Abstract
Aziz Sancar, Nobel Prize winning Turkish scientist, made several discoveries which had a major impact on molecular sciences, particularly disciplines that focus on carcinogenesis and cancer treatment, including molecular pathology. Cloning the photolyase gene, which was the initial step of his work on DNA repair mechanisms, discovery of the “Maxicell” method, explanation of the mechanism of nucleotide excision repair and transcription-coupled repair, discovery of “molecular matchmakers”, and mapping human excision repair genes at single nucleotide resolution constitute his major research topics. Moreover, Sancar discovered the cryptochromes, the clock genes in humans, in 1998, and this discovery led to substantial progress in the understanding of the circadian clock and the introduction of the concept of “chrono-chemoterapy” for more effective therapy in cancer patients. This review focuses on Aziz Sancar’s scientific studies and their reflections on molecular pathology of neoplastic diseases. While providing a new perspective for researchers working in the field of pathology and molecular pathology, this review is also an evidence of how basic sciences and clinical sciences complete each other.
Keywords: Aziz Sancar, Carcinogenesis, Molecular Pathology, Nobel prize, Neoplasia
INTRODUCTION
Henrich Rohrer, Nobel Prize winning physicist, once said that science requires constantly walking a tightrope between faithful belief and impulse to question, between common knowledge and creativity, between the defense of old territory and the decision to leave established grounds, between bias and impartiality, between expertise and fresh minds, between ambition and passion, between arrogance and self-confident conviction – in short, between human weakness and scientific standards, between today and tomorrow (1).
Scientists/researchers who have the patience to walk this tightrope are more likely to make discoveries that change the world, and one of them is Aziz Sancar, who was awarded the Nobel Prize in Chemistry with Tomas Lindahl and Paul Modrich in 2015 for their conceptual studies on DNA repair (2). Aziz Sancar was born in Savur, a small town in Southeast Turkey, in 1946. His fascination with biochemistry started in his second year of medical school when he learned about the DNA double helix for the first time (3). During his medical education at the Istanbul University Faculty of Medicine, he had the chance to work with excellent researchers including Dr. Mutahhar Yenson and Dr. Muzaffer Aksoy (4). After practicing in Mardin for about 2 years as a physician, he won a NATO fellowship and attended a PhD program at Johns Hopkins University in 1971. However, life had other plans for Sancar, and he left Johns Hopkins and returned to Turkey in June 1972. After a short break, he went to the United States in 1973, and received his PhD in 1977 at the University of Texas at Dallas (3,4). His research journey officially began while he was a PhD student. The road to success was rocky but in the end, he won the Nobel Prize, one of the most prestigious awards in the world.
Sancar’s discoveries (5–13) (Table 1) had a major impact on several scientific disciplines, particularly disciplines that focus on carcinogenesis and cancer treatment. As molecular pathology, a relatively new discipline that incorporates the morphology and molecular alterations of diseases, is among those disciplines, here we discussed the impact of Aziz Sancar’s studies on the molecular pathology of neoplastic diseases.
Table 1.
Overview of Sancar’s greatest scientific discoveries.
|
Discovery |
Highlights |
|
Cloning the photolyase gene |
· Sancar’s mentor, Dr. Claud S. Rupert, (14) discovered photolyase in 1958 (“the beginning of the scientific field of DNA repair” as referred to by Sancar himself in his Nobel lecture (65)). · Almost 20 years after this discovery, Sancar and Rupert (5) succeeded in cloning the photolyase gene, and this was the initial step of Sancar’s work on DNA repair mechanisms. |
|
Discovery of the “Maxicell” method |
· The maxicell method, discovered by Sancar (6), is used to identify plasmid-encoded proteins that use a mutant strain of E. coli that is defective in repairing DNA damage. |
|
Explanation of the mechanism of nucleotide excision repair in E. coli and humans |
· While cloning the excision repair genes uvrA, uvrB, and uvrC, Sancar and Rupp (7) found that UvrABC nuclease made dual incisions and named the enzyme “ABC excinuclease”. · Afterwards, Sancar and colleagues (8) discovered that dual incisions made during nucleotide excision repair in humans were different than in E. coli. |
|
Transcription-coupled repair: “Yunus Emre Opus” |
· Sancar and Selby (10) identified a factor that recognized and removed RNA polymerase from the damaged site while inducing the accumulation of the excision nuclease at the damage: TRCF (Transcription-Repair Coupling Factor). · Sancar describes this paper as his most aesthetically pleasing work, specifically as his “Yunus Emre Opus” (65). |
|
Discovery of “molecular matchmakers” |
· While studying DNA repair mechanisms, Sancar and Hearst discovered a class of proteins, “molecular matchmakers”, that promotes formation of a stable DNA-protein complex (9). |
|
Excision repair map of the human genome at single nucleotide resolution: “Piri Reis map” |
· Sancar and colleagues (11) mapped the sites of repair across the entire human genome, using XR sequencing and at single nucleotide resolution. · Sancar refers to this map as his Piri Reis map, while he describes it as the most satisfying accomplishment in his lab in the 2000s (4,65). |
|
Cryptochrome and the Circadian clock |
· In 1996, after reading an article about the circadian clock and jetlag in a flight magazine, Sancar began his studies on the circadian clock (4), showing that cryptochromes (named by Sancar himself) affect the clock (12,13), leading to several subsequent studies. |
FIRST SUCCESS and the LONGEST JOURNEY: CLONING the PHOTOLYASE GENE
The discovery of the photolyase enzyme, known as SANCAR’s enzyme in the literature, has taken 40 years of Sancar’s scientific life, and is very important since it contains the first information leading to the understanding of DNA damage repair in humans. The photolyase enzyme was first described in 1958 by Sancar’s mentor Dr. Rupert by observing the invigorating effect of blue light on bacteria (14). Dr. Rupert (14) showed that ultraviolet (UV) light can kill bacteria by damaging its DNA, and also revealed the presence of an enzyme that repairs DNA damage in visible light using blue light energy. UV was able to damage bacterial DNA by converting two adjacent pyrimidines, including thymines, into a CPD (cyclobutene pyrimidine dimer), while photolyase enzyme was helping to convert the abnormal thymine dimer into two normal thymine molecules using blue light energy (15–17). However, Rupert and his team could not purify the enzyme due to the low level of enzyme content in bacteria, and therefore could not demonstrate how the enzyme converted sunlight into chemical energy, in other words how it repaired DNA damage caused by UV exposure.
Sancar has achieved great success by exploring the working principles and mechanism of photolyase and has pioneered the understanding of DNA repair mechanisms. After joining Dr. Rupert and his team, Sancar primarily aimed to clone the photolyase gene and obtain the enzyme in pure form. At this point, he first showed that the photo-activated photolyase can be transferred through the plasmids in the cell and that the plasmids can help in the replication process of this enzyme (18). Subsequently, using recombinant DNA (rDNA) technology, which was newly revived at that time, he produced a mutant E. coli clone that did not have the photolyase enzyme. Using this mutant form, he enabled E. coli that could not repair its DNA to use blue light, thanks to the photolyase enzyme carried by the plasmid. In other words, his team demonstrated that they could treat the mutant E. coli by placing the photolyase gene in the normal E. coli chromosome into the plasmid (5,19). In the meantime, they were able to replicate photolyase gene many times using the plasmid and succeeded in obtaining abundant (100-fold amplification) amounts of this enzyme (5). Obtaining the enzyme easily and in abundance allowed them to investigate the repair mechanism of DNA damage by this enzyme using blue light. Therefore, in their subsequent studies, Sancar and colleagues (20–35) analyzed enzyme content and identified light-absorbing components and the working dynamics and revealed the 3D structure of the enzyme by crystallizing photolyases (36).
Sancar continues to work on the photolyase enzyme, which covers the longest period of his scientific journey, and he has also searched for its equivalent in human beings. In fact, the photolyase enzyme is very important in terms of introducing the DNA repair mechanism as a discipline from the first years it was described. Could the presence of this enzyme in bacteria but not in humans be a limitation for humanity? While there is no clear answer for this question yet, some researchers have been studying the possible use of photolyase in mRNA-based gene therapy in humans (37,38), and promising therapeutic agents containing photolyase have been reported to be used in the treatment of lesions triggered by UV rays (39,40). These agents have been combined with thermostimulation in another study (41). However, the efficacy of these topical agents containing photolyase remains controversial and further investigation is needed to explore whether these agents are truly beneficial or not.
DISCOVERY of the “MAXICELL” METHOD
Maxicell is a method developed by Aziz Sancar and colleagues (6) to identify the proteins encoded by the bacterial plasmid. This method may be used to produce any protein, and thus, has taken its place in biochemistry and molecular biology practice as a viable genetic engineering and rDNA technology method. The method uses a mutant, non-photo reactivable E. coli strain that cannot repair DNA under experimental light. Following irradiation, in about 6 hours, 80% of the bacterial chromosomal DNA breaks down and the irradiated mutant bacteria can no longer encode genes and therefore synthesize proteins. On the contrary, plasmids in the irradiated bacteria escape DNA damage due to their small size, and continue transcription and protein synthesis, and these gene products can be monitored using radioactive labeling (6). Although the irradiation limit that the plasmids can escape from is not known precisely, the maxicell method has been used up to 10x106 plasmid size (42).
