Abstract
Background and Aims
A key histopathological feature of inflammatory bowel disease is damage to the mucosa, including breakdown of the epithelial barrier. Human enteroids and colonoids are a critical bench-to-bedside tool for studying the epithelium in inflammatory bowel disease. The goal of the current study was to define transcriptional differences in healthy versus diseased subjects that are sustained in enteroids and colonoids, including from disease-spared tissue.
Methods
Biopsies and matching enteroid or colonoid cultures from pediatric patients with ileal Crohn disease (N = 6) and control subjects (N = 17) were subjected to RNA sequencing followed by bioinformatic and machine learning analyses. Late passage enteroids were exposed to cytokines to assess durable transcriptional differences.
Results
We observed substantial overlap of pathways upregulated in Crohn disease in enteroids and ileal biopsies, as well as colonoids and rectal biopsies. KEGG pathways for cytokine-cytokine receptor interaction, chemokine signaling, protein export, and Toll-like receptor signaling were upregulated in both ileal and rectal biopsies, as well as enteroids and colonoids. In vitro cytokine exposure reactivated genes previously increased in biopsies. Machine learning predicted biopsy location (100% accuracy) and donor disease status (83% accuracy). A random forest classifier generated using ileal enteroids identified rectal colonoids from ileal Crohn disease subjects with 80% accuracy.
Conclusion
We confirmed transcriptional profiles of Crohn disease biopsies are expressed in enteroids and colonoids. Furthermore, transcriptomic data from disease-spared rectal tissue can identify patients with ileal Crohn disease. Our data support the use of patient enteroids and colonoids as critical translational tools for the study of inflammatory bowel disease.
Keywords: Enteroid, Colonoid, 3D Model, Crohn’s Disease, Inflammatory Bowel Disease
Graphical abstract
Introduction
Inflammatory bowel disease (IBD) comprises 2 main histopathological features: chronic inflammation within the gastrointestinal (GI) tract and damage to the mucosa (epithelial lining of the intestine), yet currently available treatment approaches focus primarily on quenching inflammation.1,2 Quenching inflammation alleviates symptomatology of IBD, but mucosal healing is the most prominent predictor of sustained remission.3 Since initial publications in the late 2000s,4,5 intestinal or colonic murine or patient-derived enteroids and colonoids have been used to study the GI tract mucosa at homeostasis and in various diseases.6 However, different culture modalities7,8 and media configurations5,9,10 make it difficult to gauge the extent to which enteroids and colonoids recapitulate the physiology of intestinal mucosa of the individual donor.
Previous studies demonstrated that enteroids and colonoids do retain some physiologic characteristics of the diseased tissue. These studies include demonstration that the genomic and transcriptomic profiles of enteroids and colonoids are permanently altered when they are derived from actively inflamed sites.11, 12, 13 Others have revealed that enteroids and colonoids from patients with IBD share disease characteristics such as upregulated MHC (major histocompatibility complex) class II expression14 and reduced junctional proteins ZO-1, occludin, and claudin-1 in vitro.15 While these studies compared enteroids and colonoids from patients with IBD to those from control subjects, there still is a limited understanding of how enteroids and colonoids compare to patient tissue in vivo, particularly those obtained from noninflamed regions. Two studies aiming to address this gap in knowledge have been published recently.16,17 Niklinska-Schirtz et al.17 reported that enteroids from patients with Crohn’s disease (CD) share transcriptomic signatures with ileal epithelium (isolated crypts). Arnauts et al.16 demonstrated that the inflammatory signature identified in whole biopsies from patients with ulcerative colitis or control subjects is present in intestinal epithelium (isolated crypts), yet it is lost within a week of colonoid culture. Importantly, a 24-hour treatment with a cocktail of inflammatory mediators restored the signature in colonoids from either inflamed or noninflamed regions.16 This finding demonstrated that it is not necessary to biopsy directly from an inflamed region to establish colonoids to model ulcerative colitis epithelium in vitro.
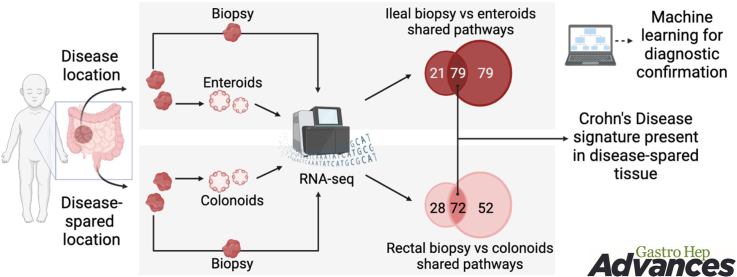
In the current study our goal was to use an unbiased approach to: (1) identify biologically relevant transcriptomic features of intestinal biopsies that are retained in enteroids/colonoids and (2) evaluate the features of tissue from disease-spared GI segments (rectum of patients with CD localized to ileum only). We analyzed whole biopsies (as opposed to isolated crypts), to minimize artifacts that can be introduced by tissue processing. For pathway analysis, we performed gene set enrichment analysis (GSEA) on the entire Kyoto Encyclopedia of Genes and Genomes (KEGG) database and found a substantial overlap in pathways enriched toward IBD in whole biopsies and enteroids/colonoids. We confirmed that cytokine treatment can elicit enhanced pro-inflammatory gene expression in colonoids from patients with CD compared to cytokine-treated control colonoids. Finally, we used random forest classification to validate retention of discriminating gene expression features relevant to intestinal segment (ileum or rectum), as well as the disease status of the donor patient. Taken together, our findings establish enteroids and colonoids as important tools in GI research and reveal the presence of disease-specific transcriptomic differences in disease-spared tissue (rectum) and colonoids derived from it.
Materials and methods
Subject enrollment and demographics
The biopsies were collected from pediatric patients aged 0–17 years (Tables 1 and A1) undergoing endoscopy at the Children’s Hospital of Philadelphia (CHOP) between December 2016 and November 2020. Prior to sample collection, informed consent was obtained in accordance with institutional review board guidelines (protocol 16-013042). Research biopsies were collected by a trained gastroenterologist from the ileum and rectum in addition to those for clinical care. Patients with a history of bone marrow transplantation or prior diagnosis of any hemorrhagic, coagulopathic, or aggregation disorders were excluded from this study. Expert pathologists confirmed IBD diagnoses. Control subjects were evaluated for GI complaints but found to have normal endoscopic and histologic findings. Thirteen IBD patients recruited were new diagnoses and received no prior IBD medications, 3 patients had existing IBD, and data regarding treatment and disease duration are presented in Table A1.
Table 1.
Subject Demographics
| Crohn's | Control | |
|---|---|---|
| Subjects, n | 6 | 17 |
| Male, n (%) | 1 (17%) | 8 (47%) |
| Age at sample collection, mean (±SD), y | 15.2 (1.3) | 12.6 (3.8) |
| Age range | 14–17 | 6–17 |
| Pre-existing IBD diagnosis, n | 0 | N/A |
| Treatment at the time of biopsy, n | 0 | N/A |
N/A, not applicable; SD, standard deviation.
