/z 858 were detected, m/z 419 and 858 corresponded to +2 ions of ALPMHIR and LSFNPTQLEEQCHI, two peptides from the C-terminal of β-lactoglobulin B, which were also observed in the MS spectrum of intact protein (Figure S1–a, Supporting Information).
Using the Scheme 1 setup, ffor microdroplet digestion of β-lactoglobulin B, two syringe pumps were used to deliver protein and enzyme solution separately at 5 μL/min flow rate. The reactants were mixed online via a Tee before spray. Microdroplet digestion was quenched immediately after reaching the quenching buffer. Overnight bulk digestion (at 37 °C) of β-lactoglobulin B was conducted as comparison. After digestion, both digests were vacuum dried and reconstituted in solvent with peptide TKPR being spiked as an internal standard (IS) for nanoESI-MS analysis. As shown in Figure 1a, after digestion, numerous peptide fragments were observed while signals corresponding to the intact protein could no longer be seen in both microdroplet and bulk digests, indicating the tryptic digestion in both conditions was complete. Moreover, by manually searching fully cleaved peptides (Figure 1a inset) based on the sequence of β-Lactoglobulin B, the same # of fully cleaved peptides were identified both in microdroplet and bulk digestion, leading to the same sequence coverage (Figure 1b). Interestingly, there were some peptides (highlighted in red, Figure 1a inset) showing higher abundance in microdroplet than bulk digestion. For example, top two long peptide VAGTWYSLAMAASDISLLDAQSAPLR and YLLFCMENSAEPEQSLACQCLVR were detected with much lower intensity in overnight bulk digest (2.64E+05 and 5.63E+05) than in microdroplet digest (5.18E+06 and 2.51E+06) (identified peptide intensities are summarized in Table S1, Supporting Information). This result indicates that long peptides might suffer hydrolysis in solution with long incubation time and elevated temperature at 37 °C while they could be well preserved during short microdroplet digestion period. This finding is consistent with the previously reported proteomic studies in which long digestion time resulted in reduced number of identified peptides and low protein sequence coverage.55–57
Figure 1.
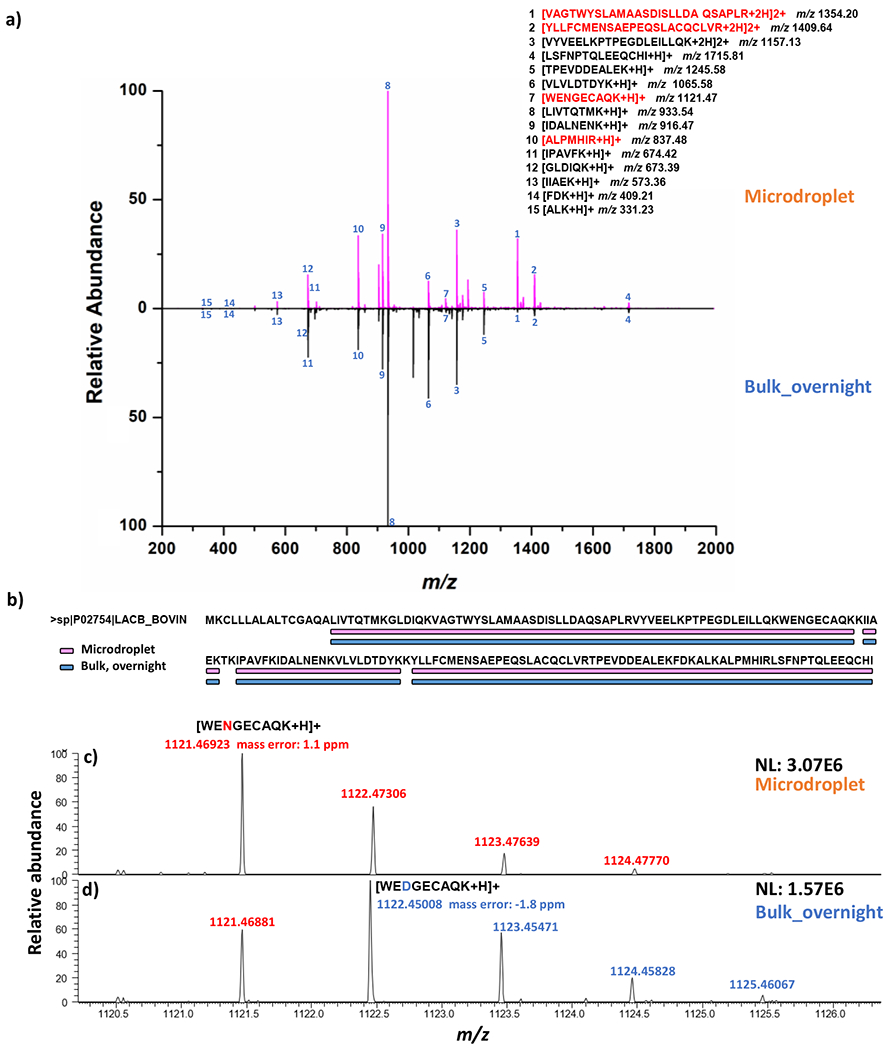
(a) Positive ion mode nanoESI mass spectra showing the microdroplet digestion (top panel) and bulk digestion (bottom panel) of β-lactoglobulin B, inset: fully cleaved tryptic peptides from β-lactoglobulin B (peptides highlighted in red showing higher abundance in microdroplet); (b) a sequence coverage plot of β-Lactoglobulin B obtained from microdroplet digestion and bulk digestion based on fully cleaved peptides; Mass spectra of unmodified WENGECAQK in microdroplet (c) and deamidated WENGECAQK in bulk overnight digest (i.e., the deamidation product peptide WEDGECAQK) (d), respectively; NL indicates normalized level (NL) intensity.