While Sancar used this method especially in his studies on photolyase (5,43–45) and DNA repair by endo/exonucleases (46–48), many other researchers adopted this method, mainly in their studies that focused on viral and/or bacterial antigens (49–54). More importantly, the maxicell method has been used to produce some hormones (55,56) and monoclonal antibodies (57–59), i.e., molecules that can be both used for diagnostic and treatment purposes. The maxicell method has also facilitated the purification of recombinant gene products and production of DNA fragments (duplicates) in a fast and easy way, representing an initial step for nucleic acid amplification methods such as polymerase chain reaction (PCR) and other quantitative techniques. Currently, PCR is among the most frequently used techniques in molecular pathology practice.
EXPLANATION of the MECHANISM of NUCLEOTIDE EXCISION REPAIR in E. COLI and HUMANS
DNA damage compromises the functional integrity of DNA (60) and occurs through multi-faceted mechanisms (60–64). When it occurs, the cells can either repair the damage, or stop the progression of the cell cycle, or induce apoptosis (65).
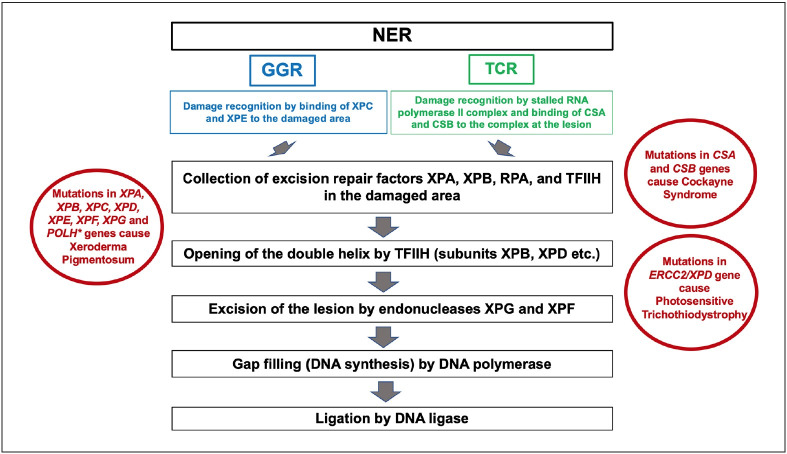
Nucleotide excision repair (NER) is a multi-component, multi-stage enzymatic system which includes recognition and elimination of a wide range of DNA damage (60–64). NER has been described in detail by Sancar et al. in prokaryotes and eukaryotes (60,61,65–67). All free-living organisms have excision repair genes (60,66). The damage is removed as a 12-13 nucleotide-long oligomer in prokaryotes and a 24-32 nucleotide-long oligomer in eukaryotes (60,63,68). NER starts with the recognition of the DNA damage, and then an oligomer, which has been formed by dual incision of the damaged area, is excised and released, and finally, the gap is filled by repair synthesis and ligation (60,67). These steps can be affected by many in vivo factors such as transcription, DNA replication, epigenetic modifications and/or binding of regulatory proteins to DNA (69). Six proteins (XPA, RPA, XPC, TFIIH, XPG and XPF-ERCC1 complex) are involved in excision repair in humans (60,61,68,70–72) (Figure 1). XPC, XPA and RPA are responsible for damage recognition, TFIIH (“Transcription factor II Human”) plays a role in DNA unwinding, and XPG and XPF-ERCC1 complex are responsible for 3’ and 5’ incisions (60).
Figure 1.
Basic steps of nucleotide excision repair (NER). While global genome repair (GGR) recognizes and repairs damages in the entire genome, transcription-coupled repair (TCR) operates on transcribed regions only (60,83,151). TFIIH: Transcription factor II Human. *POLH gene encodes DNA polymerase eta.
Repair of UVB-induced pyrimidine dimers is important for the prevention of skin cancer development by NER mechanisms (64). Loss of DNA repair capacity due to NER deficiency causes genomic instability which is a carcinogenic feature (73). NER has been the focus of interest in various studies, both for understanding the pathogenesis and for the discovery of targeted therapies. Defective NER has been associated with three rare autosomal recessive hereditary diseases: xeroderma pigmentosum (XP), Cockayne syndrome (CS), and photosensitive trichothiodystrophy (TTD) (64) (Figure 1).
Among these three syndromes, pathologists often encounter XP while signing out skin resections. In XP, a defect in NER greatly increases the lethality and mutagenicity of the DNA-damaging agents and it is characterized by deficient nucleotide excision repair, extreme sensitivity to sunlight, and early onset skin cancer in humans (60,64,65,69). The risk of sunlight-induced skin cancers is greatly increased in XP patients, compared to the normal population (65,74). This finding is not surprising considering that excision repair in humans is the only known mechanism for eliminating UV-induced lesions (60).
NER deficiency has been demonstrated to be associated with non-skin cancer as well. In a study of breast cancer patients, mRNA expression levels of the NER genes have been shown to decrease in representative tumor samples compared to normal tissue samples by microarray analysis and these results have partially been confirmed at the protein level (73). Hence, the authors have suggested that NER deficiency may contribute to the development of sporadic breast cancer and that early-stage breast cancer may be sensitive to genotoxic chemotherapeutic agents such as cisplatin, the damage of which is eliminated by NER (73). Lu et al. (75) have reported that the polymorphisms of nucleotide excision repair genes ERCC1 rs11615 and ERCC5(XPG) rs17655 are related to increased risk of laryngeal cancer but the biological effect of these polymorphisms is uncertain. Impaired NER has also been suggested to contribute to the development of head and neck squamous cell carcinomas (76). Defects in NER and base excision repair (BER), one of the main mechanisms of defense against oxidative DNA damage, have been reported to play a role in susceptibility to differentiated thyroid carcinoma (77). However, this effect appears to be due to some polymorphic genes with a weak overall effect (77).
Nevertheless, defective NER mechanisms do not contribute to the pathogenesis of some cancer types. For example, a minor relationship has been found between colorectal cancer and XRCC3 (a recombination repair gene) polymorphism, but the significance of this finding remains to be explored and NER genes are not considered to be major players in colorectal carcinogenesis (74). In another study, Gaddameedhi et al. (71) have found that melanoma cells retain their capacity for NER, and suggested that NER loss probably does not contribute much to the progression of melanoma.
In a study that measured the repair rates of UV-induced DNA damage during the differentiation of human embryonal carcinoma cells to neurons and muscle cells, NER capacity increased with the cellular differentiation level (62). Current standard anticancer therapies limit tumor growth by decreasing tumor proliferation or vascularization, but they have limited effects on cancer stem cells (78). A better understanding of the biological effect of potential NER inhibitors should facilitate the development of optimal synthetic killer combinations (78). This approach is promising not only for the individualized treatment of cancer patients with NER deficiency syndrome, but also in the treatment of patients with cancer in general (78).
TRANSCRIPTION-COUPLED REPAIR: “YUNUS EMRE OPUS”
NER works more effectively in the transcribed regions of the genome. At this point, the “transcription coupled repair mechanism” comes into play. This mechanism, which is activated when RNA polymerase encounters DNA damage, does not allow the transcription to continue before the damage is repaired, provides faster repair of the damage, and reduces the harmful effects of transcription pause in the cell (79). The mechanism of this priority repair system, which is selective to the transcribed chain of the gene, had not been illuminated for a long time. In 1985, photodimers formed in the transcribed genes in various mammals and bacteria have been shown to be removed 5-10 times faster than the non-transcribed regions in the chromosome (80–82). Aziz Sancar and Christopher Selby (83) demonstrated that the progression of the RNA polymerase was stopped by DNA damage and caused the formation of a metastable RNA polymerase elongation complex at the damaged site, but this complex inhibited the repair rather than accelerating it in their study designed with damaged DNA, purified RNA polymerase and the UvrA, UvrB and UvrC proteins in E. coli. Consequently, they purified a protein in the form of translocase, which they named as “transcription-repair coupling factor (TRCF)”, which recognizes the RNA polymerase complex, separates it from the damaged area, and enables UvrA to come to the damaged area. Thus, they succeeded in enlightening the mechanism by finding the missing factor in the system. They also discovered that this protein was encoded by the MFD (mutation frequency decline) gene, which was known to prevent UV-induced DNA damage (10,84–87).
With this discovery, studies aiming to explain the mechanism of TCR in humans by various researchers including Aziz Sancar revealed that the damage recognition step was carried out by stopped RNA polymerase II complex, differently from global repair, and that this complex collected CSB translocase and the core excision repair factors except XPC to the damaged area and the next steps continued the same as in global repair (11,60,88–91) (Figure 1).
The reflections of the studies on this topic to the clinical medicine and molecular pathology can be grouped in three categories: 1) effects on cellular aging, carcinogenesis, apoptosis; 2) effects on cisplatin susceptibility and resistance, and 3) the potential use in targeted therapy.