Generation of enteroids and colonoids from patient biopsies
A detailed list of reagents and solutions can be found in Table A2. Mucosal biopsies (ileal and rectal) were obtained endoscopically from noninflamed regions adjacent to areas of active inflammation (for CD) or from areas least affected by inflammation (for ulcerative colitis) during patient colonoscopies performed for diagnostic evaluation or disease surveillance (Table A1). Two biopsies were obtained from disease-spared segments of the terminal ileum and rectum. Biopsies were transported on ice in Advanced DMEM/F12 supplemented with 1x GlutaMAX, 10 mM HEPES, 50 μg/mL Gentamicin, and 0.5 μg/mL amphotericin B. One fragment was snap-frozen in TRIzol reagent for RNA extraction and the second was either used for crypt isolation and culture within 2 hours of tissue collection or cryopreserved for subsequent crypt isolation at a later time.
Enteroids/colonoids were generated as described previously.14,18 Briefly, the biopsies were washed twice by vortexing in 10 mL of ice-cold 1% BSA (bovine serum albumin) solution in cold DPBS (Dulbecco's phosphate-buffered saline), then washed twice in ice-cold chelating buffer (2% sorbitol, 1% sucrose, 1% BSA, 50 μg/mL gentamicin, and 0.5 μg/mL amphotericin B, in DPBS) in a well of a 6-well plate, then transferred to a 1.5 mL microcentrifuge tube containing 1 mL ice-cold chelating buffer supplemented with 10 mM EDTA (ethylenediaminetetraacetic acid), and placed to rock at 4 °C for 30 minutes. The crypts were mechanically separated from the lamina propria (via scraping), filtered through a 100 μm cell strainer, washed, and resuspended in ice-cold 1% BSA solution in DPBS for counting. Approximately 100 crypts were seeded per 30 μL dome of 80% matrigel (diluted with complete organoid growth medium), with a minimum of 2 wells per tissue sample. The enteroids and colonoids were grown in complete human IntestiCult organoid growth medium (STEMCELL Technologies), supplemented with 50 μg/mL gentamicin (10 μM Y27632 were added at the time of seeding), the media was replaced every Monday, Wednesday, and Friday. Mechanical passaging was performed as described previously.14 These enteroids and colonoids were grown for 3 days in IntestiCult media prior to collection for either: (1) cryopreservation for storage or (2) preservation in TRIzol reagent for RNA isolation.
RNA sequencing
RNA sequencing was performed on enteroids and colonoids as well as whole tissue biopsies. Enteroids and colonoids were harvested in TRIzol on day 3 of the first passage and stored at −80 °C. The whole biopsies were submerged in RNA-later, snap-frozen in liquid nitrogen, and then transferred on dry ice for storage in −80 °C. RNA-sequencing (RNA-seq) libraries were prepared at the CHOP Center for Applied Genomics. Total RNA with RIN (RNA Integrity Number) values above 6 was normalized to 400 ng, converted to cDNA, and then used to prepare each library using the Illumina TruSeq Stranded Messenger RNA Library Kit and the IDT for Illumina Unique Dual Index Kit in an automated Sciclone Liquid Handler Instrument. Final libraries were analyzed on a Perkin Elmer Labchip GX for quantification and quality control. Samples were pooled and sequenced on the Illumina NovaSeq 6000 platform (151-bp read length) at the CHOP Center for Spatial and Functional Genomics. RNA-seq data are available in the Gene Expression Omnibus (study accession number GSE228122).
Quantitative reverse transcription polymerase chain reaction
Total RNA was isolated from enteroids and colonoids using the Zymo Direct-zol RNA Microprep (Zymo Research #R2062). The cDNA was synthesized from equivalent levels of extracted RNA using TaqMan Reverse Transcription Reagents (Applied Biosystems #N8080234). Real-time polymerase chain reaction (PCR) assays were performed using the TaqMan Fast Universal PCR Master Mix (Applied Biosystems #4352042) per the manufacturer’s protocol with the QuantStudio 3 Real-Time PCR System. Gene expression was calculated using the R = 2-ddCt method, with TBP (TATA box binding protein gene) as housekeeping gene. The following TaqMan assays were used: TBP (Hs00427621_m1), CXCL3 (Hs00171061_m1), CXCL11 (Hs00171138_m1), HLA-DRA (Hs00219575_m), PF4 (Hs00427220_m1).
Bioinformatics
RNA-sequencing data preprocessing and analysis
Read counts were obtained from the transcriptome pseudoalignment program kallisto (version 0.46.2)19 with default parameters, and input into R (v4.1.3). The reference genome is GRCh38. Data were normalized, and differentially expressed genes (DEGs) were identified using the DESeq2 package (version 1.34.0).20 Approximately 20,000 genes with a total count ≥10 across all samples were considered to be expressed and reserved for downstream analysis. DEGs were defined as absolute value of log ratio between case and control expression level greater than 1.5, and P value less than .05. The full list of DEGs, P values, and false discovery rate (FDR)-adjusted P values can be found in Table A3. Top 200 DEGs (top 100 upregulated and top 100 downregulated DEGs) were analyzed using hierarchical clustering pheatmap function in R to compare the transcriptomes between organoid and biopsy samples. All the DEGs were also presented using volcano plot with a P value cutoff of .05 and log ratio cutoff of log2 (1.5). The ‘stat’ output field of DEGs from DESeq2 was then used as input for gene set enrichment analysis (GSEA)21,22 preranked analysis to identify enriched pathways from the KEGG database.
“Golden rule” gene set validation analysis
To validate the DEGs identified from our organoid samples, the same pipeline was performed on the ‘reference’ dataset downloaded from the NIH (National Institutes of Health) National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) database (accession number PRJNA643576).17 A total of 1004 upregulated DEGs identified in CD patients from the ‘reference’ dataset were used as the ‘golden rule’ gene set. The GSEA was then performed to compare the upregulated DEG set from our CD patients with the ‘golden rule’ gene set. Normalized enrichment score and P value were measured to show if our upregulated DEGs from CD patients match with the ‘golden rule’ gene set.
Random forest classifier
A random forest-based classifier was built using the randomForest R package, which is based on the algorithm of Breiman and Cutler.23 Random forest is a learning method that constructs numerous decision trees. The predicted class of the input instance will be decided upon majority vote. To test whether organoid recapitulate the biology of original tissue, a total of 27 control ileal enteroid and rectal colonoid samples were used as a training dataset and a total of 30 control biopsy samples were used as test dataset. A total of 1690 DEGs between ileal enteroids and rectal colonoids were identified as classifier features. The model was trained on these 27 enteroid + colonoid samples, with 5000 trees and 10 variables to select randomly for each tree. Prediction accuracy was measured on the testing dataset containing the same ileal and rectal gene expression data. The confusion matrix was provided to show the results.
Support vector machine classifier
A support vector machine (SVM) classifier was built using the caret R package.24 This is a supervised learning model used for classification, regression, and outlier detection. As a proof of concept, we attempted to use random forest-based feature selection package Boruta,25 followed by a SVM classifier, to test whether enteroids retain CD-relevant transcriptional profiles from the biopsy, sufficiently to predict the tissue of origin. In this case, 43 enteroid samples (20 Crohn’s and 23 control) were used as a training dataset, and 44 biopsy samples (26 Crohn’s and 18 control) were used as a testing dataset. 879 DEGs were selected as candidates from Crohn’s vs control samples, and 33 final genes were reserved after the feature selection using the Boruta package, with 5000 tress and P value cutoff of .05. Then the SVM model was trained on the dataset from 43 enteroid samples containing expression data for only the 33 final genes with repeated 3-fold cross-validation method. Prediction accuracy was measured on the testing dataset comprising 44 ileal biopsy samples containing the expression data for the same 33 genes from Crohn’s and control biopsies. The results are presented as a confusion matrix.