In addition, we also examined the digestion-induced chemical modification of asparagine deamidation of our detected peptides (Figure 1a) in microdroplet and bulk digests. Deamidation as an important PTM during protein degradation, has been widely studied and known to be accelerated at high temperatures and high pH values,58 Indeed, one peptide from β-Lactoglobulin B, WENGECAQK, had much higher deamidation level observed in bulk overnight digest. As shown in Figure 1c where only native form of WENGECAQK was detected at m/z 1121 from MD (theoretical mass m/z 1121.46803, mass error: 1.1 ppm), in contrast, the deamidated form WEDGECAQK with 1 Da shift (m/z 1122.45008, theoretical mass m/z 1122.45205, mass error: 1.8 ppm) was observed in bulk digest (Figure 1d) with relatively higher intensity as native peptide and well resolved by our high-resolution MS. This result indicated that the ultra-fast microdroplet digestion (ca. 250 μs) at room temperature also tends to prevent the digested peptides from deamidation, besides preserving long peptides.
As reproducibility is important for protein digestion, in this study, we repeated the microdroplet digestion with three individual runs using the online mixing setup described above (Scheme 1). The nanoESI MS spectra for triplicate digestion of microdroplet (a-c) and bulk overnight (d-f) are shown in Figure S2 (Supporting Information). Similar profile across digestion replicates were observed for each digestion condition. In general, microdroplet digestion showed high reproducibility between three individual runs in terms of the sequence coverage. For most of the fully cleaved peptides, the standard deviation among three digestion replicates is acceptable (Figure S3, Supporting Information).
In our microdroplet digestion setup (Scheme 1), in order to reduce the loss of peptides from collection of the sprayed microdroplets, the collection vial was covered with a lid and the emitter tip was inserted into the vial through the lid via a pre-drilled hole. As a test, standard peptide KVPRNQDWL solution was sprayed and collected using the setup shown in Scheme 1. By comparing the sprayed peptide signals before and after the spray, the collection recovery was measured to be 95% (details shown in Supporting Information, Table S2), indicating that the peptide loss during microdroplet spray collection was small.
After examining microdroplet digestion with β-Lactoglobulin B as an example, we moved forward to investigate the microdroplet digestion of proteins in mixture. No digestion of multiple proteins in microdroplet was examined before. In our experiment, NIST 8671 mAb was mixed with HCP protein PLBL2 at 100:1 ratio to mimic a protein mixture where one protein is dominant and the other one is a minor species. PLBL2 is a 66KDa catalytic HCP protein that co-purifies frequently in mAb preparations produced in Chinese hamster ovary (CHO) cells, which might cause the unwanted immunogenic effects in patients and affect the drug stability due to its enzymatic degradation of polysorbates 20 and 80, common adjuvants present in formulated drug substance (DS).59, 60 Therefore, PLBL2 needs to be removed from mAb preparations down to acceptable levels and therefore it is necessary to monitor the presence of PLBL2 in IgG antibodies. In this study, we attempted to examine how the microdroplet digestion would occur to a low-level PLBL2 protein and a high-level concentration of large mAb antibody. On the other hand, we evaluated if chemical modifications to antibody like asparagine/glutamine deamidation could be reduced during the short digestion time of MD.
Microdroplet digestion and bulk digestion of the mixture of NIST 8671 mAb and PLBL2 were conducted as described in experimental section. Both microdroplet digest and bulk overnight digest were prepared as 3 digestion replicates and then subject to LC/MS analysis. Following LC/MS/MS analysis, peptide identification and quantification were performed to evaluate the quality of the digested samples between the two different digestion conditions. The digestion efficiency was studied in terms of the determination of sequence coverage, missed cleavage, semi-specific cleavage and non-specific cleavage as the metrics. As shown in Figure 2a, comparable sequence coverage of antibody heavy chain was attained for both microdroplet and bulk digestions. A slightly higher sequence coverage of antibody light chain and lower abundance protein PLBL2 in microdroplet was observed compared with bulk digestion. Upon further investigating the identified peptides containing zero, one, or two missed tryptic cleavages (Figure 2b, c and d), microdroplet and bulk overnight digestion showed comparable trypsin digestion efficiency for mAb heavy chain. However, for mAb light chain and low abundance PLBL2, microdroplet showed less miss cleaved peptides than bulk.
Figure 2.
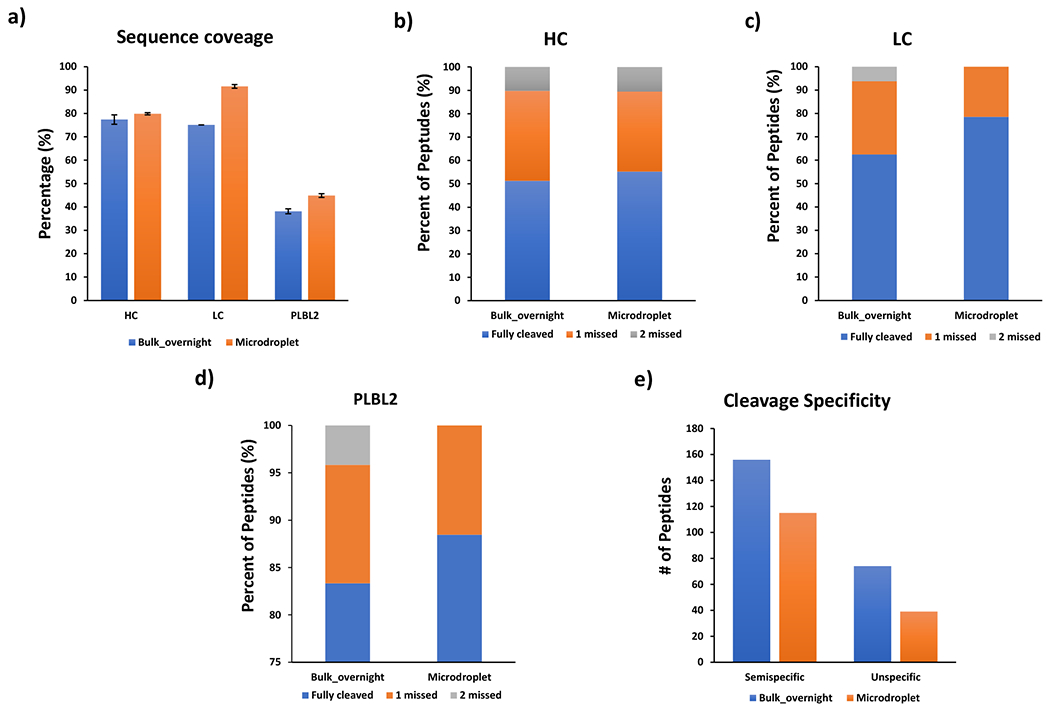
(a) Sequence coverages of mAb heavy chain, light chain and PLBL2. The percentages of identified peptides containing 0, 1, or 2 missed tryptic cleavages from mAb heavy chain (HC, b), mAb light chain (LC, c) and PLBL2 (d). Numbers of semi-specific cleaved peptides and unspecific cleaved peptides (e) identified under different digestion conditions.