There is evidence that the RNA polymerase II complex activates P53, initiates apoptosis, and acts as the primary sensor in all DNA damage response reactions (92). Mutations that inactivate TCR are known to cause Cockayne Syndrome in humans (11,93). Mutations occurring in the CSB/ERCC2 gene encoding TRCF and some mutations that disrupt repair function in XP genes cause Cockayne Syndrome (85) (Figure 1). Damaged TCR system causes the DNA damage to stop transcription, resulting in impaired cell function, premature aging, and cell death. The effects of TCR damage differ from tissue to tissue depending on factors such as tissue metabolism, activity of antioxidant systems, and other repair systems. This explains the clinical picture seen in Cockayne Syndrome, which is the most well-known TCR-associated disorder, and is characterized by cellular aging in tissues consisting of non-proliferating or slow proliferating cells such as Schwann cells and neurons, resulting in progressive neurodevelopmental disorders. The delicate balance between global repair disorders that cause cancer development and TCR disorders that cause premature aging is critical in preventing both cancer and premature aging. The TCR system, which is still intact in global repair disorder, ensures cell survival and delayed cellular aging, but results in accumulation of DNA damage in the non-transcribed genes and the non-transcribed chain of active genes, thereby causing mutagenesis and an increased risk of cancer. On the other hand, cells that cannot be repaired by the TCR system die due to transcriptional stress providing a strong protection against cancer (94). From this point of view, the use of TCR-inhibiting or -blocking agents is promising as an adjunctive treatment approach to chemotherapy. In this way, it is thought that tumor cells will become more sensitive to the lethal effects of chemotherapy and at the same time, surviving tumor cells will not carry the treatment-induced mutations (95).
Resistance to chemotherapy is an important problem in cancer treatment. The sensitivity to cisplatin-based treatments, which are commonly used in cancer management, has been reported to be increased in cells with TCR damage. It has been stated that detection of TCR disorders in cancer cells may be helpful in predicting resistance to cisplatin treatment (96). Cisplatin resistance is thought to be related to increased DNA repair capacity, P53 mutations, or loss of DNA mismatch repair capacity. There is evidence that reduced TCR capacity increases the susceptibility of tumor cells to apoptosis induced by cisplatin even in cell lines with p53 mutation and DNA mismatch defects, and that the TCR system may be a potential target in overcoming cisplatin resistance in cancer treatment (97). For example, in experimental studies in chronic lymphocytic leukemia (CLL), TCR inhibition, in addition to the treatment regimen, has been shown to induce cell death regardless of the previous treatment and to have synergistic effects with the treatment regimen, revealing that it is a mechanism that can be used in refractory disease to re-sensitize CLL cells (98,99).
DISCOVERY of “MOLECULAR MATCHMAKERS”
Sancar and Hearst discovered “molecular matchmakers” while studying DNA repair mechanisms. They defined the “molecular matchmakers” as a class of proteins that make conformational (structural) changes in at least one of the DNA-binding protein pairs to increase the formation of the DNA-protein complex (9). Structural changes facilitate and stabilize the protein-DNA complex formation. The degree of change enhances the specificity and stability of the complex (100). The molecular matchmaker is a protein that combines two compatible but also solitary macromolecules with an ATP-dependent reaction, promotes their fusion, and then leaves the new complex to continue its processes (9,101). Molecular-matchmakers are not only involved in DNA-protein binding but also in other molecular interactions, such as RNA-protein, RNA-DNA and macromolecule-small ligands (101).
Protein interactions are essential in all stages of homeostasis. Therefore, elaboration of these interactions provides unique opportunities to understand the molecular basis of diseases, and to develop better diagnostic and treatment strategies. It is crucial to characterize biochemical, physical and functional aspects of protein interactions (102,103). Mutations that affect protein structure may cause impaired protein-DNA interactions, protein misfolding, new unwanted protein interactions, or pathogen-host protein interactions (9,102). For instance, mutations in P53’s DNA-binding domain impair its ability to bind to target DNA sequences, blocking several tumor suppressing mechanisms such as apoptosis, genetic stability etc. (102). It has already been demonstrated that misfolded and aggregated proteins may cause several diseases, either by disruption of specific binding abilities, formation of unwanted proteins, and/or over-accumulation of the impaired protein (104–106). Viruses that can integrate their genetic material to the host genome such as the human papilloma virus (HPV) and hepatitis B virus (HBV) can initiate and/or promote carcinogenesis via pathogen-host protein interactions (107,108).
Studies on protein interactions help to predict genotype-phenotype associations, and new diagnostic tools can be created from these associations. Identifying involved pathways serves as a key to discover new diagnostic and prognostic tools. The links of diseases and proteins help us find key areas as potential drug targets and provide information for drug design (102). For example, the heterogeneous nuclear ribonucleoprotein (hnRNP) A2/B1, which is an RNA matchmaker (109), has been shown to promote carcinogenesis, invasion and metastasis in pancreatic ductal adenocarcinoma, glioblastoma and lung cancer, but to play an inhibitory role in metastasis in breast cancer, making it a promising prognostic biomarker and a potential molecular therapeutic target for breast cancer (110).
EXCISION REPAIR MAP of the HUMAN GENOME at SINGLE NUCLEOTIDE RESOLUTION: "PIRI REIS MAP"
Sancar and colleagues (11) have recently mapped the sites of repair across the entire human genome at single nucleotide resolution using XR (excision repair) sequencing. Sancar refers to this map as his “Piri Reis map” (4,65) showing a “new world of repair genes”, referring to Piri Reis, an Ottoman admiral and cartographer who drew one of the oldest maps of the New World. During excision repair, a single strand with 30 nucleotides that includes the lesion is removed from the DNA after dual incisions. XR-sequencing basically depends on capturing and sequencing this excised strand, allowing to create a genome-wide map of human excision repair at single-nucleotide resolution and provides valuable data about the effects of genomic position and chromatin status on DNA damage and repair (11). Using the same sequencing method, they then conducted other studies investigating the effects of anti-cancer drugs such as cisplatin on DNA damage and repair (111–113). For example, in a study aiming to understand DNA damage caused by cisplatin and its repair in the mouse liver by using XR-sequencing, Sancar and colleagues (111) showed that the repair of the transcribed strand is dominant in the first two days after cisplatin injection and then the repair of the non-transcribed strand becomes dominant, an information that may be useful for designing chemotherapy regimens. Another XR-sequencing-based study in colon cancer cell lines demonstrated up-regulation of membrane transport pathways in the oxaliplatin-resistant cells (113). Such studies using the XR sequencing method may provide an improvement in the way of illuminating the cellular response or resistance mechanisms to anti-cancer drugs.
ROLE of the CIRCADIAN CLOCK in CARCINOGENESIS and CHRONOTHERAPY: A NEW APPROACH in CANCER TREATMENT, “CHRONOCHEMOTHERAPY”
The circadian rhythm (also called the “biological clock” (“circa” meaning “approximately”, “diem” meaning “day” in Latin) is a mechanism regulating physiological and metabolic events in living creatures in the form of daily endogenous rhythms over a 24-hour period. Although it demonstrates an endogenous oscillation, the circadian rhythm uses some signals received from the environment, such as light and nutrition, as determinants in the regulation of rhythms. The circadian clock is a universal regulatory system that creates daily rhythms in multiple physiological periods interfering with other regulatory systems and pathways in mammals in order to ensure the stabilization of homeostasis (114). It influences the physiological rhythm and metabolic events significantly (111).
In humans, the circadian clock has been determined to be formed under four gene controls called CLOCK, BMAL1, Cryptochrome and Period (114). The cryptochrome genes discovered by Sancar and Miyamoto (115,116) regulate the circadian clock in plants and animals. Sancar (12) named the cryptochromes (CRY1 and CRY2) based on their resemblance to the plant blue light photoreceptors which also had sequence similarity to photolyase. In this cycle, as the transcriptional activators, CLOCK and BMAL1 proteins bind to promoters of the Cryptochrome and Period genes to activate their transcription, followed by the inhibition of CLOCK-BMAL1-activated transcription (117). The CLOCK – BMAL1 complex has been shown to affect up to 10% of the entire transcript in the circadian cycle (118). Loss of several tumor suppressor mechanisms caused by an impaired circadian rhythm has been suggested to contribute to carcinogenesis (119–121). Deregulated expression of numerous circadian cycle proteins has been found to be associated with poor prognosis and aggressive behavior in several malignancies (122–127). It has also been shown that single nucleotide polymorphisms in the CRY2 and CLOCK genes increase the risk of breast (128) and colorectal cancer (129).
On the other hand, whether the disruption of the circadian clock is a risk for cancer remains unclear due to paucity of supporting data from well-controlled genetic studies on mice (114) and the presence of findings indicating otherwise (130), despite the supporting evidence from experimental studies (131,132). In the few limited epidemiological studies that investigated the relationship between circadian rhythm and cancer development, a higher risk of breast, prostate and colorectal carcinoma have been reported in people who work in the night shift (133–135). As of 2019, the International Agency for Research on Cancer (IARC) defines night work as “group 2A, probably carcinogenic to humans” based on the limited evidence on the subject (136). Hence, further investigation is required to fully elucidate the relationship between the circadian rhythm and carcinogenesis.
Nevertheless, the potential use of therapeutic agents to re-regulate the circadian rhythm to cure human diseases and to develop anti-cancer drugs targeting circadian clock genes and proteins are still hot research topics (137–139). Moreover, administration of chemotherapy based on the circadian rhythm (“chrono-chemotherapy”) has opened a new era. Dysregulation of the circadian clock can affect cancer susceptibility by regulating DNA damage repair mechanisms and apoptosis. The NER mechanism has also been demonstrated to be regulated by the circadian rhythm. Sancar et al. (140) have observed that NER increased gradually in the morning and reached its highest level in the evening, and that mice exposed to UVB radiation in the morning hours developed four times more invasive skin cancer compared to the evening hours. Subsequently, they investigated the most convenient time during the day for administration of cisplatin, a chemotherapeutic agent, in order to reduce cisplatin’s adverse effects as the damage caused by cisplatin on DNA is repaired by the NER system, and suggested that it can be administered when DNA repair in normal tissues is the highest during the day, i.e., in accordance with the circadian rhythm (111,141). Other researchers have also reported promising results for the use of chrono-chemotherapy (142,143). Although evidence on the efficiency of chrono-chemotherapy remains to be fully elucidated, Sancar and colleagues have suggested that further research may provide information on the efficacy and feasibility of timed administration of cisplatin (141), and their detailed research on this topic is ongoing.