Statistical analyses
For correlation analysis, normalized gene expression matrices were obtained across all the organoid and biopsy samples. Correlation scores (R2) were obtained by individually comparing each enteroid or colonoid sample with all the biopsy samples using linear regression model from stats package (v3.6.2). Summaries of Welch two sample t tests along with boxplots and heatmaps are provided to compare the correlation score between subject-matched enteroid/colonoid-biopsy pairs and random pairs. In GSEA, the enrichment of different gene set from KEGG database is compared between organoid and biopsy data. Fisher exact test was performed on the number of genes enriched toward disease samples vs control samples for each of the gene sets, to test if the DEGs within each gene set were from the same distribution between the organoid and biopsy data. The statistics for gene expression changes measured by qRT-PCR were calculated in GraphPad Prizm 9 software.
Results
Enteroids and colonoids from patients with ileal CD exhibit disease-specific changes in gene expression
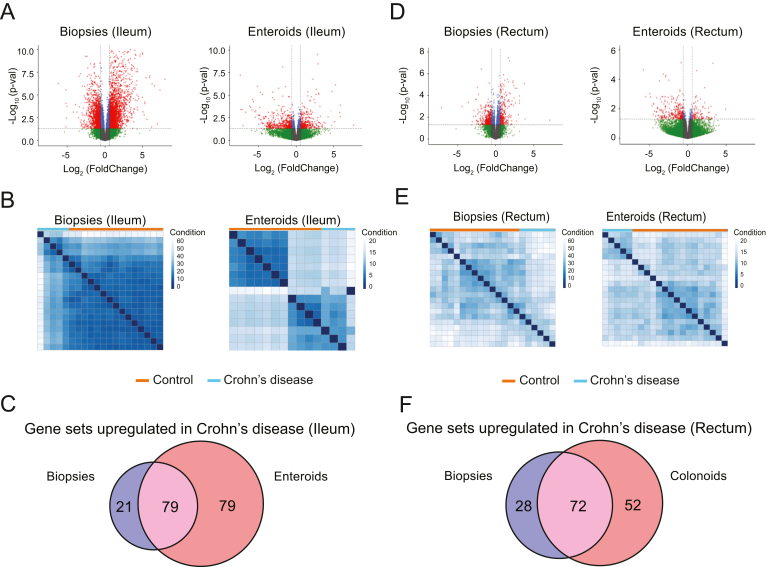
To define disease-specific gene expression signatures retained in enteroid or colonoid cultures, we evaluated samples from 17 control subjects (no pathological diagnosis in lower GI tract), 6 subjects with ileal CD and 7 subjects with ulcerative colitis. Subject demographics are listed in Table 1. Four biopsy fragments were collected from each subject (2 from terminal ileum and 2 from rectum), and from each location one fragment was used to extract RNA, while an enteroid or colonoid line was established from the second fragment. The summary of assays conducted for each subject is presented in Table A2. We performed DEG analysis on bulk RNA-seq data from whole biopsies and early-passage enteroids and colonoids using DESeq2 R package. Durable, long-term transcriptional changes in the epithelium are more likely to be retained in the stem and progenitor cell populations; therefore, we opted to maintain the enteroids and colonoids in expansion medium and harvested for RNA extraction on day 3 of passage 1. We compared the entire transcriptomes (18,913 protein-coding genes from the hg38/GRCh38 reference genome) from ileal biopsies of CD or control subjects and detected 3481 DEGs (Table A3). Likewise, we compared the entire transcriptomes from CD or control ileal enteroids and detected 693 DEGs (Table A3). The top 100 DEGs from both upregulated and downregulated categories were subjected to hierarchical clustering. We detected 2 independent clusters for the enteroids, while for ileal biopsies we also detected 2 clusters, with exception of 2 CD cases. These data indicate preservation of CD-specific transcriptional patterns in standard expansion enteroid culture (Figure 1A and B).
Figure 1.
Enteroids and colonoids retain transcriptomic signatures from biopsies. (A, D) Volcano plots demonstrating the changes in gene expression between the control and Crohn’s disease samples from ileal biopsies and enteroids (A) or rectal biopsies and colonoids (D). Red and green dots represent the genes that are significantly upregulated or downregulated (fold change ≥ 1.5 and P ≤ .05) in Crohn’s disease samples compared to controls, respectively. Blue dots represent the genes with P ≤ .05, but fold change <1.5. Grey dots represent the genes with no significant expression differences. (B, E) Hierarchical clustering of top 200 differentially expressed genes selected (Crohn’s disease vs control), generated using the 100 differentially expressed genes from the upregulated gene list and 100 from downregulated gene list. (C) Venn diagram of the number of functional gene sets upregulated in Crohn’s disease in both ileal biopsies and enteroids (purple), gene sets that are upregulated in Crohn’s disease only in enteroids (red), and gene sets that are upregulated in Crohn’s disease only in ileal biopsies (blue). (F) Venn diagram of the number of functional gene sets upregulated in Crohn’s disease in both rectal biopsies and colonoids (purple), gene sets that are upregulated in Crohn’s disease only in colonoids (red), and gene sets that are upregulated in Crohn’s disease only in rectal biopsies (blue). N = 15 (control, ileal biopsies), 14 (control, enteroids), 5 (Crohn’s, ileal biopsies), 4 (Crohn’s, enteroids), 15 (control, rectal biopsies), 13 (control, colonoids), 6 (Crohn’s, rectal biopsies), 5 (Cronh’s, colonoids).
We validated our approach by comparing our data to a recently published study comparing enteroids to isolated ileal crypts from patients with CD or control subjects.17 We applied our data processing pipeline to the raw data generated by Niclinska-Schirtz et al. and used these DEGs as a reference gene set. GSEA of our DEGs demonstrated significant enrichment toward the reference gene set (P = .00; normalized enrichment score = 1.59) (Figure A1). This indicates that our gene expression analyses are consistent with findings from independent groups.
To determine the extent to which transcriptomic profiles are similar between enteroids and whole biopsies, we first compared CD-specific DEGs from whole ileal biopsies or enteroids. We found 208 genes were shared between biopsies and enteroids, corresponding to 6% or 30% of total DEGs in biopsies or enteroids, respectively. The paucity of shared DEGs was expected, since we are comparing enteroids, which are purified epithelial cultures, to whole biopsies, which comprise multiple cell types. Because of the lack of overlap at the gene-level, we instead focused on evaluating whether biologically relevant transcriptional profiles from intestinal mucosa are retained in enteroids. We conducted gene set enrichment analyses comparing CD-specific DEGs from whole biopsies or enteroids to pathways (gene sets) in the KEGG Pathway database.26 Out of pathways upregulated in CD (P < .05; FDR < 0.25), the shared pathways represented 50% of all pathways upregulated in enteroids or 79% of all pathways upregulated in biopsies (Figure 1C and Table A4). Out of pathways downregulated in CD in the ileum, shared pathways represented 5% or 16% of all pathways downregulated in biopsies and colonoids, respectively.
Rectal colonoids retain select transcriptomic features of ileal biopsies and enteroids from ileal CD patients
In the rectum (disease-spared tissue), biopsies of CD or control subjects revealed 623 DEGs (Table A3). Likewise, we compared CD or control rectal colonoids and detected 364 DEGs (Table A3). The top 100 DEGs from both upregulated and downregulated categories were subjected to hierarchical clustering, which resulted in 2 independent clusters for both biopsies and colonoids, with the exception of one control sample in each group. These results reveal the presence of Crohn’s-specific transcriptional patterns in the biopsies, as well as standard expansion colonoid culture, from disease-spared tissue (Figure 1D and E).