Aside from the missed cleavages, we also checked the semi-specific cleavage (a specific cleavage at one terminus and a non-tryptic cleavage site at the other end) and non-specific cleavage (enzymatic cleavage of C-terminal to a residue other than Lys or Arg for both termini), since microdroplet digestion was conducted in a new reaction environment with unknown reaction mechanism which may affect the enzyme specificity. As shown in Figure 2e, bulk overnight digestion showed higher level of semi-specific cleavages with 156 peptides identified, whereas the number of identified semi-specific peptides from microdroplet digest was 115. This trend continued when comparing non-specific cleavage in Figure 2e, a lower-level non-specific cleavage was observed in microdroplet digestion with 39 peptides identified whereas 74 non-specific cleaved peptides were found from bulk overnight digestion. All these results suggested that microdroplet digestion has comparable digestion efficiency with bulk and no significant increment of semi-specific cleavage as well as non-specific cleavage was observed.
It is noteworthy that a significant number of semi-specific and nonspecific tryptic peptides were identified in the case of antibody sample digestion. However, most of these peptides were of relatively low-abundance compared to fully tryptic peptides. For example, as shown in Figure S4a (Supporting Information), four semi-tryptic peptides were detected corresponding to fully tryptic peptides VYACEVTHQGLSSPVTK (amino acid 190-206) from light chain. However, the relative abundance of those semi-tryptides were lower than 1%. Similar result was observed for another peptide TPEVTCVVVDVSHEDPEVK (amino acid 259-277) from heavy chain (Figure S4b, Supporting Information). Although increased number of semi-tryptic peptides were detected, the overall relative abundance corresponding to the fully tryptic peptides were low. The intensity distribution of semi-specific and unspecific peptides was shown in Figure S4c and d (Supporting Information) whereby the major peptides were detected at low abundance (lower than 1E6). Indeed, in previous studies, nonspecific trypsin cleavages have been frequently reported and showed a high percentage especially in standard protein digest.61, 62 Several potential sources for generating partially tryptic peptides has been reported, including possible chymotrypsin contamination in trypsin, pseudo-trypsin activity due to partial autolysis of trypsin, proteolytic products due to endogenous proteases, and in-source fragmentation of fully tryptic peptides.62 Besides, it has been shown to be related to a number of digestion conditions and pre-digestion treatment.63–66 In this work, to prevent the autolysis of trypsin before spray, trypsin was reconstituted in 0.1% acetic acid. Although mildly acidic environment was often recommended by the manufacturer to prevent trypsin autolysis, a significantly increased level of nonspecific cleavages during the trypsin digestion process has been reported before.67
In addition, we quantitatively compared each fully cleaved peptide based on the peak intensities reported from Maxquant. Interestingly, the abundance of different fully cleaved tryptic peptides was found to be quite distinct under the two digestion conditions. For example, as shown in Figure 3a and b, several long peptides showed much lower intensities in bulk overnight digest than in microdroplet digest such as VTNMDPADTATYYCAR, WQQGNVFSCSVMHEAL-HNHYTQK, ESGPALVKPTQTLTLTCTFSGFSLSTAGMSVGWIR, TPEVTCVVVDVSHEDPEVK, and THTCPPCPAPELLGGPSVFLFPPKPK. It is likely that these peptides are long and more susceptible to hydrolysis during bulk overnight digestion at 37 °C, whereas microdroplet digestion could significantly minimize the hydrolysis due to the fast digestion speed at room temperature. However, the other long peptides from mAb heavy chain like DMIFNFYFDVWGQGTTVTVSSASTK and GFYPSDIAVEWESNGQPENNYK showed higher abundance in bulk overnight digest than microdroplet.
Figure 3.
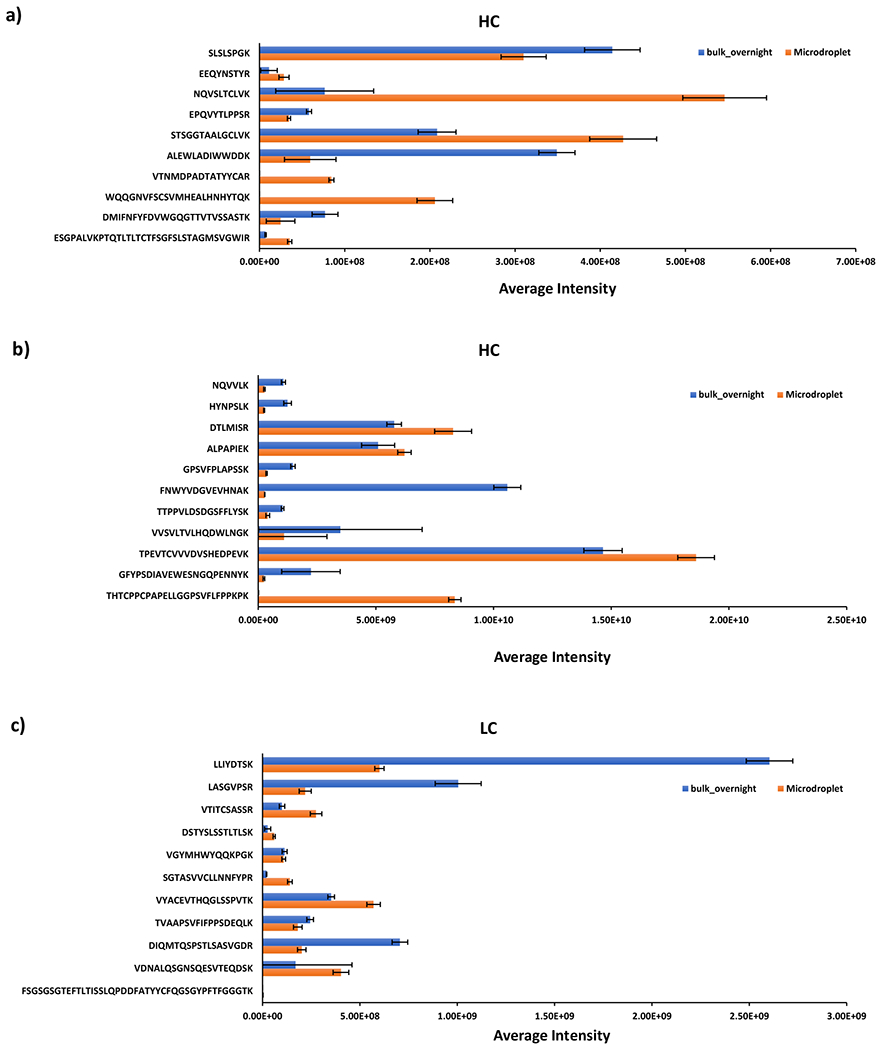
Averaged intensities of fully cleaved peptides identified from mAb heavy chain (a and b) and light chain (c) under different digestion conditions (the fully cleaved peptides identified from mAb heavy chain were grouped in (a), and (b) due to the large difference in scale of the averaged intensity across different peptides). Standard deviations from the reported mean shown are for n = 3 digestion replicates.