FUTURE REMARKS
Aziz Sancar’s studies had a remarkable impact on other disciplines, especially on cancer research. His discoveries on photolyase (144) and NER have already led to studies on carcinogenesis (74–76,145), and targeted therapy options are being investigated based on his work on NER (146,147), TCR (96,98), cryptochromes, and the circadian clock (114). The future of targeted therapy, at least in part, seems to be built around some of his studies.
Sancar is a living proof of the saying “Science knows no country, because knowledge belongs to humanity, and is the torch which illuminates the world.” (148). Who would have thought someone from a small town in Turkey would go all the way to the Nobel Prize? But then, life is all about surprises. And now, it is time for junior scientists/researchers to work on their take-home messages from Sancar’s studies. To date, only few pathologists have won the Nobel Prize (and in different categories). Some pathologists have claimed that pathologists could not receive the Nobel Prize, because morphology alone was not enough, and with the emergence of molecular pathology, this obstacle may be overcome (149). Regardless, there is a bigger reward that science promises to researchers: “The emotional thrill of being the first person in the history of the world to see or to understand something (150)”, and Sancar’s success is an inspiration for junior researchers.
CONFLICT of INTEREST
The authors declare no conflict of interest.
FUNDING
The authors received no specific funding.
Acknowledgments
The authors would like to thank to Professor Sulen Sarioglu and Associate Professor S. Mehtat Unlu for their guidance and valuable contributions during the writing of this article, and Professor Aziz Sancar, for the critical reading of the manuscript.
References
- Rohrer H. Science-"Walking a Tightrope" in trust and confidence in scientific research. In: Hermeren G, , Sahlin K, Sahlin N, , editors. Kungl Vitterhets Historie. Stockholm: [Google Scholar]
- uuuu [ Dec 14; 2019 ];The Nobel Prize in Chemistry 2015. https://www.nobelprize.org/prizes/chemistry/2015/summary/
- uuuu [ Dec 14; 2019 ];Aziz Sancar - Biographical. https://www.nobelprize.org/prizes/chemistry/2015/sancar/biographical/
- Bursali O. Aziz Sancar ve Nobel'in Öyküsü. Kırmızı Kedi Yayınevi; Istanbul: [Google Scholar]
- Sancar A., Rupert C. S. Cloning of the phr gene and amplification of photolyase in Escherichia coli. Dec;1978 Gene. 4:295–308. doi: 10.1016/0378-1119(78)90047-1. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. Jan;1979 J Bacteriol. 137:692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Rupp W. D. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. May;1983 Cell. 33:249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- Sibghatullah null, Husain I., Carlton W., Sancar A. Human nucleotide excision repair in vitro: repair of pyrimidine dimers, psoralen and cisplatin adducts by HeLa cell-free extract. Jun;1989 Nucleic Acids Res. 17:4471–4484. doi: 10.1093/nar/17.12.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hearst J. E. Molecular matchmakers. Mar;1993 Science. 259:1415–1420. doi: 10.1126/science.8451638. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Gene- and strand-specific repair in vitro: partial purification of a transcription-repair coupling factor. Sep;1991 Proc Natl Acad Sci U S A. 88:8232–8236. doi: 10.1073/pnas.88.18.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Jinchuan, Adar Sheera, Selby Christopher P., Lieb Jason D., Sancar Aziz. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. May;2015 Genes Dev. 29:948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu D. S., Zhao X., Zhao S., Kazantsev A., Wang R. P., Todo T., Wei Y. F., Sancar A. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Nov;1996 Biochemistry. 35:13871–13877. doi: 10.1021/bi962209o. [DOI] [PubMed] [Google Scholar]
- Vitaterna M. H., Selby C. P., Todo T., Niwa H., Thompson C., Fruechte E. M., Hitomi K., Thresher R. J., Ishikawa T., Miyazaki J., Takahashi J. S., Sancar A. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Oct;1999 Proc Natl Acad Sci U S A. 96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert C. S., Goodgal S. H., Herriott R. M. Photoreactivation in vitro of ultraviolet-inactivated Hemophilus influenzae transforming factor. Jan;1958 J Gen Physiol. 41:451–471. doi: 10.1085/jgp.41.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert C. S. Photoenzymatic repair of ultraviolet damage in DNA. I. Kinetics of the reaction. Mar;1962 J Gen Physiol. 45:703–724. doi: 10.1085/jgp.45.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert C. S. Photoenzymatic repair of ultraviolet damage in DNA. II. Formation of an enzyme-substrate complex. Mar;1962 J Gen Physiol. 45:725–741. doi: 10.1085/jgp.45.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff D. L., Rupert C. S. Disappearance of thymine photodimer in ultraviolet irradiated DNA upon treatment with a photoreactivating enzyme from baker's yeast. Apr;1962 Biochem Biophys Res Commun. 7:237–240. doi: 10.1016/0006-291x(62)90181-x. [DOI] [PubMed] [Google Scholar]
- Sancar A., Rupert C. S. Determination of plasmid molecular weights from ultraviolet sensitivities. Mar;1978 Nature. 272:471–472. doi: 10.1038/272471a0. [DOI] [PubMed] [Google Scholar]
- Sancar A. 1977A study of photoreactivating enzyme (DNA photolyase) of Escherichia coli.
- Sancar G. B., Smith F. W., Reid R., Payne G., Levy M., Sancar A. Action mechanism of Escherichia coli DNA photolyase. I. Formation of the enzyme-substrate complex. Jan;1987 J Biol Chem. 262:478–485. [PubMed] [Google Scholar]
- Jorns M. S., Baldwin E. T., Sancar G. B., Sancar A. Action mechanism of Escherichia coli DNA photolyase. II. Role of the chromophores in catalysis. Jan;1987 J Biol Chem. 262:486–491. [PubMed] [Google Scholar]
- Sancar G. B., Jorns M. S., Payne G., Fluke D. J., Rupert C. S., Sancar A. Action mechanism of Escherichia coli DNA photolyase. III. Photolysis of the enzyme-substrate complex and the absolute action spectrum. Jan;1987 J Biol Chem. 262:492–498. [PubMed] [Google Scholar]
- Heelis P. F., Payne G., Sancar A. Photochemical properties of Escherichia coli DNA photolyase: selective photodecomposition of the second chromophore. Jul;1987 Biochemistry. 26:4634–4640. doi: 10.1021/bi00389a007. [DOI] [PubMed] [Google Scholar]
- Payne G., Heelis P. F., Rohrs B. R., Sancar A. The active form of Escherichia coli DNA photolyase contains a fully reduced flavin and not a flavin radical, both in vivo and in vitro. Nov;1987 Biochemistry. 26:7121–7127. doi: 10.1021/bi00396a038. [DOI] [PubMed] [Google Scholar]
- Husain I., Sancar G. B., Holbrook S. R., Sancar A. Mechanism of damage recognition by Escherichia coli DNA photolyase. Sep;1987 J Biol Chem. 262:13188–13197. [PubMed] [Google Scholar]
- Johnson J. L., Hamm-Alvarez S., Payne G., Sancar G. B., Rajagopalan K. V., Sancar A. Identification of the second chromophore of Escherichia coli and yeast DNA photolyases as 5,10-methenyltetrahydrofolate. Apr;1988 Proc Natl Acad Sci U S A. 85:2046–2050. doi: 10.1073/pnas.85.7.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. F., Sancar A. Active site of Escherichia coli DNA photolyase: mutations at Trp277 alter the selectivity of the enzyme without affecting the quantum yield of photorepair. Jun;1990 Biochemistry. 29:5698–5706. doi: 10.1021/bi00476a009. [DOI] [PubMed] [Google Scholar]
- Li Y. F., Heelis P. F., Sancar A. Active site of DNA photolyase: tryptophan-306 is the intrinsic hydrogen atom donor essential for flavin radical photoreduction and DNA repair in vitro. Jun;1991 Biochemistry. 30:6322–6329. doi: 10.1021/bi00239a034. [DOI] [PubMed] [Google Scholar]
- Kim S. T., Heelis P. F., Okamura T., Hirata Y., Mataga N., Sancar A. Determination of rates and yields of interchromophore (folate----flavin) energy transfer and intermolecular (flavin----DNA) electron transfer in Escherichia coli photolyase by time-resolved fluorescence and absorption spectroscopy. Nov;1991 Biochemistry. 30:11262–11270. doi: 10.1021/bi00111a011. [DOI] [PubMed] [Google Scholar]
