Abstract
We used pulsed-field gel electrophoresis (PFGE) to study the genetic relatedness of 235 isolates of Shigella flexneri and Shigella sonnei collected in Hong Kong (97 isolates from 1986 and 1987 and 138 isolates from 1994 and 1995). Altogether, 13 gels were run with bacteriophage lambda ladder DNA (Pharmacia) as an external reference in every sixth lane, standardized reagents and methods, and isolates randomized for species and years. For quantitative illustration of the relationships within a large body of isolates, computer-generated dendrograms were used to determine the number of isolates in pulsotypes at Dice coefficients of similarity of 75% (PT75) and 50% (PT50). For S. flexneri, there was a significant difference in the distribution of isolates collected during the two periods in both PT75 and PT50, with 68% of isolates collected in 1994 and 1995 sharing a coefficient of similarity of ≥68%. For S. sonnei, a significant difference was observed in PT50 only. We also used Upholt’s formula for an approximation of the fraction of nucleotide difference between isolates and Molecular Evolutionary Genetics Analysis to determine relative genetic distances. For both species, the relative genetic distances between isolates of the earlier collection period were significantly greater (P < 0.0001), i.e., they were further apart and therefore more diverse than those of the later period. We conclude that it is possible for a typical clinical laboratory to analyze a large amount of PFGE information on Shigella isolates obtained under controlled conditions. Such data analysis should enhance surveillance capabilities and give indications of further work to be done on various aspects of bacterial pathogenicity of the species.
Infections caused by Shigella spp. are major causes of diarrheal disease in both developing and developed countries. We studied the antimicrobial resistance of Shigella flexneri and Shigella sonnei isolates collected in Hong Kong from 1986 to 1995 and found that there was a significant increase in the proportion of S. flexneri isolates showing resistance to four or more agents. No such increase was seen in S. sonnei isolates (1). There is little information on the genetic relatedness of Shigella isolates correlated over time. We report here our analysis of genetic relatedness based on data obtained by pulsed-field gel electrophoresis (PFGE), which shows a greater genetic diversity for both species among isolates obtained in 1986 and 1987 than among those obtained 10 years later.
MATERIALS AND METHODS
Bacterial strains.
In total, we studied 63 S. flexneri isolates collected in 1986 and 1987 and 100 collected in 1994 and 1995 and 34 S. sonnei isolates collected in 1986 and 1987 and 38 collected in 1994 and 1995. Details of these isolates were described by Chu et al. (1). The isolates were obtained from clinical stool samples of individual patients at the Prince of Wales Hospital or the Princess Margaret Hospital. For S. flexneri, the proportions of isolates resistant to ampicillin, amoxicillin-clavulanate, chloramphenicol, tetracycline, trimethoprim, or sulfamethoxazole were significantly higher in the 1994 to 1995 collection than in the 1986 to 1987 collection. One antibiogram showed resistance to seven antimicrobials in 44 of the 1994 to 1995 isolates. The proportions of S. sonnei isolates showing resistance to four or more agents were not statistically different between the two collections (1).
In addition to the isolates described above, three epidemiologically linked isolates of S. flexneri of serotype II were obtained in 1996. These came from three separate stool samples received on the same day from the same patient.
Serotyping.
Serotyping was carried out according to the manufacturer’s instructions (Denka Seiken, Tokyo, Japan).
PFGE.
PFGE was carried out as described by Kaufmann and Pitt (7). Colonies were obtained from an overnight culture on nutrient agar and were emulsified and suspended in sodium chloride-EDTA buffer to obtain a turbidity of about 1 × 109 CFU/ml.
PFGE genomic fingerprints were generated with a Clamped Homogeneous Electric Fields electrophoretic apparatus (Mapper, BioRad). The restriction enzyme XbaI was used to digest in situ the agarose-embedded Shigella genomic DNA. Certified PFGE-grade agarose from BioRad was used at 1% (wt/vol) for the electrophoretic separation of the digested fragments. An electric field of 6 V/cm was applied at an included field angle of 120° with pulsed time ramping from 1 to 40 sec over 35 h. Tris-borate-EDTA buffer at half strength (50 mM Tris, 40 mM borate, and 0.5 mM EDTA) was used as running buffer and was maintained at 14°C. After electrophoresis, the gel was stained with ethidium bromide (1 μg/ml) for 10 min. The DNA was visualized with a UV transilluminator, and images of the illuminated gels were digitized with a gel documentation device (Ultra Violet Products Life Sciences, Cambridge, England). The images were then processed with the GelCompar software (Applied Maths, Kortrijk, Belgium). Dice coefficients of similarity were calculated as follows: F = 2nxy/(nx + ny), where nx is the total number of fragments from isolate X, ny is the total number of fragments from isolate Y, and nxy is the number of fragments shared by the two isolates. Comparison matrices were generated, and dendrograms were constructed by the unweighed pair group method with arithmetic averages (UPGMA).