The same trend was observed for mAb light chain (Figure 3c) wherein long peptides like SGTASVVCLLNNFYPR, VYACEVTHQGLSSPVTK, VDNALQSGNSQESVTEQDSK and FSGSGSGTEFTLTISSLQPDDFATYYCFQGSGYPFTFGGGTK, were detected with lower intensities in bulk overnight digest than microdroplet digest. For low abundance protein PLBL2 (Figure 4), most of the fully cleaved peptides showed a relatively higher intensity in microdroplet than bulk, indicating that microdroplet might have a slightly higher digestion efficiency of low abundance proteins. It has been reported that the bias digestion exists between low and high abundance proteins due to the fact that the rate of trypsin digestion is dependent on the concentration of each individual protein in a mixture sample.68 Common strategies to address the abundance challenge include affinity depletion and enrichment of proteins with antibody arrays or ligand libraries and prefractionation of proteins.68 In our microdroplet and bulk digestions of PLBL2, the sequence coverage was about 40% (Figure 2a). To further improve the sequence coverage of spiked PLBL2, the prefractionation or affinity enrichment could be the solution.
Figure 4.
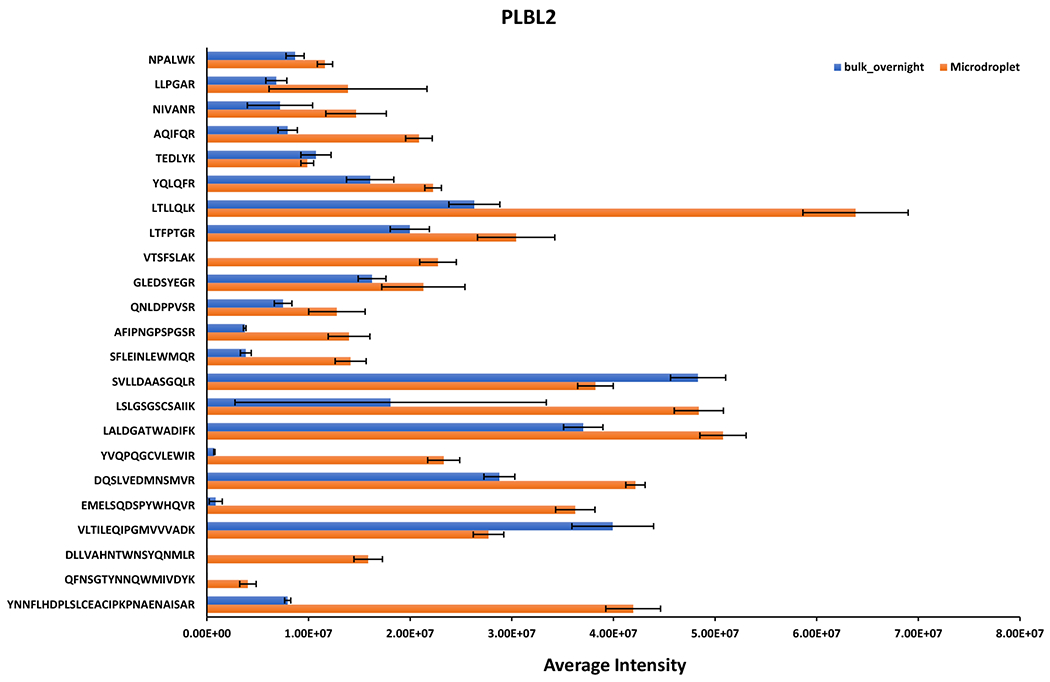
Averaged intensities of fully cleaved peptides identified from PLBL2 under different digestion conditions. Standard deviations from the reported mean shown are for n = 3 digestion replicates.
Upon further investigation of peptides that contain asparagine deamidation site as shown in Figure 5a. For example, deamidation sites like N385, and N469 from PLBL2 and N136 and N151 from light chain as well as N300, N289 (279), N387, N364, N78(Q79), and N318 from heavy chain were identified and relative levels are quantified, as shown in Figure 5a (denotation of N289(279) means either N289 or N279 underwent deamidation, the same for N78(Q79)). Overall, much higher level of deamidation was observed in bulk overnight digest than microdroplet. As the rate of deamidation also depends on the adjacent amino acid residue’s size (smaller size reduces steric hindrance for the initial cyclization reaction of the deamidation process),44 the level of deamidation varies among different peptides. For instance, Gly is considered as the most favorable amino acid promoting deamidation, followed by Ser, Thr and Asn,70, 71 which is in line with the detected deamidation rate displayed in Figure 5a in which high deamidation level site like N385 from PLBL2, N387 and N318 from heavy chain are followed by Gly, whereas N136 from light chain is adjacent to Asn. All these results indicated that reducing the digestion time without reducing digestion efficiency is preferable to minimize the digestion-induced deamidation.