- Langenbacher Thomas, Zhao Xiaodong, Bieser Gero, Heelis Paul F., Sancar Aziz, Michel-Beyerle Maria Elisabeth. Substrate and Temperature Dependence of DNA Photolyase Repair Activity Examined with Ultrafast Spectroscopy. Oct;1997 [ Nov 03; 2022 ];J Am Chem Soc. 119:10532–10536. doi: 10.1021/ja9721024. [DOI] [Google Scholar]
- Payne G., Sancar A. Absolute action spectrum of E-FADH2 and E-FADH2-MTHF forms of Escherichia coli DNA photolyase. Aug;1990 Biochemistry. 29:7715–7727. doi: 10.1021/bi00485a021. [DOI] [PubMed] [Google Scholar]
- Okamura Tadashi, Sancar Aziz, Heelis Paul F., Begley Tadhg P., Hirata Yoshinori, Mataga Noboru. Picosecond laser photolysis studies on the photorepair of pyrimidine dimers by DNA photolyase. 1. Laser photolysis of photolyase-2-deoxyuridine dinucleotide photodimer complex. Apr;1991 [ Nov 03; 2022 ];J Am Chem Soc. 113:3143–3145. doi: 10.1021/ja00008a050. [DOI] [Google Scholar]
- Kavakli I. Halil, Sancar Aziz. Analysis of the role of intraprotein electron transfer in photoreactivation by DNA photolyase in vivo. Dec;2004 Biochemistry. 43:15103–15110. doi: 10.1021/bi0478796. [DOI] [PubMed] [Google Scholar]
- Sancar Aziz. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Jun;2003 Chem Rev. 103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- Sancar Aziz. Structure and function of photolyase and in vivo enzymology: 50th anniversary. Nov;2008 J Biol Chem. 283:32153–32157. doi: 10.1074/jbc.R800052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. W., Kim S. T., Sancar A., Deisenhofer J. Crystal structure of DNA photolyase from Escherichia coli. Jun;1995 Science. 268:1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- Boros Gábor, Karikó Katalin, Muramatsu Hiromi, Miko Edit, Emri Eszter, Hegedűs Csaba, Emri Gabriella, Remenyik Éva. Transfection of Human Keratinocytes with Nucleoside-Modified mRNA Encoding CPD-Photolyase to Repair DNA Damage. 2016Methods Mol Biol. 1428:219–228. doi: 10.1007/978-1-4939-3625-0_14. [DOI] [PubMed] [Google Scholar]
- Boros Gábor, Miko Edit, Muramatsu Hiromi, Weissman Drew, Emri Eszter, Rózsa Dávid, Nagy Georgina, Juhász Attila, Juhász István, Horst Gijsbertus, Horkay Irén, Remenyik Éva, Karikó Katalin, Emri Gabriella. Transfection of pseudouridine-modified mRNA encoding CPD-photolyase leads to repair of DNA damage in human keratinocytes: a new approach with future therapeutic potential. Dec;2013 J Photochem Photobiol B. 129:93–99. doi: 10.1016/j.jphotobiol.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig Susana, Granger Corinne, Garre Aurora, Trullàs Carles, Sanmartin Onofre, Argenziano Giuseppe. Review of Clinical Evidence over 10 Years on Prevention and Treatment of a Film-Forming Medical Device Containing Photolyase in the Management of Field Cancerization in Actinic Keratosis. Jun;2019 Dermatol Ther (Heidelb) 9:259–270. doi: 10.1007/s13555-019-0294-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puviani M., Barcella A., Milani M. Efficacy of a photolyase-based device in the treatment of cancerization field in patients with actinic keratosis and non-melanoma skin cancer. Dec;2013 G Ital Dermatol Venereol. 148:693–698. [PubMed] [Google Scholar]
- Laino Luigi, Elia Fulvia, Desiderio Flora, Scarabello Alessandra, Sperduti Isabella, Cota Carlo, DiCarlo Aldo. The efficacy of a photolyase-based device on the cancerization field: a clinical and thermographic study. Aug;2015 J Exp Clin Cancer Res. 34:84–84. doi: 10.1186/s13046-015-0203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudega Bauke, Mooi Frits R. The Use of Minicells and Maxicells to Detect the Expression of Cloned Genes. In: Walker John M., Gaastra Wim., editors. Techniques in Molecular Biology. Springer Netherlands; Dordrecht: [ Nov 03; 2022 ]. pp. 239–255. [DOI] [Google Scholar]
- Sancar A., Sancar G. B. Escherichia coli DNA photolyase is a flavoprotein. Jan;1984 J Mol Biol. 172:223–227. doi: 10.1016/s0022-2836(84)80040-6. [DOI] [PubMed] [Google Scholar]
- Sancar A., Smith F. W., Sancar G. B. Purification of Escherichia coli DNA photolyase. May;1984 J Biol Chem. 259:6028–6032. [PubMed] [Google Scholar]
- Jorns M. S., Sancar G. B., Sancar A. Identification of a neutral flavin radical and characterization of a second chromophore in Escherichia coli DNA photolyase. Jun;1984 Biochemistry. 23:2673–2679. doi: 10.1021/bi00307a021. [DOI] [PubMed] [Google Scholar]
- Sancar A., Wharton R. P., Seltzer S., Kacinski B. M., Clarke N. D., Rupp W. D. Identification of the uvrA gene product. May;1981 J Mol Biol. 148:45–62. doi: 10.1016/0022-2836(81)90234-5. [DOI] [PubMed] [Google Scholar]
- Sancar A., Clarke N. D., Griswold J., Kennedy W. J., Rupp W. D. Identification of the uvrB gene product. May;1981 J Mol Biol. 148:63–76. doi: 10.1016/0022-2836(81)90235-7. [DOI] [PubMed] [Google Scholar]
- Sancar A., Kacinski B. M., Mott D. L., Rupp W. D. Identification of the uvrC gene product. Sep;1981 Proc Natl Acad Sci U S A. 78:5450–5454. doi: 10.1073/pnas.78.9.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Feb;1985 Proc Natl Acad Sci U S A. 82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah K. C., Sankar S., Porter A. G. Expression and processing of human rhinovirus type 14 polypeptide precursors in Escherichia coli maxicells. Sep;1988 Gene. 69:265–274. doi: 10.1016/0378-1119(88)90437-4. [DOI] [PubMed] [Google Scholar]
- Kay A., Mandart E., Trepo C., Galibert F. The HBV HBX gene expressed in E. coli is recognised by sera from hepatitis patients. May;1985 EMBO J. 4:1287–1292. doi: 10.1002/j.1460-2075.1985.tb03774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baez Luis. Jan;1993 Studies on Hepatitis a Virus: I. Expression of Viral Capsid Peptides in Escherichia Coli and Their Application for an Immunoassay. II. Development of an Assay for Viral Detection by Anti-Hav Antibodies. https://digitalcommons.lsu.edu/gradschool_disstheses/5482
- Neidhardt F. C., Wirth R., Smith M. W., Van Bogelen R. Selective synthesis of plasmid-coded proteins by Escherichia coli during recovery from chloramphenicol treatment. Jul;1980 J Bacteriol. 143:535–537. doi: 10.1128/jb.143.1.535-537.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L. T., Foxall P. A., Russell R., Mobley H. L. Purification of recombinant Helicobacter pylori urease apoenzyme encoded by ureA and ureB. Jul;1992 Infect Immun. 60:2657–2666. doi: 10.1128/iai.60.7.2657-2666.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal T. S., Lowry P. J., Savva D. Expression and partial purification of human pro-opiomelanocortin in Escherichia coli. Sep;1989 J Mol Endocrinol. 3:105–112. doi: 10.1677/jme.0.0030105. [DOI] [PubMed] [Google Scholar]
- Gilbert M. S., Lowry P. J., Castro M. G., Woods R. J., Savva D. Expression and partial purification of human prolactin in Escherichia coli. 1991Int J Biochem. 23:107–114. doi: 10.1016/0020-711x(91)90016-g. [DOI] [PubMed] [Google Scholar]
- Hoffman P. S., Butler C. A., Quinn F. D. Cloning and temperature-dependent expression in Escherichia coli of a Legionella pneumophila gene coding for a genus-common 60-kilodalton antigen. Jun;1989 Infect Immun. 57:1731–1739. doi: 10.1128/iai.57.6.1731-1739.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam R., Seilhamer J. J., Pritchard A. E., Cummings D. J. Identification of Paramecium mitochondrial proteins using antibodies raised against fused mitochondrial gene products. 1986Gene. 49:129–138. doi: 10.1016/0378-1119(86)90392-6. [DOI] [PubMed] [Google Scholar]
- Holley M. C. A simple in vitro method for raising monoclonal antibodies to cochlear proteins. 1992Tissue Cell. 24:613–624. doi: 10.1016/0040-8166(92)90032-3. [DOI] [PubMed] [Google Scholar]
- Reardon Joyce T., Sancar Aziz. Nucleotide excision repair. 2005Prog Nucleic Acid Res Mol Biol. 79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- Reardon Joyce T., Sancar Aziz. Purification and characterization of Escherichia coli and human nucleotide excision repair enzyme systems. 2006Methods Enzymol. 408:189–213. doi: 10.1016/S0076-6879(06)08012-8. [DOI] [PubMed] [Google Scholar]