As expected, based upon our findings in the ileum, comparison of CD-specific DEGs from whole rectal biopsies or colonoids revealed minor overlap, 35 genes were shared between the biopsies and colonoids, corresponding to 6% or 10% of total DEGs in biopsies or colonoids, respectively. Conversely, GSEA revealed that out of pathways upregulated in CD (P < .05; FDR < 0.25), shared pathways represented 72% or 58% of all pathways upregulated in biopsies or colonoids, respectively (Figure 1F and Table A4). Out of pathways downregulated in CD, shared pathways represented 39% or 53% of all pathways enriched in colonoids or biopsies, respectively.
GSEA reveals pathway retention in enteroids and colonoids from ileal CD patients
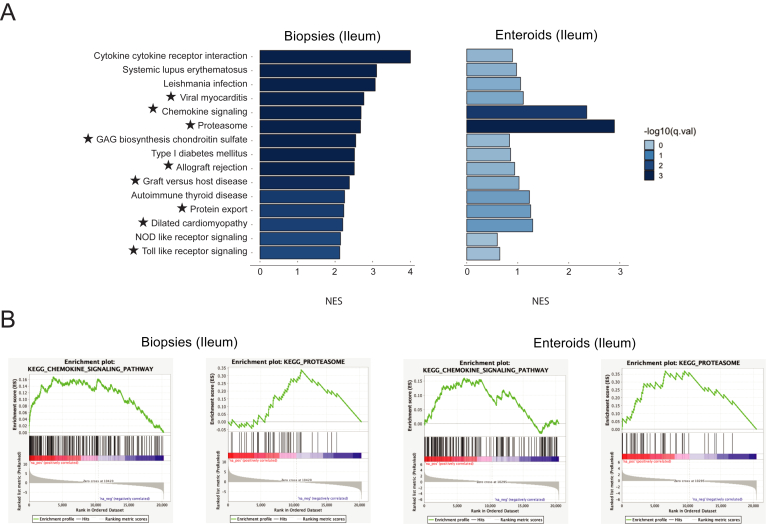
We have validated whether the pathways (gene sets) most upregulated in CD biopsies are also significantly upregulated in CD enteroids. Using the Fisher's exact test, we confirmed enrichment of 9 gene sets from the KEGG database: viral myocarditis, chemokine signaling, proteasome, glucosaminoglycan biosynthesis chondroitin sulfate, allograft rejection, graft vs host disease, protein export, dilated cardiomyopathy, and Toll-like receptor signaling (Figure 2 and Table A5). These findings suggest that physiologically relevant signaling events from whole tissue are retained in enteroids.
Figure 2.
Ileal enteroids retain transcriptomic signatures of biopsies in Crohn’s disease. (A) Pathway enrichment bar plot demonstrating the 15 KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways most upregulated in ileal biopsies from Crohn’s disease patients, in the biopsies, as well as in enteroids. ★ indicates a pathway deemed significantly upregulated in both biopsies and enteroids by Fisher's exact test. (B) Gene set enrichment analysis plots for the chemokine signaling and proteasome KEGG pathways, in both ileal biopsies and enteroids.
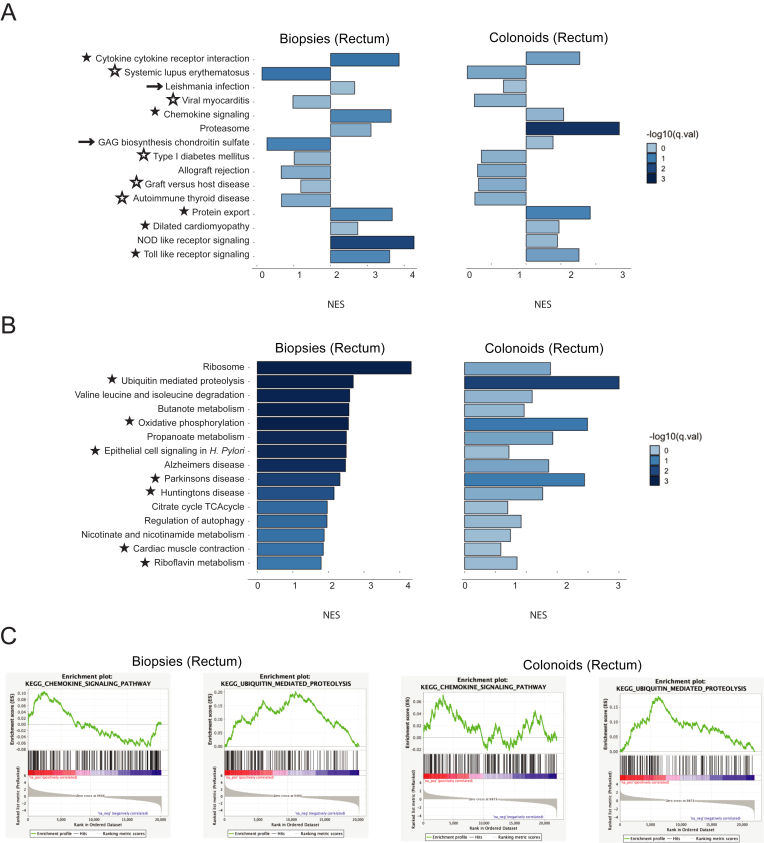
We next evaluated which of the 15 pathways most upregulated in ileal biopsies from ileal CD patients were also upregulated in rectal biopsies and colonoids. Fisher's exact test validated that 5 pathways (cytokine-cytokine receptor interaction, chemokine signaling, protein export, pilated cardiomyopathy, and Toll-like receptor signaling) were significantly upregulated in rectal biopsies and colonoids from CD patients (Figure 3A–C and Table A5). Interestingly, 6 of the gene sets upregulated in ileal biopsies were downregulated in both rectal biopsies and colonoids (systemic lupus erythematosus, viral myocarditis, type I diabetes mellitus, allograft rejection, graft vs host disease, and autoimmune thyroid disease).
Figure 3.
Biopsies and colonoids from disease-spared tissue (rectum) carry Crohn's disease-specific gene expression changes. (A) The 15 KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways most upregulated in ileal biopsies from Crohn’s disease patients (Figure 2A), in rectal biopsies and colonoids. (B) Pathway enrichment bar plot demonstrating the 15 KEGG pathways most upregulated in rectal biopsies (but not in ileal biopsies) from Crohn’s disease patients, in biopsies and colonoids. (A, B) ★ indicates a pathway deemed significantly upregulated in both rectal biopsies and colonoids by Fisher's exact test. ☆ indicates a pathway deemed significantly downregulated in both rectal biopsies and colonoids by Fisher's exact test. → indicates a pathway with opposite normalized enrichment score values in biopsies vs colonoids, for which Fisher's exact test still indicates same direction of enrichment in Crohn’s. (C) Gene set enrichment analysis plots for the chemokine signaling and ubiquitin-mediated proteolysis KEGG pathways, in both rectal biopsies and colonoids.