Figure 5.
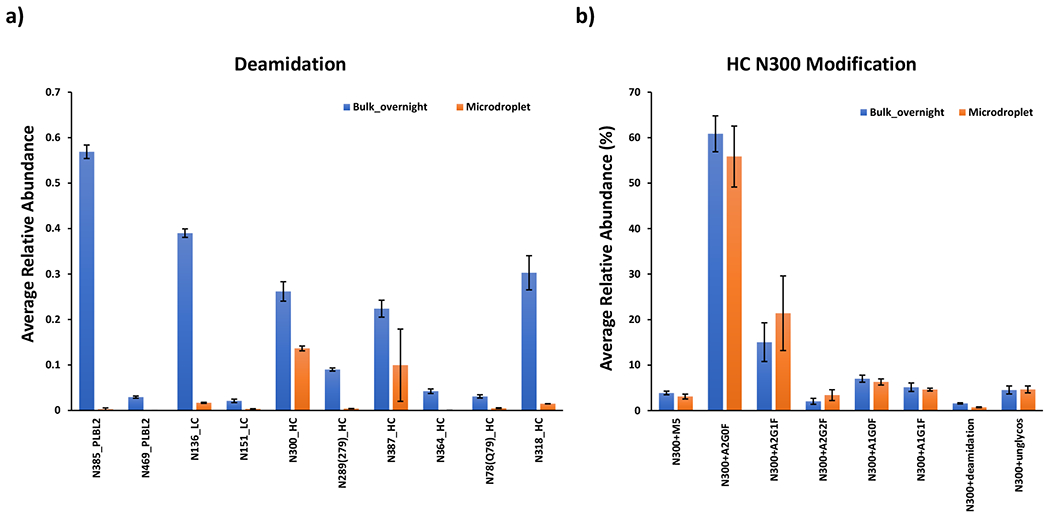
Averaged relative abundances of (a) deamidation for mAb and PLBL2 and (b) glycosylation for mAb heavy chain N300 under different digestion conditions. Averaged relative abundance was calculated as the intensity ratio of one modified peptide to the intensity sum of modified and native peptides. Peak intensities of native and modified peptides were recorded after manually searching of +2 charge forms of each peptide from MS1 spectra with 5ppm mass accuracy tolerance. Standard deviations from the reported mean shown are for n = 3 digestion replicates.
Interestingly, EEQY300NSTYR containing one glycan site at N300 was detected with slightly higher deamidation rate in bulk overnight than microdroplet (Figure 5a). Upon further investigation of glycans, deamidated and unmodified peptides of EEQY300NSTYR in Figure 5b, we found that the relative abundance of unmodified peptides in microdroplet and bulk were similar, indicating deamidation rate is relevant to the glycan loss. Indeed, this result is consistent with the previous report that glycan loss in solution is accompanied by the conversion of Asn to Asp.72 Thus, a lower level of deamidation rate observed in microdroplet digestion might prevent glycan shedding that occurs during overnight digestion.
Finally, glycosylation was also assessed for the glycan profiles on heavy chain N300 site, as shown in Figure 5b. Most abundant glycan forms including complex biantennary oligosaccharides containing from 0 to 2 non-reducing galactoses with fucose attached to the reducing end of N-acetylglucosamine (A2G0F, A2G1F, A2G2F, A1G0F, A1G1F) along with high mannose structures (M5) were searched. The ratio of the relative abundances of most glycan forms was consistent between bulk and microdroplet digestions, suggesting that microdroplet digestion could be applied to monitor PTMs for mAb.
In addition, in the microdroplet digestion experiments described above, the flow rate for protein injection was low. We further explored the possibility of using a high flow injection rate like 100 μL/min for each pump. In our test, 100 μL of 1 μg/μL β-lactoglobulin B in ABC buffer was loaded in one syringe and 100 μL of 0.05 μg/μL trypsin in 0.1% acetic acid was loaded in the other syringe for microdroplet spray (using the apparatus shown in Scheme 1). The flow rate of both syringes was 100 μL/min and N2 was set at 120 psi. The microdroplet was collected into a vial containing a quenching solution of 50 μL H2O/ACN/FA (50:50:1, v/v) for just 10 s. MS spectrum (Figure S5, Supporting Information) showed β-lactoglobulin B was still well digested under such a high flow rate. The tolerance of high sample flow rate for microdroplet digestion would further speed up the total collection procedure.
Overall, the data presented in this study demonstrate that microdroplet digestion has the potential to significantly speed up the bottom-up LC/MS analysis of mAb, stemming from the efficient digestion, even for the low abundant proteins. Moreover, ultrafast digestion performed at room temperature allows to minimize artificial modification like asparagine/glutamine deamidation.
Conclusions
Fast enzymatic reactions of proteins and antibodies in microdroplet were investigated with detailed peptide analysis in this study. Some interesting features of MD were found. First, MD is fast, resulting in the preservation of long peptides in digest. Second, fast microdroplet digest could reduce deamidation of asparagine or glutamine residues, in comparison to bulk digestion. Third, MD is applicable to digest both high- and low-abundance proteins in mixture. Fourth, no significant difference between MD and bulk digestion was observed for trypsin digestion specificity based on examination of semi- and unspecific-cleaved peptides. Fifth, similar glycan profiles on heavy chain N300 site was observed in microdroplet and lower level of deamidation rate might prevent the glycans shedding during digestion. Sixth, microdroplet digestion tolerates a high sample injection flow rate, which could reduce the peptide collection time. Our study demonstrates that microdroplet could be considered as an emerging, fast and simple protein digestion approach and has the potential for the routine usage in bottom-up protein characterization such as peptide mapping for mAbs.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (1R15GM137311-01) for the financial support of this work.