Bacteriophage lambda ladder DNA (Pharmacia) was used as an external reference in every sixth lane, and all test fingerprint images were accordingly normalized. Individual gels contained randomly chosen isolates obtained in different years and representing both Shigella species. The additional three epidemiologically linked isolates from the same patient were included on two different gels. The same batches of agarose gel, chemicals for buffers, lambda ladder, and restriction enzyme were used throughout the study.
Inference of relatedness.
The approximate degree of nucleotide sequence similarity of chromosomal DNA in the S. flexneri and S. sonnei isolates was examined with the computer programs GelCompar (see above) and Molecular Evolutionary Genetics Analysis (MEGA), version 1.01 (8).
The relative genetic distances between isolates were estimated by using the modification of the methods of Nei and Li (15) and Upholt (21) applied by El-Adhami et al. (2) and Jorgensen et al. (6). GelCompar constructed the matrix based on the Dice coefficients of similarity. The fraction of nucleotide difference between isolates was then calculated by using Upholt’s formula: p = 1 − {[(F2 + 8F)1/2 − F]/2}1/n, where F is the Dice coefficient of similarity and n has a value of 6 for XbaI (21). The MEGA program was used on the pair-wise matrix of p values to draw a dendrogram by the neighbor-joining method (17). The relative genetic distances (P) between isolates collected during the same period were calculated by adding together the successive internodal values obtained by MEGA. The distribution of P values for the two collection periods were compared by the Mann-Whitney U test (SPSS).
RESULTS
Of the 63 S. flexneri isolates from 1986 and 1987, 21 were serotype I, 31 were serotype II, 9 were serotype III, 1 was serotype IV, and 1 was serotype VI. Of the 100 S. flexneri isolates from 1994 and 1995, 3 were serotype I, 90 were serotype II, 4 were serotype III, and 3 were serotype IV. The number of serotype II isolates increased significantly from 49% (31 of 63) among the isolates from 1986 and 1987 to 90% (90 of 100) among the isolates from 1994 and 1995 (chi-square test, P < 0.001).
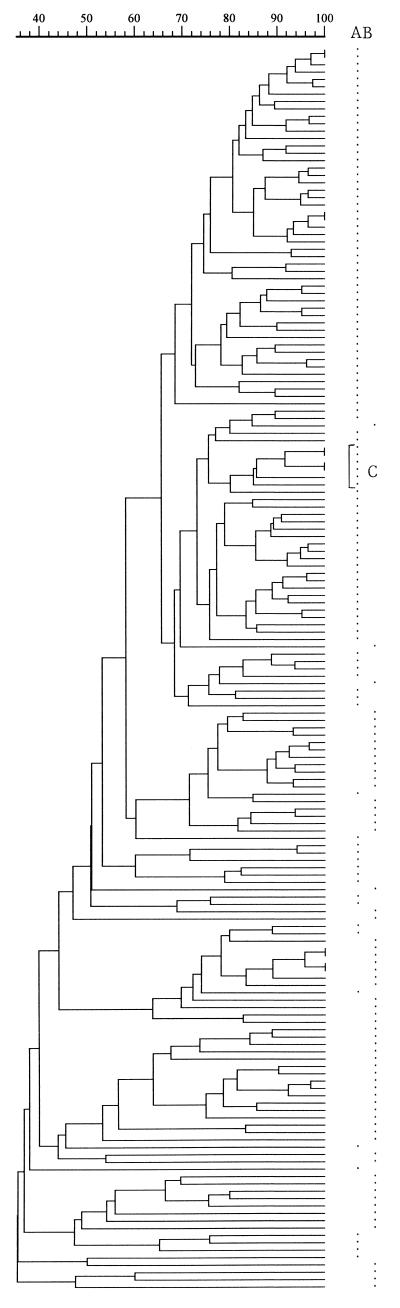
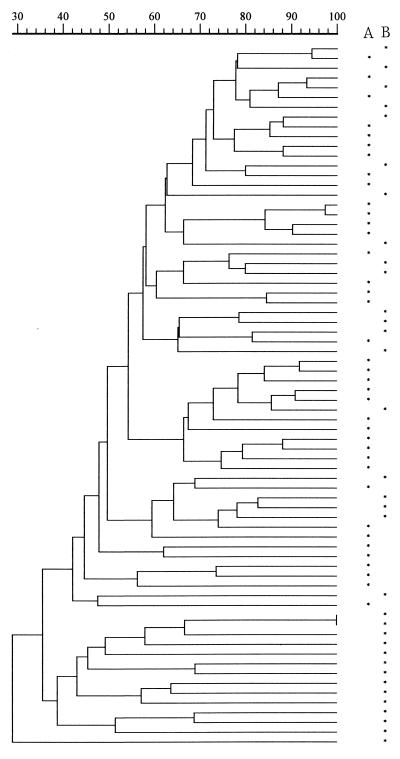
Altogether, 10 gels with 20 to 24 test lanes and five to six lambda ladders and 3 gels with 12 test lanes and three lambda ladders were run. Of the 67 lanes of lambda DNA, 1 lane from each of 7 gels was not included as a reference track because of an unsatisfactory image. Of the remaining 60 lanes, 59 shared a coefficient of similarity of ≥90% and 1 shared 85%. These 60 lanes were used as reference tracks for both Shigella species. Figures 1 and 2 are the dendrograms of S. flexneri and S. sonnei, respectively, constructed with GelCompar. Two of the three S. flexneri isolates from 1996 clustered at 100% on the same gel and at 90% with their duplicates on a separate gel. All three isolates clustered with their duplicates at 85% (Fig. 1).