We also found pathways significantly upregulated in rectal, but not ileal, biopsies from ileal CD patients. Fisher's exact test revealed that 7 of those gene sets (ubiquitin-mediated proteolysis, oxidative phosphorylation, Parkinson’s disease, Huntington’s disease, nicotine and nicotinamide metabolism, cardiac muscle contraction, and riboflavin metabolism) are also upregulated in colonoids (Figure 3B and C and Table A5). The presence of neurodegenerative disease-associated pathways on the list was surprising, but we hypothesized that the genes comprising these KEGG pathways may be overlapping with other larger gene sets. Indeed, leading-edge analysis (part of the GSEA package) revealed that Parkinson’s disease, Huntington’s disease, and Alzheimer’s disease pathways shared a large fraction of upregulated genes with the oxidative phosphorylation pathway (Figure A3). These findings suggest that despite the lack of endoscopic or histological signs of disease, rectum of ileal Crohn’s patients is transcriptionally different from rectum of control subjects.
Transcriptomic features are retained in enteroid/colonoid-biopsy pairs matched by subject
As an independent measure of whether the enteroids/colonoids recapitulate the transcriptional profiles from original tissue, we compared correlation factors for normalized gene expression matrices from enteroids/colonoids and biopsies in random pairs or matched by subject. In the ileum, the correlation coefficient was significantly higher (by 20%, 0.5124 vs 0.4277; P = .025) for subject-matched pairs of enteroids and biopsies (Figure A2A and B). In the rectum, the correlation scores were even further separated, with R2 = 33% higher (0.6463 vs 0.4866; P = 2E-16) for subject-matched pairs of colonoids and biopsies (Figure A2C and D).
Exposure to inflammation ex vivo allows to model CD features in late-passage enteroids
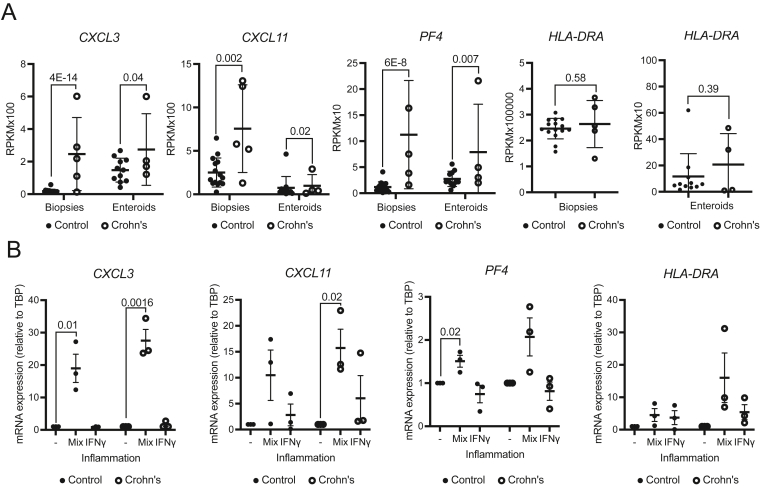
To test how well the enteroids and colonoids retain transcriptional changes in continuous culture, we focused on 3 genes from the chemokine signaling pathway (CXCL3, CXCL11, and PF4) that were significantly upregulated in CD in both biopsies and enteroids, as well as the HLA-DRA (Major Histocompatibility Complex, Class II, DR Alpha) gene, which we previously reported to be upregulated in IBD colonoids14 (Figure 4A). When we set out to assess the expression level of these genes in enteroids at passage >8 (Table A1), we found no consistent trend of expression change in CD enteroids, likely due to interexperimental variability, and a normalization of phenotype due to long-term culture in the absence of inflammatory milieu. Thus, we opted to focus on the gene expression changes when an inflammatory milieu was introduced in vitro by exposing the enteroids to a previously described cocktail of TNFα, IL-1β, and flagellin (100 ng/mL, 20 ng/mL, and 1 μg/mL, respectively),16 or to IFNγ (50 ng/mL), a prominent cytokine upstream of the JAK-STAT signaling pathway in IBD and a master-regulator of HLA family genes. CXCL3 was upregulated in both control and Crohn’s enteroids in response to the cocktail (P = .01 and .0016, respectively). Interestingly, the mean magnitude of the change differed between the 2 groups, with 19- and 28-fold increase in expression observed for control or Crohn’s enteroids, respectively, although this was not statistically significant (Figure 4B). In contrast, exposure to IFNγ resulted in negligible upregulation (Figure 4B). CXCL11 was significantly upregulated only in Crohn’s enteroids in response to the inflammatory cocktail (P = .02) (Figure 4B). PF4 was upregulated in both control and Crohn's enteroids when exposed to the cocktail (P = .02) (Figure 4B), while the mean magnitude of change was similar for control and Crohn’s enteroids. Finally, there was a trend of upregulation in HLA-DRA expression observed in response to both treatments, with the upregulation in response to the cytokine cocktail being more specific to CD. Specifically, control enteroids showed a 5-fold increase in HLA-DRA expression while Crohn’s enteroids showed a 16-fold increase. In comparison, IFNγ resulted in a roughly 4- and 6-fold upregulation for control and Crohn’s enteroids, respectively (Figure 4B). We reported previously that HLA-DRA is increased to a varying degree in IFNγ—treated colonoids—a subset of patients with IBD exhibited significantly increased HLA-DRA whereas others exhibited modest effects. Our current cohort appears to represent the latter. Taken together, these findings indicate that reintroduction of inflammation in vitro can bring forth transcriptional events observed in mucosal tissue, even in late passage enteroids.
Figure 4.
Re-exposure to inflammation restores disease-specific gene expression changes in late-passage enteroids. (A) Reads per kilobase per million (RPKM) values from RNA-sequencing. Data presented as mean ± SD. N = 15 (biopsies, control), 5 (biopsies, Crohn’s), 11 (enteroids, control), 4 (enteroids, Crohn’s). P values generated via Wald test (part of DESeq2 package). (B) Relative gene expression (RT-PCR) in untreated enteroids or in the presence of inflammation (a mix of TNFa + IL-1b + flagellin, or IFNg alone). N = 3 lines from individual subjects per condition, representative of 2–3 independent experiments is shown. Data presented as mean ± SEM. P values generated via unpaired 2-tailed Student’s t test. RT-PCR, real-time polymerase chain reaction; SD, standard deviation; SEM, standard error of mean.
Machine learning identifies discriminating features of intestinal segment and disease state
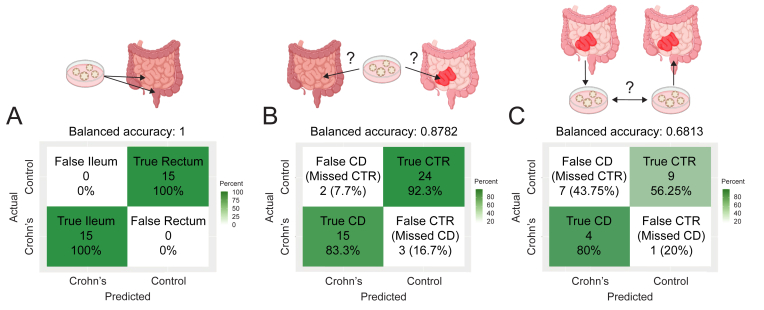
We next used machine learning (random forest classification) to test in an unbiased manner whether enteroids and colonoids recapitulate the biology of original tissue. We generated a reference dataset from ileal enteroids and rectal colonoids (control subjects, N = 11 and 16, respectively), yielding a classifier comprising 1690 DEGs. The top 30 genes are listed (importance ranked by mean decrease accuracy) (Table S6 and Figure A4A). When applied to a testing dataset containing gene expression data from ileal and rectal biopsies (control subjects, N = 15 and 15, respectively), this classifier predicted the tissue of origin of the biopsy with 100% accuracy (P = 9.313 E-10; Figure 5A). These results indicate that transcriptional profiles of enteroids and colonoids recapitulate the differences between GI tract segments.