- Li Wentao, Liu Wenjie, Kakoki Ayano, Wang Rujin, Adebali Ogun, Jiang Yuchao, Sancar Aziz. Nucleotide excision repair capacity increases during differentiation of human embryonic carcinoma cells into neurons and muscle cells. Apr;2019 J Biol Chem. 294:5914–5922. doi: 10.1074/jbc.RA119.007861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Jun-Hyuk, Kim So-Young, Kim Sook-Kyung, Kemp Michael G., Sancar Aziz. An Integrated Approach for Analysis of the DNA Damage Response in Mammalian Cells: NUCLEOTIDE EXCISION REPAIR, DNA DAMAGE CHECKPOINT, AND APOPTOSIS. Nov;2015 J Biol Chem. 290:28812–28821. doi: 10.1074/jbc.M115.690354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibeling Diana, Laspe Petra, Emmert Steffen. Nucleotide excision repair and cancer. Sep;2006 J Mol Histol. 37:225–238. doi: 10.1007/s10735-006-9041-x. [DOI] [PubMed] [Google Scholar]
- uuuu [ Dec 18; 2019 ];Aziz Sancar - Nobel Lecture. https://www.nobelprize.org/prizes/chemistry/2015/sancar/lecture/
- Canturk Fazile, Karaman Muhammet, Selby Christopher P., Kemp Michael G., Kulaksiz-Erkmen Gulnihal, Hu Jinchuan, Li Wentao, Lindsey-Boltz Laura A., Sancar Aziz. Nucleotide excision repair by dual incisions in plants. Apr;2016 Proc Natl Acad Sci U S A. 113:4706–4710. doi: 10.1073/pnas.1604097113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Jinchuan, Selby Christopher P., Adar Sheera, Adebali Ogun, Sancar Aziz. Molecular mechanisms and genomic maps of DNA excision repair in Escherichia coli and humans. Sep;2017 J Biol Chem. 292:15588–15597. doi: 10.1074/jbc.R117.807453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp Michael G., Sancar Aziz. DNA excision repair: where do all the dimers go? Aug;2012 Cell Cycle. 11:2997–3002. doi: 10.4161/cc.21126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Jinchuan, Li Wentao, Adebali Ogun, Yang Yanyan, Oztas Onur, Selby Christopher P., Sancar Aziz. Genome-wide mapping of nucleotide excision repair with XR-seq. Jan;2019 Nat Protoc. 14:248–282. doi: 10.1038/s41596-018-0093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adar Sheera, Hu Jinchuan, Lieb Jason D., Sancar Aziz. Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Apr;2016 Proc Natl Acad Sci U S A. 113:E2124–2133. doi: 10.1073/pnas.1603388113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddameedhi Shobhan, Kemp Michael G., Reardon Joyce T., Shields Janiel M., Smith-Roe Stephanie L., Kaufmann William K., Sancar Aziz. Similar nucleotide excision repair capacity in melanocytes and melanoma cells. Jun;2010 Cancer Res. 70:4922–4930. doi: 10.1158/0008-5472.CAN-10-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey-Boltz Laura A., Kemp Michael G., Reardon Joyce T., DeRocco Vanessa, Iyer Ravi R., Modrich Paul, Sancar Aziz. Coupling of human DNA excision repair and the DNA damage checkpoint in a defined in vitro system. Feb;2014 J Biol Chem. 289:5074–5082. doi: 10.1074/jbc.M113.542787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latimer Jean J., Johnson Jennifer M., Kelly Crystal M., Miles Tiffany D., Beaudry-Rodgers Kelly A., Lalanne Nancy A., Vogel Victor G., Kanbour-Shakir Amal, Kelley Joseph L., Johnson Ronald R., Grant Stephen G. Nucleotide excision repair deficiency is intrinsic in sporadic stage I breast cancer. Dec;2010 Proc Natl Acad Sci U S A. 107:21725–21730. doi: 10.1073/pnas.0914772107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort R., Mo L., McEwan C., Melton D. W. Lack of involvement of nucleotide excision repair gene polymorphisms in colorectal cancer. Jul;2003 Br J Cancer. 89:333–337. doi: 10.1038/sj.bjc.6601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Baocai, Li Jing, Gao Qingzu, Yu Wenfa, Yang Qinghui, Li Xiaoyu. Laryngeal cancer risk and common single nucleotide polymorphisms in nucleotide excision repair pathway genes ERCC1, ERCC2, ERCC3, ERCC4, ERCC5 and XPA. May;2014 Gene. 542:64–68. doi: 10.1016/j.gene.2014.02.043. [DOI] [PubMed] [Google Scholar]
- Sliwinski T., Markiewicz L., Rusin P., Kabzinski J., Dziki L., Milonski J., Olszewski J., Blaszczyk J., Szemraj J., Majsterek I. Impaired nucleotide excision repair pathway as a possible factor in pathogenesis of head and neck cancer. Nov;2011 Mutat Res. 716:51–58. doi: 10.1016/j.mrfmmm.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Cipollini Monica, Figlioli Gisella, Maccari Giuseppe, Garritano Sonia, De Santi Chiara, Melaiu Ombretta, Barone Elisa, Bambi Franco, Ermini Stefano, Pellegrini Giovanni, Cristaudo Alfonso, Foddis Rudy, Bonotti Alessandra, Romei Cristina, Vivaldi Agnese, Agate Laura, Molinari Eleonora, Barale Roberto, Forsti Asta, Hemminki Kari, Elisei Rossella, Gemignani Federica, Landi Stefano. Polymorphisms within base and nucleotide excision repair pathways and risk of differentiated thyroid carcinoma. May;2016 DNA Repair (Amst) 41:27–31. doi: 10.1016/j.dnarep.2016.03.011. [DOI] [PubMed] [Google Scholar]
- Liakos Anastasios, Lavigne Matthieu D., Fousteri Maria. Nucleotide Excision Repair: From Neurodegeneration to Cancer. 2017Adv Exp Med Biol. 1007:17–39. doi: 10.1007/978-3-319-60733-7_2. [DOI] [PubMed] [Google Scholar]
- Kulaksiz G, Sancar A. Nucleotide excision repair and cancer. 2007Turkish Journal of Biochemistry. 32:104–111. [Google Scholar]
- Bohr V. A., Smith C. A., Okumoto D. S., Hanawalt P. C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Feb;1985 Cell. 40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Oct;1987 Cell. 51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Mellon I., Hanawalt P. C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nov;1989 Nature. 342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. Dec;1990 J Biol Chem. 265:21330–21336. [PubMed] [Google Scholar]
- Selby C. P., Witkin E. M., Sancar A. Escherichia coli mfd mutant deficient in "mutation frequency decline" lacks strand-specific repair: in vitro complementation with purified coupling factor. Dec;1991 Proc Natl Acad Sci U S A. 88:11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Sep;1994 Microbiol Rev. 58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby C. P., Drapkin R., Reinberg D., Sancar A. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Feb;1997 Nucleic Acids Res. 25:787–793. doi: 10.1093/nar/25.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A. Mechanisms of DNA excision repair. Dec;1994 Science. 266:1954–1956. doi: 10.1126/science.7801120. [DOI] [PubMed] [Google Scholar]
- Huang J. C., Svoboda D. L., Reardon J. T., Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5' and the 6th phosphodiester bond 3' to the photodimer. Apr;1992 Proc Natl Acad Sci U S A. 89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda D. L., Taylor J. S., Hearst J. E., Sancar A. DNA repair by eukaryotic nucleotide excision nuclease. Removal of thymine dimer and psoralen monoadduct by HeLa cell-free extract and of thymine dimer by Xenopus laevis oocytes. Jan;1993 J Biol Chem. 268:1931–1936. [PubMed] [Google Scholar]
- Wood R. D. Nucleotide excision repair in mammalian cells. Sep;1997 J Biol Chem. 272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- Kemp Michael G., Reardon Joyce T., Lindsey-Boltz Laura A., Sancar Aziz. Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. Jun;2012 J Biol Chem. 287:22889–22899. doi: 10.1074/jbc.M112.374447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey-Boltz Laura A., Sancar Aziz. RNA polymerase: the most specific damage recognition protein in cellular responses to DNA damage? Aug;2007 Proc Natl Acad Sci U S A. 104:13213–13214. doi: 10.1073/pnas.0706316104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver James E., Lam Ernest T., Revet Ingrid. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nov;2009 Nat Rev Genet. 10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- Marteijn Jurgen A., Lans Hannes, Vermeulen Wim, Hoeijmakers Jan H. J. Understanding nucleotide excision repair and its roles in cancer and ageing. Jul;2014 Nat Rev Mol Cell Biol. 15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- Cleaver James E. Transcription coupled repair deficiency protects against human mutagenesis and carcinogenesis: Personal Reflections on the 50th anniversary of the discovery of xeroderma pigmentosum. Oct;2017 DNA Repair (Amst) 58:21–28. doi: 10.1016/j.dnarep.2017.08.004. [DOI] [PubMed] [Google Scholar]