References:
- 1.Basuri P; Gonzalez LE; Morato NM; Pradeep T; Cooks RG, Accelerated microdroplet synthesis of benzimidazoles by nucleophilic addition to protonated carboxylic acids. Chem. Sci 2020, 11 (47), 12686–12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bain RM; Pulliam CJ; Cooks RG, Accelerated Hantzsch electrospray synthesis with temporal control of reaction intermediates. Chem. Sci 2015, 6 (1), 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee S; Zare RN, Syntheses of isoquinoline and substituted quinolines in charged microdroplets. Angew. Chem 2015, 127 (49), 15008–15012. [DOI] [PubMed] [Google Scholar]
- 4.Yan X, Emerging microdroplet chemistry for synthesis and analysis. Int. J. Mass Spectrom 2021, 468, 116639. [Google Scholar]
- 5.Wei Z; Li Y; Cooks RG; Yan X, Accelerated reaction kinetics in microdroplets: Overview and recent developments. Annu. Rev. Phys. Chem 2020, 77, 31–51. [DOI] [PubMed] [Google Scholar]
- 6.Yan X; Bain RM; Cooks RG, Organic reactions in microdroplets: Reaction acceleration revealed by mass spectrometry. Angew. Chem., Int. Ed 2016, 55 (42), 12960–12972. [DOI] [PubMed] [Google Scholar]
- 7.Mondal S; Acharya S; Biswas R; Bagchi B; Zare RN, Enhancement of reaction rate in small-sized droplets: A combined analytical and simulation study. J. Chem. Phys 2018, 148 (24), 244704. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Z; Yan X; Lai Y-H; Zare RN, Fluorescence polarization anisotropy in microdroplets. J. Phys. Chem. Lett 2018, 9 (11), 2928–2932. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee S; Zare RN, Influence of inlet capillary temperature on the microdroplet chemistry studied by mass spectrometry. J. Phys. Chem. A . 2019, 123 (36), 7704–7709. [DOI] [PubMed] [Google Scholar]
- 10.Hao H; Leven I; Head-Gordon T, Can electric fields drive chemistry for an aqueous microdroplet? Nat. Commun 2022, 13 (1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girod M; Moyano E; Campbell DI; Cooks RG, Accelerated bimolecular reactions in microdroplets studied by desorption electrospray ionization mass spectrometry. Chem. Sci 2011, 2 (3), 501–510. [Google Scholar]
- 12.Tang S; Fan L; Cheng H; Yan X, Incorporating electro-epoxidation into electrospray ionization mass spectrometry for simultaneous analysis of negatively and positively charged unsaturated glycerophospholipids. J. Am. Soc. Mass Spectrom 2020, 32 (9), 2288–2295. [DOI] [PubMed] [Google Scholar]
- 13.Heiss DR; Badu-Tawiah AK, In-Source Microdroplet Derivatization Using Coaxial Contained-Electrospray Mass Spectrometry for Enhanced Sensitivity in Saccharide Analysis. Anal. Chem 2021, 93 (50), 16779–16786. [DOI] [PubMed] [Google Scholar]
- 14.Wei Z; Zhang X; Wang J; Zhang S; Zhang X; Cooks RG, High yield accelerated reactions in nonvolatile microthin films: chemical derivatization for analysis of single-cell intracellular fluid. Chem. Sci 2018, 9 (40), 7779–7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JK; Kim S; Nam HG; Zare RN, Microdroplet fusion mass spectrometry for fast reaction kinetics. Proc. Natl. Acad. Sci 2015, 112 (13), 3898–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wleklinski M; Loren BP; Ferreira CR; Jaman Z; Avramova L; Sobreira TJ; Thompson DH; Cooks RG, High throughput reaction screening using desorption electrospray ionization mass spectrometry. Chem. Sci 2018, 9 (6), 1647–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkar D; Mahitha MK; Som A; Li A; Wleklinski M; Cooks RG; Pradeep T, Metallic nanobrushes made using ambient droplet sprays. Adv. Mater 2016, 28 (11), 2223–2228. [DOI] [PubMed] [Google Scholar]
- 18.He X; Gan Z; Fisenko S; Wang D; El-Kaderi HM; Wang W-N, Rapid formation of metal–organic frameworks (MOFs) based nanocomposites in microdroplets and their applications for CO2 photoreduction. ACS Appl. Mater. Interfaces 2017, 9 (11), 9688–9698. [DOI] [PubMed] [Google Scholar]
- 19.Li X; Cao Y; Luo K; Sun Y; Xiong J; Wang L; Liu Z; Li J; Ma J; Ge J, Highly active enzyme–metal nanohybrids synthesized in protein–polymer conjugates. Nat. Catal 2019, 2 (8), 718–725. [Google Scholar]
- 20.Zhong X; Chen H; Zare RN, Ultrafast enzymatic digestion of proteins by microdroplet mass spectrometry. Nat. Commun 2020, 11 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao P; Gunawardena HP; Zhong X; Zare RN; Chen H, Microdroplet ultrafast reactions speed antibody characterization. Anal. Chem 2021, 93 (8), 3997–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rainer T; Eidelpes R; Tollinger M; Müller T, Microdroplet Mass Spectrometry Enables Extremely Accelerated Pepsin Digestion of Proteins. J. Am. Soc. Mass Spectrom 2021, 32 (7), 1841–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng H; Tang S; Yang T; Xu S; Yan X, Accelerating Electrochemical Reactions in a Voltage–Controlled Interfacial Microreactor. Angew. Chem., Int. Ed 2020, 59 (45), 19862–19867. [DOI] [PubMed] [Google Scholar]
- 24.Burris BJ; Badu-Tawiah AK, Enzyme-Catalyzed Hydrolysis of Lipids in Immiscible Microdroplets Studied by Contained-Electrospray Ionization. Anal. Chem 2021, 93 (38), 13001–13007. [DOI] [PubMed] [Google Scholar]
- 25.Gong C; Li D; Li X; Zhang D; Xing D; Zhao L; Yuan X; Zhang X, Spontaneous Reduction-Induced Degradation of Viologen Compounds in Water Microdroplets and Its Inhibition by Host–Guest Complexation. J. Am. Chem. Soc 2022. [DOI] [PubMed] [Google Scholar]