Figure 5.
Enteroid/colonoid-informed machine learning approach identifies discriminating features of intestinal segment epithelium at homeostasis and in Crohn’s disease. (A) Outcome of testing the enteroid/colonoid-informed random forest model “tissue of origin classifier” on the dataset comprising biopsy-derived data. (B) Outcomes of testing the enteroid-informed support vector machine model “disease state classifier” on the dataset comprising the biopsy-derived data. (C) Outcomes of testing the enteroid-informed support vector machine model “disease state classifier” on the dataset comprising the rectal colonoid-derived data. CD, Crohn’s disease.
Next, we used random forest-based feature selection followed by a support vector machine (SVM) classifier to generate a classifier from enteroid data that would predict the donor subject’s disease status based on biopsy data. To increase our reference dataset beyond our own CD cohort, we incorporated recently published data from another group.17 This expanded reference dataset, containing data from 20 CD and 23 control enteroid lines, yielded a classifier comprising 35 genes (Table A6 and Figure A4B). We applied this tool to a testing dataset containing data from 26 CD and 18 control biopsies, demonstrating a balanced accuracy of 93.38% (P = 4.32 e-7; Figure 5B). Specifically, out of 26 actual control biopsies, 24 (92.3%) were predicted correctly, while 2 (7.7%) control subjects were missed and falsely predicted as CD (missed control subjects). Of 18 CD biopsies, 15 (83.3%) were predicted correctly, while 3 (16.7%) were falsely predicted as control (missed CD subjects). Increased sample size and refinement of the classifier may support the use of transcriptional changes retained in enteroids for predictive value in CD.
When the above classifier (informed by ileal enteroids) was applied to rectal colonoid data, this tool worked with the balanced accuracy of 68.13% (P = .9577; Figure 5C). Specifically, of 16 actual control colonoids, 9 (56.25%) were predicted correctly, while 7 (43.75%) control subjects were missed and falsely predicted as having CD (missed control subjects). Of 5 rectal colonoids from CD subjects, 4 (80%) were correctly predicted as Crohn’s, while 1 (20%) was falsely predicted as control (a missed CD subject). The high P value in this analysis does not reflect the accuracy of prediction but is a consequence of an imbalance in our test cohort, as we have significantly more controls than CD samples. These results indicate that rectal colonoids from subjects with ileal CD retain disease-specific features, despite originating from a disease-spared intestinal segment.
Colonoids from patients with ulcerative colitis exhibit disease-specific changes in gene expression
We also collected rectal biopsies from patients with ulcerative colitis (Figure A5A) and routed them for bulk RNA-seq or colonoid generation followed by bulk RNA-seq. DESeq2 analysis of RNA-seq data from rectal biopsies of ulcerative colitis or control subjects revealed 4454 DEGs (Table A3). Likewise, we compared gene expression data from ulcerative colitis or control rectal colonoids and detected 459 DEGs (Table A3). The top 100 DEGs from both upregulated and downregulated categories were subjected to hierarchical clustering, and the resulting data indicate preservation of ulcerative colitis-specific transcriptional patterns in standard expansion colonoid culture (Figure A5B and C).
As with CD, ulcerative colitis-specific DEGs from whole rectal biopsies or colonoids revealed minor overlap, 179 genes were shared between the biopsies and colonoids, corresponding to 4% or 39% of total in biopsies or colonoids, respectively. We also compared correlation coefficients for normalized gene expression matrices from colonoids and biopsies in random pairs or matched by subject. We found the correlation coefficient was significantly higher (by 11%, 0.5573 vs 0.5038; P = .017) for subject-matched pairs of enteroids and biopsies (Figure A5D). GSEA revealed that out of pathways upregulated in ulcerative colitis (P < .05; FDR < 0.25), shared pathways represented 62% or 70% of all pathways upregulated in biopsies or colonoids, respectively. Out of pathways downregulated in ulcerative colitis, shared pathways represented 72% or 64% of all pathways downregulated in biopsies and colonoids, respectively (Table A4).
Discussion
The goal of this work was to probe the extent to which enteroids and colonoids transcriptionally recapitulate the original tissue. We found that the enteroids from patients with CD retain upregulation of several pathways previously implicated in CD, such as Toll-like receptor signaling, and chemokine signaling (Figure 2A). Toll-like receptors belong to the family of pattern recognition receptors, which are implicated in susceptibility to IBD.27 For example, TLR5 promotes pathogenic response to the bacterial protein flagellin associated with Crohn’s pathogenesis.28,29 Chemokine pathways have been implicated in IBD,30 and here we report sustained upregulation in culture of previously reported chemokine genes in enteroids from CD patients. PF4 encodes platelet factor 4, which was reported to be elevated in CD patients’ plasma31 and positively correlates with disease activity index.32 CXCL3 and CXCL11 encode C-X-C motif chemokine ligands 3 and 11, respectively, also known as growth-regulated protein gamma (GROγ) and interferon-inducible T-cell alpha chemoattractant (ITAC); both genes have an NFκB response element and are upregulated in IBD.33 Taken together, the present data are consistent with multiple previous studies in CD.
Among the less expected findings was upregulation of the proteasome pathway in biopsies from CD patients, which is retained in enteroids. Upregulation of proteasome subunits beta1i and beta2i was previously reported in colonic epithelium of CD but not ulcerative colitis patients.34 Intriguingly, we observed CD-specific transcriptomic signatures in disease-spared tissue (rectum of patients with ileal Crohn’s), and in the colonoids derived from it. Our results suggest that there may be underappreciated transcriptional signals in the Crohn’s rectum even in the absence of mucosal pathology that could identify the presence of disease in the ileum. Another implication of this finding is that gene expression data from normal-appearing mucosa may support diagnostic assignments when IBD class is indeterminate. Discriminating between colonic CD and ulcerative colitis in pediatric patients, for example, is often a challenge, requiring repeat endoscopies to demonstrate the chronicity in support of CD, yet making this distinction is important, because of the implications for treatment approaches. This is particularly true in case of severe disease where an incorrect ulcerative colitis assignment followed by colectomy can lead to long-term morbidity in patients with underlying CD. Additional, larger cohorts of patients are needed to verify the use of rectal biopsy classifiers to support CD diagnosis when the totality of clinical, endoscopic, and histologic data are indeterminate, but if proven true, the ability to use a less invasive procedure (flexible sigmoidoscopy) could improve diagnosis and ultimately the patients’ quality of life.
We observed upregulation of the oxidative phosphorylation pathway in biopsies and rectum of CD patients. This initially seemed to contradict previous studies that reported suppression of mitochondrial function in IBD. However, prior studies relied on transcriptomes from whole biopsies taken from actively inflamed tissue,35 whereas, we focused on epithelium from locations that were inflammation-adjacent. Furthermore, the crypt orientation in immunohistological staining for OXPHOS complexes presented in other reports,35,36 does not consistently offer insight into the crypt base. We utilized stem cell-biased culture conditions, which may have revealed transcriptional events that would remain unnoticed when examining the intestinal epithelium broadly. Interestingly, a dedicated investigation focusing on Lgr5+ crypt-base columnar cells revealed increased metabolic activity as a feature of intestinal stem cells.37 Our findings, along with published studies, underscore the complexity of intestinal epithelium and support the use of single cell analyses including in organoids.