- Furuta Takahisa, Ueda Takahiro, Aune Gregory, Sarasin Alain, Kraemer Kenneth H., Pommier Yves. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Sep;2002 Cancer Res. 62:4899–4902. [PubMed] [Google Scholar]
- Stubbert Lawton J., Smith Jennifer M., McKay Bruce C. Decreased transcription-coupled nucleotide excision repair capacity is associated with increased p53- and MLH1-independent apoptosis in response to cisplatin. May;2010 BMC Cancer. 10:207–207. doi: 10.1186/1471-2407-10-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann G., Vasyutina E., Bloehdorn J., Reinart N., Schneider J. I., Babu V., Knittel G., Crispatzu G., Mayer P., Prinz C., Muenzner J. K., Biersack B., Efremov D. G., Chessa L., Herling C. D., Stilgenbauer S., Hallek M., Schobert R., Reinhardt H. C., Schumacher B., Herling M. Targeting transcription-coupled nucleotide excision repair overcomes resistance in chronic lymphocytic leukemia. May;2017 Leukemia. 31:1177–1186. doi: 10.1038/leu.2016.294. [DOI] [PubMed] [Google Scholar]
- Takagi Kazutaka, Kawai Yasukazu, Yamauchi Takahiro, Iwasaki Hiromichi, Ueda Takanori. Synergistic effects of combination with fludarabine and carboplatin depend on fludarabine-mediated inhibition of enhanced nucleotide excision repair in leukemia. Oct;2011 Int J Hematol. 94:378–389. doi: 10.1007/s12185-011-0930-8. [DOI] [PubMed] [Google Scholar]
- Pandey Preeti, Hasnain Sabeeha, Ahmad Shandar. Protein-DNA interactions. In: Ranganathan Shoba, Gribskov Michael, Nakai Kenta, Schönbach Christian., editors. Encyclopedia of Bioinformatics and Computational Biology. Academic Press; Oxford: pp. 142–154. [Google Scholar]
- Orren D. K., Selby C. P., Hearst J. E., Sancar A. Post-incision steps of nucleotide excision repair in Escherichia coli. Disassembly of the UvrBC-DNA complex by helicase II and DNA polymerase I. Jan;1992 J Biol Chem. 267:780–788. [PubMed] [Google Scholar]
- Gonzalez Mileidy W., Kann Maricel G. Chapter 4: Protein interactions and disease. 2012PLoS Comput Biol. 8:e1002819–e1002819. doi: 10.1371/journal.pcbi.1002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan Daniel P., Matthews Jacqueline M. Protein-protein interactions in human disease. Aug;2005 Curr Opin Struct Biol. 15:441–446. doi: 10.1016/j.sbi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Soto Claudio, Estrada Lisbell D. Protein misfolding and neurodegeneration. Feb;2008 Arch Neurol. 65:184–189. doi: 10.1001/archneurol.2007.56. [DOI] [PubMed] [Google Scholar]
- Van Drie John H. Protein folding, protein homeostasis, and cancer. Feb;2011 Chin J Cancer. 30:124–137. doi: 10.5732/cjc.010.10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden Emma, Logue Susan E., Healy Sandra J., Manie Serge, Samali Afshin. The role of the unfolded protein response in cancer progression: From oncogenesis to chemoresistance. Jan;2019 Biol Cell. 111:1–17. doi: 10.1111/boc.201800050. [DOI] [PubMed] [Google Scholar]
- Scheffner Martin, Whitaker Noel J. Human papillomavirus-induced carcinogenesis and the ubiquitin-proteasome system. Feb;2003 Semin Cancer Biol. 13:59–67. doi: 10.1016/s1044-579x(02)00100-1. [DOI] [PubMed] [Google Scholar]
- Tarocchi Mirko, Polvani Simone, Marroncini Giada, Galli Andrea. Molecular mechanism of hepatitis B virus-induced hepatocarcinogenesis. Sep;2014 World J Gastroenterol. 20:11630–11640. doi: 10.3748/wjg.v20.i33.11630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith Emily K., Balas Maggie M., Sindy Karla, Haislop Krystal, Johnson Aaron M. An RNA matchmaker protein regulates the activity of the long noncoding RNA HOTAIR. Jul;2016 RNA. 22:995–1010. doi: 10.1261/rna.055830.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Yu, Li Huan, Liu Fan, Gao Li-Bin, Han Rong, Chen Chen, Ding Xue, Li Shuang, Lu Kun, Yang Ling, Tian Hui-Min, Chen Bin-Bin, Li Xiao, Xu Dong-Hui, Deng Xiao-Ling, Shi Song-Lin. Heterogeneous nuclear ribonucleoprotein A2/B1 is a negative regulator of human breast cancer metastasis by maintaining the balance of multiple genes and pathways. Jan;2020 EBioMedicine. 51:102583–102583. doi: 10.1016/j.ebiom.2019.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Yanyan, Liu Zhenxing, Selby Christopher P., Sancar Aziz. Long-term, genome-wide kinetic analysis of the effect of the circadian clock and transcription on the repair of cisplatin-DNA adducts in the mouse liver. Aug;2019 J Biol Chem. 294:11960–11968. doi: 10.1074/jbc.RA119.009579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimit Askar, Adebali Ogun, Sancar Aziz, Jiang Yuchao. Differential damage and repair of DNA-adducts induced by anti-cancer drug cisplatin across mouse organs. Jan;2019 Nat Commun. 10:309–309. doi: 10.1038/s41467-019-08290-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn Courtney M., Selby Christopher P., Yang Yanyan, Hsu David S., Sancar Aziz. Genome-wide single-nucleotide resolution of oxaliplatin-DNA adduct repair in drug-sensitive and -resistant colorectal cancer cell lines. May;2020 J Biol Chem. 295:7584–7594. doi: 10.1074/jbc.RA120.013347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar Aziz, Lindsey-Boltz Laura A., Gaddameedhi Shobhan, Selby Christopher P., Ye Rui, Chiou Yi-Ying, Kemp Michael G., Hu Jinchuan, Lee Jin Hyup, Ozturk Nuri. Circadian clock, cancer, and chemotherapy. Jan;2015 Biochemistry. 54:110–123. doi: 10.1021/bi5007354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y., Sancar A. Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. May;1998 Proc Natl Acad Sci U S A. 95:6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar Aziz. Regulation of the mammalian circadian clock by cryptochrome. Aug;2004 J Biol Chem. 279:34079–34082. doi: 10.1074/jbc.R400016200. [DOI] [PubMed] [Google Scholar]
- Ye Rui, Selby Cristopher P., Chiou Yi-Ying, Ozkan-Dagliyan Irem, Gaddameedhi Shobhan, Sancar Aziz. Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock. Sep;2014 Genes Dev. 28:1989–1998. doi: 10.1101/gad.249417.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda Satchidananda, Antoch Marina P., Miller Brooke H., Su Andrew I., Schook Andrew B., Straume Marty, Schultz Peter G., Kay Steve A., Takahashi Joseph S., Hogenesch John B. Coordinated transcription of key pathways in the mouse by the circadian clock. May;2002 Cell. 109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Davis Kate, Roden Laura C., Leaner Virna D., Watt Pauline J. The tumour suppressing role of the circadian clock. Jul;2019 IUBMB Life. 71:771–780. doi: 10.1002/iub.2005. [DOI] [PubMed] [Google Scholar]
- Gauger Michele A., Sancar Aziz. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Aug;2005 Cancer Res. 65:6828–6834. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
- Mteyrek Ali, Filipski Elisabeth, Guettier Catherine, Oklejewicz Malgorzata, Horst Gijsbertus T. J., Okyar Alper, Lévi Francis. Critical cholangiocarcinogenesis control by cryptochrome clock genes. Jun;2017 Int J Cancer. 140:2473–2483. doi: 10.1002/ijc.30663. [DOI] [PubMed] [Google Scholar]
- Cadenas Cristina, Sandt Leonie, Edlund Karolina, Lohr Miriam, Hellwig Birte, Marchan Rosemarie, Schmidt Marcus, Rahnenführer Jörg, Oster Henrik, Hengstler Jan G. Loss of circadian clock gene expression is associated with tumor progression in breast cancer. 2014Cell Cycle. 13:3282–3291. doi: 10.4161/15384101.2014.954454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt Pauline J., Maske Christopher P., Hendricks Denver T., Parker M. Iqbal, Denny Lynette, Govender Dhirendra, Birrer Michael J., Leaner Virna D. The Karyopherin proteins, Crm1 and Karyopherin beta1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. Apr;2009 Int J Cancer. 124:1829–1840. doi: 10.1002/ijc.24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Chia-Ming, Shay Jacqueline, Zeng Ting-Chuan, Chou Jian-Liang, Huang Tim H.-M., Lai Hung-Cheng, Chan Michael W. Y. Epigenetic silencing of ARNTL, a circadian gene and potential tumor suppressor in ovarian cancer. Nov;2014 Int J Oncol. 45:2101–2107. doi: 10.3892/ijo.2014.2627. [DOI] [PubMed] [Google Scholar]