- 26.Nie H; Wei Z; Qiu L; Chen X; Holden DT; Cooks RG, High-yield gram-scale organic synthesis using accelerated microdroplet/thin film reactions with solvent recycling. Chem. Sci 2020, 11 (9), 2356–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bark SJ; Muster N; Yates JR; Siuzdak G, High-temperature protein mass mapping using a thermophilic protease. J. Am. Chem. Soc 2001, 123 (8), 1774–1775. [DOI] [PubMed] [Google Scholar]
- 28.Basile F; Hauser N, Rapid online nonenzymatic protein digestion combining microwave heating acid hydrolysis and electrochemical oxidation. Anal. Chem 2011, 83 (1), 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carvalho LB; Capelo-Martinez J-L; Lodeiro C; Wisniewski JR; Santos HM, Ultrasonic-Based Filter Aided Sample Preparation as the General Method to Sample Preparation in Proteomics. Anal. Chem 2020, 92 (13), 9164–9171. [DOI] [PubMed] [Google Scholar]
- 30.Ji J; Zhang Y; Zhou X; Kong J; Tang Y; Liu B, Enhanced protein digestion through the confinement of nanozeolite-assembled microchip reactors. Anal. Chem 2008, 80 (7), 2457–2463. [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Ferrer D; Capelo J; Vazquez J, Ultra fast trypsin digestion of proteins by high intensity focused ultrasound. J. Proteome Res 2005, 4 (5), 1569–1574. [DOI] [PubMed] [Google Scholar]
- 32.Mouchahoir T; Schiel JE, Development of an LC-MS/MS peptide mapping protocol for the NISTmAb. Anal. Bioanal. Chem 2018, 410 (8), 2111–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivera-Albarran ME; Ray SJ, A Novel Combined Microstrip Resonator/Nanospray Ionization Source for Micro wave-Assisted Trypsin Digestion of Proteins. J. Am. Soc. Mass Spectrom 2020, 31 (8), 1684–1696. [DOI] [PubMed] [Google Scholar]
- 34.Russell WK; Park Z-Y; Russell DH, Proteolysis in mixed organic– aqueous solvent systems: applications for peptide mass mapping using mass spectrometry. Anal. Chem 2001, 73 (11), 2682–2685. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y; Zhang W; Ouyang Z, Fast protein analysis enabled by high-temperature hydrolysis. Chem. Sci 2020, 11 (38), 10506–10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplon H; Reichert JM, Antibodies to watch in 2021. MAbs 2021, 13 (1), 1860476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zitvogel L; Kroemer G, Antibodies regulate antitumour immunity. Nature 2015, 521 (7550), 35–37. [DOI] [PubMed] [Google Scholar]
- 38.Chen L; Han X, Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J. Clin. Invest 2015, 125 (9), 3384–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen DS; Mellman I, Oncology meets immunology: the cancer-immunity cycle, immunity 2013, 39 (1), 1–10. [DOI] [PubMed] [Google Scholar]
- 40.Tsumoto K; Isozaki Y; Yagami H; Tomita M, Future perspectives of therapeutic monoclonal antibodies. Immunotherapy 2019, 11 (2), 119–127. [DOI] [PubMed] [Google Scholar]
- 41.Liu H; Gaza-Bulseco G; Faldu D; Chumsae C; Sun J, Heterogeneity of monoclonal antibodies. J. Pharm. Sci 2008, 97 (7), 2426–2447. [DOI] [PubMed] [Google Scholar]
- 42.Jenkins N; Murphy L; Tyther R, Post-translational modifications of recombinant proteins: significance for biopharmaceuticals. Mol. Biotechnol 2008, 39 (2), 113–118. [DOI] [PubMed] [Google Scholar]
- 43.Manning MC; Chou DK; Murphy BM; Payne RW; Katayama DS, Stability of protein pharmaceuticals: an update. Pharm. Res 2010, 27 (4), 544–575. [DOI] [PubMed] [Google Scholar]
- 44.Gupta S; Jiskoot W; Schöneich C; Rathore AS, Oxidation and Deamidation of Monoclonal Antibody Products: Potential Impact on Stability, Biological Activity, and Efficacy. J. Pharm. Sci 2021. [DOI] [PubMed] [Google Scholar]
- 45.Timm V; Gruber P; Wasiliu M; Lindhofer H; Chelius D, Identification and characterization of oxidation and deamidation sites in monoclonal rat/mouse hybrid antibodies. J. Chromatogr. B. 2010, 878 (9-10), 777–784. [DOI] [PubMed] [Google Scholar]
- 46.Bertolotti-Ciarlet A; Wang W; Lownes R; Pristatsky P; Fang Y; McKelvey T; Li Y; Li Y; Drummond J; Prueksaritanont T, Impact of methionine oxidation on the binding of human IgGl to FcRn and Fcγ receptors. Mol. Immunol 2009, 46 (8-9), 1878–1882. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y; Li X; Liu Y-H; Richardson D; Li H; Shameem M; Yang X In Simultaneous monitoring of oxidation, deamidation, isomerization, and glycosylation of monoclonal antibodies by liquid chromatography-mass spectrometry method with ultrafast tryptic digestion, MAbs, Taylor & Francis: 2016; pp 1477–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boll B. r.; Bessa J; Folzer E; Ríos Quiroz A; Schmidt R; Bulau P; Finkler C; Mahler H-C; Huwyler J. r.; Iglesias A, Extensive chemical modifications in the primary protein structure of IgG1 subvisible particles are necessary for breaking immune tolerance. Mol. Pharmaceutics 2017, 14 (4), 1292–1299. [DOI] [PubMed] [Google Scholar]
- 49.Pavon JA; Li X; Chico S; Kishnani U; Soundararajan S; Cheung J; Li H; Richardson D; Shameem M; Yang X, Analysis of monoclonal antibody oxidation by simple mixed mode chromatography. J. Chromatogr. A 2016, 1431, 154–165. [DOI] [PubMed] [Google Scholar]
- 50.Rogers RS; Nightlinger NS; Livingston B; Campbell P; Bailey R; Balland A In Development of a quantitative mass spectrometry multi-attribute method for characterization, quality control testing and disposition of biologies, MAbs, Taylor & Francis: 2015; pp 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stroop SD, A modified peptide mapping strategy for quantifying site-specific deamidation by electrospray time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom 2007, 21 (6), 830–836. [DOI] [PubMed] [Google Scholar]