We used a machine learning approach (random forest classifier) as an alternative strategy to determine the extent to which enteroid/colonoid culture recapitulates the original tissue. Though it was not our goal for this experiment to derive conclusions about the biology of intestinal epithelium in CD, two discriminating features identified for CD deserve to be discussed here. CEACAM5, the gene with highest score for distinguishing CD from control (Figure A4B and C and Table A6), has been previously reported as upregulated during active CD, and is likely driven by IL-22.38 Another study reported a different finding, demonstrating that CEACAM5 expression is unchanged in ulcerative colitis, yet downregulated (by mRNA and protein39) in CD. These findings suggest an essential role for CEACAM5 in biology of Crohn’s and follow-up studies are needed to reveal the specific mechanism. Finally, among the distinguishing features for CD we found two chemokine-encoding genes: CXCL5 and CCL28 (Figure A4B and Table A6). Increased levels of CXCL5 chemokine were reported in the serum of IBD patients40 and increased serum levels of CCL28 were reported in ulcerative colitis subjects.41 Chemokines have not been compared specifically in CD vs ulcerative colitis, and by disease severity, but considering that these genes were selected as discriminating features for Crohn’s, it is intriguing to investigate the protein profiles in the patients’ serum as biomarkers.
There are limitations to our machine learning-based experiment. For example, while we report 100% accuracy in predicting whether the biopsy came from rectum or ileum (Figure 5A), it is important to keep in mind that we have not tested this approach outside of this simplistic binary scenario. A more relevant validation would be whether the classifier would successfully predict a duodenal biopsy as both “false ileum” and “false rectum”. This same caveat applies to our disease state classifier (Figure 5B)—would it be able to predict a biopsy from a patient with acute ileitis as both “false Crohn’s disease” and “false control”? Another possible limitation is that male subjects are under-represented in our CD cohort (17%). Data suggest that there is male predominance in pediatric CD, and these subjects are more likely to have complications related to IBD.42 Future studies with higher numbers of patients can be done to address these limitations. Interestingly, the random forest classifier for predicting the subject’s disease status worked well for calling CD (but not its absence) in colonoids from disease-spared tissue (Figure 5C). This could be because even though the classifier was built from ileal enteroid data, the CD features are retained in rectal colonoids. We propose that the classifier did not work well for predicting controls, because the discriminating features for controls were informed by ileum, and the biology between the two intestinal segments is too different. Additional refinement could improve the accuracy of the classifier when using rectal colonoids to predict ileal Crohn’s.
When comparing enteroids or colonoids to original tissue, we detected minor overlap at the DEG-level in Crohn’s compared control subjects. This is not surprising, since biopsies comprise a higher degree of cell type heterogeneity, while enteroids and colonoids are epithelial-only structures, further enriched toward stem and progenitor cell compartments due to culture conditions. We opted for sequencing the biopsies whole, as opposed to enriching for epithelium via crypt scraping or antigen-based sorting, to minimize excessive damage to the epithelial cells. Furthermore, our goal was to evaluate the conditions most available to investigators, regardless of their proximity to a clinical center or a collaborator with properly cryopreserved biopsies. Flash-frozen whole biopsies are the most accessible source of RNA from human intestinal mucosa, and we wanted to see how it compares to transcriptomes of epithelial-only enteroids and colonoids. We used bulk RNA sequencing, as opposed to single cell transcriptomics, for a similar reason, these are robust tools widely available to investigators. The substantial overlap we detected in KEGG pathways for both ileal and colonic biopsies and enteroids or colonoids, respectively (Figures 2 and 3), indicates that these epithelial cultures retain the disease-relevant biological functions. Yet, it is important to remember that a change in gene expression does not always translate to a change in protein abundance and function, and thus all the findings reported herein must be validated in functional studies prior to drawing any conclusions about the biology of intestinal epithelium in CD.
When comparing the analyses from ileum and rectum, we found much fewer DEGs (in Crohn’s vs control) in rectal biopsies, than in ileal (a 5.6-fold decrease: 3481 vs 623 genes, respectively). However, in the enteroids there were only 1.9-fold more DEGs than in colonoids (693 vs 364). From this observation, one may suggest that the key CD-related transcriptional changes in the rectum are occurring in the epithelium, specifically in the intestinal stem cell and transit amplifying cell compartments, which are over-represented in the expansion culture of enteroids and colonoids utilized in this study.
In summary, we provide evidence that enteroids retain disease-relevant transcriptomic signatures of the ileal biopsies from CD patients. Furthermore, we demonstrate the presence of disease-relevant transcriptomic changes in rectal biopsies from patients with no evidence of colonic inflammation, and in colonoids derived from these biopsies. These findings validate patient-derived enteroids and colonoids as valuable models for the study of intestinal mucosa from patients with IBD, and the possibility of utilizing rectal biopsies or colonoids for screening or diagnosing ileal CD could be of great benefit to patients.
Acknowledgments:
The authors thank the Biorepository Resource Center and the Center for Applied Genomics at the Children’s Hospital of Philadelphia (CHOP) Research Institute. We are grateful to the nurses at the CHOP gastrointestinal endoscopic suite for supporting our research; and to Gary D. Wu, M.D., Jennifer Radke, Catharine S. Kang, and the members of Hamilton lab for discussions.
Authors' Contributions:
Tatiana Karakasheva, Yusen Zhou, Gloria Soto, Daana Roach, and Kathryn Hamilton wrote the manuscript. Noor Nema, Chizoba Umeweni, and Lauren Dolinsky procured and analyzed patient data. Tatiana Karakasheva, Yusen Zhou, Hongbo Xie, Louis Ghanem, Natalie Terry, Amanda Muir, and Kathryn Hamilton conceived ideas, analyzed and interpreted data. Gloria Soto, Tiana Jhonson, Madison Stoltz, James Pippin, Andrew Wells, and Struan Grant collected and analyzed data. Louis Ghanem, Natalie Terry, Amanda Muir, and Kathryn Hamilton obtained funding. All authors read and agreed to the submitted version of the manuscript.
Footnotes
Conflicts of Interest: The authors disclose no conflicts.
Funding: The following NIH grants: NIHR01-DK124369 (K.E.H.), Penn Center for Molecular Studies in Digestive and Liver Diseases NIHP30-DK050306 (K.E.H., A.B.M.). The following institutional grants: Children's Hospital of Philadelphia Institutional Development Funds and Gastrointestinal Epithelium Modeling Program (A.B.M., K.E.H.).
Ethical Statement: The corresponding author, on behalf of all authors, jointly and severally, certifies that their institution has approved the protocol for any investigation involving humans or animals and that all experimentation was conducted in conformity with ethical and humane principles of research.
Data Transparency Statement: Data have been uploaded to the Gene Expression Omnibus (GEO accession number GSE228122).
Reporting Guidelines: SAGER.
Material associated with this article can be found in the online version at https://doi.org/10.1016/j.gastha.2023.05.003.