- Yu Hongyan, Meng Xiangqi, Wu Jiangxue, Pan Changchuan, Ying Xiaofang, Zhou Yi, Liu Ranyi, Huang Wenlin. Cryptochrome 1 overexpression correlates with tumor progression and poor prognosis in patients with colorectal cancer. 2013PLoS One. 8:e61679–e61679. doi: 10.1371/journal.pone.0061679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoccoli Gianluigi, Colangelo Tommaso, Panza Anna, Rubino Rosa, De Cata Angelo, Tiberio Cristiana, Valvano Maria Rosa, Pazienza Valerio, Merla Giuseppe, Augello Bartolomeo, Trombetta Domenico, Storlazzi Clelia Tiziana, Macchia Gemma, Gentile Annamaria, Tavano Francesca, Vinciguerra Manlio, Bisceglia Giovanni, Rosato Valeria, Colantuoni Vittorio, Sabatino Lina, Piepoli Ada. Deregulated expression of cryptochrome genes in human colorectal cancer. Jan;2016 Mol Cancer. 15:6–6. doi: 10.1186/s12943-016-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habashy Deena Mohamed, Eissa Deena Samir, Aboelez Mona Mahmoud. Cryptochrome-1 Gene Expression is a Reliable Prognostic Indicator in Egyptian Patients with Chronic Lymphocytic Leukemia: A Prospective Cohort Study. Aug;2018 Turk J Haematol. 35:168–174. doi: 10.4274/tjh.2017.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman Aaron E., Zheng Tongzhang, Yi Chun-Hui, Stevens Richard G., Ba Yue, Zhang Yawei, Leaderer Derek, Holford Theodore, Hansen Johnni, Zhu Yong. The core circadian gene Cryptochrome 2 influences breast cancer risk, possibly by mediating hormone signaling. Apr;2010 Cancer Prev Res (Phila) 3:539–548. doi: 10.1158/1940-6207.CAPR-09-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhopuro Pia, Björklund Mikael, Sammalkorpi Heli, Turunen Mikko, Tuupanen Sari, Biström Mia, Niittymäki Iina, Lehtonen Heli J., Kivioja Teemu, Launonen Virpi, Saharinen Juha, Nousiainen Kari, Hautaniemi Sampsa, Nuorva Kyösti, Mecklin Jukka-Pekka, Järvinen Heikki, Orntoft Torben, Arango Diego, Lehtonen Rainer, Karhu Auli, Taipale Jussi, Aaltonen Lauri A. Mutations in the circadian gene CLOCK in colorectal cancer. Jul;2010 Mol Cancer Res. 8:952–960. doi: 10.1158/1541-7786.MCR-10-0086. [DOI] [PubMed] [Google Scholar]
- Ozturk Nuri, Lee Jin Hyup, Gaddameedhi Shobhan, Sancar Aziz. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Feb;2009 Proc Natl Acad Sci U S A. 106:2841–2846. doi: 10.1073/pnas.0813028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiannakopoulos Thales, Bauer Matthew R., Davidson Shawn M., Heimann Megan, Subbaraj Lakshmipriya, Bhutkar Arjun, Bartlebaugh Jordan, Vander Heiden Matthew G., Jacks Tyler. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Aug;2016 Cell Metab. 24:324–331. doi: 10.1016/j.cmet.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Susie, Donehower Lawrence A., Herron Alan J., Moore David D., Fu Loning. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. Jun;2010 PLoS One. 5:e10995–e10995. doi: 10.1371/journal.pone.0010995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn Lani R., Tamimi Rulla M., Rosner Bernard A., Brown Susan B., Stevens Richard G., Eliassen A. Heather, Laden Francine, Willett Walter C., Hankinson Susan E., Schernhammer Eva S. Rotating Night-Shift Work and the Risk of Breast Cancer in the Nurses' Health Studies. Sep;2017 Am J Epidemiol. 186:532–540. doi: 10.1093/aje/kwx140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens Thomas, Rabstein Sylvia, Wichert Katharina, Erbel Raimund, Eisele Lewin, Arendt Marina, Dragano Nico, Brüning Thomas, Jöckel Karl-Heinz. Shift work and the incidence of prostate cancer: a 10-year follow-up of a German population-based cohort study. Nov;2017 Scand J Work Environ Health. 43:560–568. doi: 10.5271/sjweh.3666. [DOI] [PubMed] [Google Scholar]
- Papantoniou Kyriaki, Devore Elizabeth E., Massa Jennifer, Strohmaier Susanne, Vetter Céline, Yang Lin, Shi Yan, Giovannucci Edward, Speizer Frank, Schernhammer Eva S. Rotating night shift work and colorectal cancer risk in the nurses' health studies. Dec;2018 Int J Cancer. 143:2709–2717. doi: 10.1002/ijc.31655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC Monographs Vol 124 group Carcinogenicity of night shift work. Aug;2019 Lancet Oncol. 20:1058–1059. doi: 10.1016/S1470-2045(19)30455-3. [DOI] [PubMed] [Google Scholar]
- Chun Sung Kook, Chung Sooyoung, Kim Hee-Dae, Lee Ju Hyung, Jang Jaebong, Kim Jeongah, Kim Doyeon, Son Gi Hoon, Oh Young J., Suh Young-Ger, Lee Cheol Soon, Kim Kyungjin. A synthetic cryptochrome inhibitor induces anti-proliferative effects and increases chemosensitivity in human breast cancer cells. Nov;2015 Biochem Biophys Res Commun. 467:441–446. doi: 10.1016/j.bbrc.2015.09.103. [DOI] [PubMed] [Google Scholar]
- Zhou Lei, Yu Yueming, Sun Shiwei, Zhang Tieqi, Wang Minghai. Cry 1 Regulates the Clock Gene Network and Promotes Proliferation and Migration Via the Akt/P53/P21 Pathway in Human Osteosarcoma Cells. 2018J Cancer. 9:2480–2491. doi: 10.7150/jca.25213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiramoto Akira, Yamane Takashi, Tsumiyama Ken, Yoshida Kohsuke, Komai Koichiro, Yamada Hiroyuki, Yamazaki Fumiyoshi, Doi Masao, Okamura Hitoshi, Shiozawa Shunichi. Mammalian clock gene Cryptochrome regulates arthritis via proinflammatory cytokine TNF-alpha. Feb;2010 J Immunol. 184:1560–1565. doi: 10.4049/jimmunol.0903284. [DOI] [PubMed] [Google Scholar]
- Gaddameedhi Shobhan, Selby Christopher P., Kaufmann William K., Smart Robert C., Sancar Aziz. Control of skin cancer by the circadian rhythm. Nov;2011 Proc Natl Acad Sci U S A. 108:18790–18795. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Yanyan, Adebali Ogun, Wu Gang, Selby Christopher P., Chiou Yi-Ying, Rashid Naim, Hu Jinchuan, Hogenesch John B., Sancar Aziz. Cisplatin-DNA adduct repair of transcribed genes is controlled by two circadian programs in mouse tissues. May;2018 Proc Natl Acad Sci U S A. 115:E4777–E4785. doi: 10.1073/pnas.1804493115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K. Q., Jin F., Jiang H., Wu W. L., Li Y. Y., Long J. H., Luo X. L., Gong X. Y., Chen X. X., Liu L. N., Gan J. Y., Zhou J. J. Analysis of follow-up results of chrono-chemotherapy or conventional chemotherapy combined with intensity modulated radiotherapy in locally advanced nasopharyngeal carcinoma. Feb;2020 Zhonghua Zhong Liu Za Zhi. 42:133–138. doi: 10.3760/cma.j.issn.0253-3766.2020.02.009. [DOI] [PubMed] [Google Scholar]
- Gou Xiao-Xia, Jin Feng, Wu Wei-Li, Long Jin-Hua, Li Yuan-Yuan, Gong Xiu-Yun, Chen Guo-Yan, Chen Xiao-Xiao, Liu Li-Na. Induction chronomodulated chemotherapy plus radiotherapy for nasopharyngeal carcinoma: A Phase II prospective randomized study. 2018J Cancer Res Ther. 14:1613–1619. doi: 10.4103/jcrt.JCRT_883_17. [DOI] [PubMed] [Google Scholar]
- Deshmukh Jayesh, Pofahl Ruth, Haase Ingo. Epidermal Rac1 regulates the DNA damage response and protects from UV-light-induced keratinocyte apoptosis and skin carcinogenesis. Mar;2017 Cell Death Dis. 8:e2664–e2664. doi: 10.1038/cddis.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C., Bond J. P., Burns D. K., Cheo D. L., Greenblatt M. S., Meira L. B., Nahari D., Reis A. M. Defective nucleotide excision repair in xpc mutant mice and its association with cancer predisposition. Mar;2000 Mutat Res. 459:99–108. doi: 10.1016/s0921-8777(99)00068-3. [DOI] [PubMed] [Google Scholar]
- Blasiak Janusz. DNA-Damaging Anticancer Drugs - A Perspective for DNA Repair- Oriented Therapy. 2017Curr Med Chem. 24:1488–1503. doi: 10.2174/0929867324666170124145557. [DOI] [PubMed] [Google Scholar]
- Poletto Mattia, Legrand Arnaud J., Dianov Grigory L. DNA Base Excision Repair: The Achilles' Heel of Tumour Cells and their Microenvironment? 2017Curr Pharm Des. 23:4758–4772. doi: 10.2174/1381612823666170710123602. [DOI] [PubMed] [Google Scholar]
- Pasteur Louis. uuuu [ Dec 19; 2019 ];Science knows no country, because knowledge belongs to humanity, and is the torch which illuminates the world. https://media.nationalgeographic.org/assets/file/breakthrough_quote_4.pdf
- AbdullGaffar Badr. Nobel Prize and Surgical Pathology. Mar;2017 Adv Anat Pathol. 24:110–111. doi: 10.1097/PAP.0000000000000141. [DOI] [PubMed] [Google Scholar]
- uuuu [ Dec 19; 2019 ]; https://4kyws.ua.edu/GAPOSCHKIN.html
- Sancar Aziz, Reardon Joyce T. Nucleotide excision repair in E. coli and man. 2004Adv Protein Chem. 69:43–71. doi: 10.1016/S0065-3233(04)69002-4. [DOI] [PubMed] [Google Scholar]