- 52.Vlasak J; Ionescu R, Heterogeneity of monoclonal antibodies revealed by charge-sensitive methods. Curr. Pharm. Biotechnol 2008, 9 (6), 468–481. [DOI] [PubMed] [Google Scholar]
- 53.Hirabayashi A; Sakairi M; Koizumi H, Sonic spray ionization method for atmospheric pressure ionization mass spectrometry. Anal. Chem 1994, 66 (24), 4557–4559. [Google Scholar]
- 54.Dulay MT; Chamberlayne CF; Zare RN, Optimizing Coaxial Sonic Spray Geometry for Generating Water Microdroplets. Anal. Chem 2022. [DOI] [PubMed] [Google Scholar]
- 55.Kamat M; Basso KB, Surprising Increase in Distinct Tryptic Peptide Counts at Low Temperature and Short Digestion Times. In 69th ASMS, Pennsylvania Convention Center; | Philadelphia, PA, USA, 2021. [Google Scholar]
- 56.Proc JL; Kuzyk MA; Hardie DB; Yang J; Smith DS; Jackson AM; Parker CE; Borchers CH, A quantitative study of the effects of chaotropic agents, surfactants, and solvents on the digestion efficiency of human plasma proteins by trypsin. J. Proteome Res 2010, 9 (10), 5422–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Somiari RI; Renganathan K; Russell S; Wolfe S; Mayko F; Somiari SB, A colorimetric method for monitoring tryptic digestion prior to shotgun proteomics. Int. J. Proteomics 2014, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dick LW Jr; Mahon D; Qiu D; Cheng K-C, Peptide mapping of therapeutic monoclonal antibodies: Improvements for increased speed and fewer artifacts. J. Chromatogr. B. 2009, 877 (3), 230–236. [DOI] [PubMed] [Google Scholar]
- 59.Hall T; Sandefur SL; Frye CC; Tuley TL; Huang L, Polysorbates 20 and 80 degradation by group XV lysosomal phospholipase A2 isomer X1 in monoclonal antibody formulations. J. Pharm. Sci 2016, 105 (5), 1633–1642. [DOI] [PubMed] [Google Scholar]
- 60.Labrenz SR, Ester hydrolysis of polysorbate 80 in mAb drug product: evidence in support of the hypothesized risk after the observation of visible particulate in mAb formulations. J. Pharm. Sci 2014, 103 (8), 2268–2277. [DOI] [PubMed] [Google Scholar]
- 61.Picotti P; Aebersold R; Domon B, The implications of proteolytic background for shotgun proteomics. Mol. Cell. Proteomics 2007, 6 (9), 1589–1598. [DOI] [PubMed] [Google Scholar]
- 62.Kim J-S; Monroe ME; Camp DG; Smith RD; Qian W-J, In-source fragmentation and the sources of partially tryptic peptides in shotgun proteomics. J. Proteome Res 2013, 12 (2), 910–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alves G; Yu Y-K, Improving peptide identification sensitivity in shotgun proteomics by stratification of search space. J. Proteome Res 2013, 12 (6), 2571–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fang P; Liu M; Xue Y; Yao J; Zhang Y; Shen H; Yang P, Controlling nonspecific trypsin cleavages in LC-MS/MS-based shotgun proteomics using optimized experimental conditions. Analyst 2015, 140 (22), 7613–7621. [DOI] [PubMed] [Google Scholar]
- 65.Alves P; Arnold RJ; Clemmer DE; Li Y; Reilly JP; Sheng Q; Tang H; Xun Z; Zeng R; Radivojac P, Fast and accurate identification of semi-tryptic peptides in shotgun proteomics. Bioinformatics 2008, 24 (1), 102–109. [DOI] [PubMed] [Google Scholar]
- 66.Nigam A; Subramanian M; Koiram Rajanna P, Non-specific digestion artifacts of bovine trypsin exemplified with surrogate peptides for endogenous protein quantitation. Chromatographia 2018, 81 (1), 57–64. [Google Scholar]
- 67.Niu B; Martinelli M II; Jiao Y; Wang C; Cao M; Wang J; Meinke E, Nonspecific cleavages arising from reconstitution of trypsin under mildly acidic conditions. PloS one 2020, 15 (7), e0236740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fonslow BR; Stein BD; Webb KJ; Xu T; Choi J; Park SK; Yates JR, Digestion and depletion of abundant proteins improves proteomic coverage. Nat. Methods 2013, 10 (1), 54–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang L; Wang N; Mitchell CE; Brownlee T; Maple SR; De Felippis MR, A novel sample preparation for shotgun proteomics characterization of HCPs in antibodies. Anal. Chem 2017, 89 (10), 5436–5444. [DOI] [PubMed] [Google Scholar]
- 70.Ohtake S; Shalaev E, Effect of water on the chemical stability of amorphous pharmaceuticals: I. Small molecules. J. Pharm. Sci 2013, 102 (4), 1139–1154. [DOI] [PubMed] [Google Scholar]
- 71.Sydow JF; Lipsmeier F; Larraillet V; Hilger M; Mautz B; Mølhøj M; Kuentzer J; Klostermann S; Schoch J; Voelger HR, Structure-based prediction of asparagine and aspartate degradation sites in antibody variable regions. PloS one 2014, 9 (6), e100736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qian J; Yearley E; Tian S; Jing L; Balsaraf A; Lo Surdo P; Huang Y; Chandramouli S; Bottomley MJ; Moniotte N; Wang Z, Non-Enzymatic and Site-Specific Glycan Shedding: A Novel Protein Degradation Pathway Observed in a Stabilized Form of RSV Prefusion F Protein. Anal. Chem 2018, 90 (18), 10897–10902. [DOI] [PubMed] [Google Scholar]
- 73.Wang W; Vlasak J; Li Y; Pristatsky P; Fang Y; Pittman T; Roman J; Wang Y; Prueksaritanont T; Ionescu R, Impact of methionine oxidation in human IgG1 Fc on serum half-life of monoclonal antibodies. Mol. Immunol 2011, 48 (6-7), 860–866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