Supplementary materials
References
- 1.Fakhoury M., Negrulj R., Mooranian A., et al. Inflammatory bowel disease: clinical aspects and treatments. J Inflamm Res. 2014;7:113–120. doi: 10.2147/JIR.S65979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conrad M.A., Rosh J.R. Pediatric inflammatory bowel disease. Pediatr Clin North Am. 2017;64:577–591. doi: 10.1016/j.pcl.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Rath T., Atreya R., Bodenschatz J., et al. Intestinal barrier healing is superior to endoscopic and histologic remission for predicting major adverse outcomes in inflammatory bowel disease: the prospective ERIca trial. Gastroenterology. 2022;164:241–255. doi: 10.1053/j.gastro.2022.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Sato T., Vries R.G., Snippert H.J., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 5.Sato T., Stange D.E., Ferrante M., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 6.Günther C., Winner B., Neurath M.F., et al. Organoids in gastrointestinal diseases: from experimental models to clinical translation. Gut. 2022;71:1892–1908. doi: 10.1136/gutjnl-2021-326560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin W., Wu A., Min S., et al. Spatiotemporal gradient and instability of Wnt induce heterogeneous growth and differentiation of human intestinal organoids. iScience. 2020;23 doi: 10.1016/j.isci.2020.101372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan C.W., Hirokawa Y., Burgess A.W. Analysis of Wnt signalling dynamics during colon crypt development in 3D culture. Sci Rep. 2015;5 doi: 10.1038/srep11036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii M., Matano M., Toshimitsu K., et al. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell. 2018;23:787–793.e6. doi: 10.1016/j.stem.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 10.Boonekamp K.E., Dayton T.L., Clevers H. Intestinal organoids as tools for enriching and studying specific and rare cell types: advances and future directions. J Mol Cell Biol. 2020;12:562–568. doi: 10.1093/jmcb/mjaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki K., Murano T., Shimizu H., et al. Single cell analysis of Crohn's disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties. J Gastroenterol. 2018;53:1035–1047. doi: 10.1007/s00535-018-1437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howell K.J., Kraiczy J., Nayak K.M., et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology. 2018;154:585–598. doi: 10.1053/j.gastro.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dotti I., Mora-Buch R., Ferrer-Picón E., et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 2017;66:2069–2079. doi: 10.1136/gutjnl-2016-312609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelsen J.R., Dawany N., Conrad M.A., et al. Colonoids from patients with pediatric inflammatory bowel disease exhibit decreased growth associated with inflammation severity and durable upregulation of antigen presentation genes. Inflamm Bowel Dis. 2021;27:256–267. doi: 10.1093/ibd/izaa145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.d'Aldebert E., Quaranta M., Sébert M., et al. Characterization of human colon organoids from inflammatory bowel disease patients. Front Cell Dev Biol. 2020;8:363. doi: 10.3389/fcell.2020.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnauts K., Verstockt B., Ramalho A.S., et al. Ex vivo Mimicking of inflammation in organoids derived from patients with ulcerative colitis. Gastroenterology. 2020;159:1564–1567. doi: 10.1053/j.gastro.2020.05.064. [DOI] [PubMed] [Google Scholar]
- 17.Niklinska-Schirtz B.J., Venkateswaran S., Anbazhagan M., et al. Ileal derived organoids from Crohn's disease patients show unique transcriptomic and secretomic signatures. Cell Mol Gastroenterol Hepatol. 2021;12:1267–1280. doi: 10.1016/j.jcmgh.2021.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahe M.M., Sundaram N., Watson C.L., et al. Establishment of human epithelial enteroids and colonoids from whole tissue and biopsy. J Vis Exp. 2015 doi: 10.3791/52483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bray N.L., Pimentel H., Melsted P., et al. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 20.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mootha V.K., Lindgren C.M., Eriksson K.F., et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 24.Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28:1–26. [Google Scholar]
- 25.Kursa M.B., Rudnicki W.R. Feature selection with the Boruta package. J Stat Softw. 2010;36:1–13. [Google Scholar]
- 26.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abraham C., Abreu M.T., Turner J.R. Pattern recognition receptor signaling and cytokine networks in microbial defenses and regulation of intestinal barriers: implications for inflammatory bowel disease. Gastroenterology. 2022;162:1602–1616.e6. doi: 10.1053/j.gastro.2021.12.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodes M.J., Cong Y., Elson C.O., et al. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gewirtz A.T., Vijay-Kumar M., Brant S.R., et al. Dominant-negative TLR5 polymorphism reduces adaptive immune response to flagellin and negatively associates with Crohn's disease. Am J Physiol Gastrointest Liver Physiol. 2006;290:1157–1163. doi: 10.1152/ajpgi.00544.2005. [DOI] [PubMed] [Google Scholar]
- 30.Trivedi P.J., Adams D.H. Chemokines and chemokine receptors as therapeutic targets in inflammatory bowel disease; pitfalls and promise. J Crohns Colitis. 2018;12 doi: 10.1093/ecco-jcc/jjy130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simi M., Leardi S., Tebano M.T., et al. Raised plasma concentrations of platelet factor 4 (PF4) in Crohn's disease. Gut. 1987;28:336–338. doi: 10.1136/gut.28.3.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vrij A.A., Rijken J., Van Wersch J.W., et al. Platelet factor 4 and β-thromboglobulin in inflammatory bowel disease and giant cell arteritis. Eur J Clin Invest. 2000;30:188–194. doi: 10.1046/j.1365-2362.2000.00616.x. [DOI] [PubMed] [Google Scholar]
- 33.Fang K., Grisham M.B., Kevil C.G. Application of comparative transcriptional genomics to identify molecular targets for pediatric IBD. Front Immunol. 2015;6:165. doi: 10.3389/fimmu.2015.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visekruna A., Joeris T., Schmidt N., et al. Comparative expression analysis and characterization of 20S proteasomes in human intestinal tissues: the proteasome pattern as diagnostic tool for IBD patients. Inflamm Bowel Dis. 2009;15:526–533. doi: 10.1002/ibd.20805. [DOI] [PubMed] [Google Scholar]
- 35.Haberman Y., Karns R., Dexheimer P.J., et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat Commun. 2019;10:1–13. doi: 10.1038/s41467-018-07841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider A.M., Özsoy M., Zimmermann F.A., et al. Expression of oxidative phosphorylation complexes and mitochondrial mass in pediatric and adult inflammatory bowel disease. Oxid Med Cell Longev. 2022;2022:9151169. doi: 10.1155/2022/9151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez-Colman M.J., Schewe M., Meerlo M., et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature. 2017;543:424–427. doi: 10.1038/nature21673. [DOI] [PubMed] [Google Scholar]
- 38.Glover L., Bowers B., Kelly C., et al. CEACAM5 is upregulated with inflammation in Crohn's colitis in response to IL-22P-207. Inflamm Bowel Dis. 2011;17:S73–S74. [Google Scholar]
- 39.Roda G., Dahan S., Mezzanotte L., et al. The defect in CEACAM family member expression in Crohn's disease IECs is regulated by the transcription factor SOX9. Inflamm Bowel Dis. 2009;15:1775. doi: 10.1002/ibd.21023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh U.P., Singh N.P., Murphy E.A., et al. Chemokine and cytokine levels in inflammatory bowel disease patients. Cytokine. 2016;77:44–49. doi: 10.1016/j.cyto.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee D.S., Lee K.L., Jeong J.B., et al. Expression of chemokine CCL28 in ulcerative colitis patients. Gut Liver. 2021;15:70–76. doi: 10.5009/gnl19273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashton J.J., Cullen M., Afzal N.A., et al. Is the incidence of paediatric inflammatory bowel disease still increasing? Arch Dis Child. 2018;103:1093–1094. doi: 10.1136/archdischild-2018-315038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.