FIG. 1.
Dendrogram based on Dice coefficients of similarity for 100 S. flexneri isolates collected in 1994 and 1995 and 63 collected in 1986 and 1987, together with duplicates of the 3 isolates collected in 1996. The dendrogram was constructed by UPGMA (GelCompar). Asterisks in column A and column B indicate isolates collected in 1994 and 1995 and those collected in 1986 and 1987, respectively. The bracketed isolates (C) are duplicates of the three 1996 isolates.
FIG. 2.
Dendrogram based on Dice coefficients of similarity for 34 S. sonnei isolates collected in 1986 and 1987 and 38 isolates collected in 1994 and 1995. The dendrogram was constructed by UPGMA (GelCompar). Asterisks in column A and column B indicate isolates collected in 1994 and 1995 and those collected in 1986 and 1987, respectively.
Isolates that showed a coefficient of similarity ≥75% (a difference of four bands) were grouped as one pulsotype (PT75) and labeled numerically. Similarly, those showing a coefficient of similarity ≥50% (a difference of seven to eight bands or more) were grouped as one pulsotype and labeled PT50s. Table 1 shows the distribution of S. flexneri isolates and their corresponding serotypes in different PT75s. With two exceptions, all PT75s contained one serotype only. Comparing the distribution of isolates from the two collection periods, there was a significant difference in PT75s as well as in PT50s (Mann-Whitney U test, P = 0.0001). Of the 27 PT75s among the isolates from 1994 and 1995, PT75s 1, 2, and 4 contained 67 of the 100 isolates, sharing a coefficient of similarity of ≥68% (a difference of four to five bands) (Fig. 1). Of the 44 isolates from 1994 and 1995 showing the same pattern of antimicrobial resistance to seven agents (1), 36 (82%) were also found in these three PT75s. For S. sonnei isolates, a similar comparison shows there was a significant difference in the distribution of isolates in different pulsotypes in PT50 only (P = 0.0007) (Table 2).
TABLE 1.
Correlation between serotype and pulsotype distribution of 163 S. flexneri isolates
| Isolate charac- teristics | No. of S. flexneri isolates in PT75 pulsotype:
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7a | 8a | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21–40b | 41–47b | Total | |
| Collection period | |||||||||||||||||||||||
| 1986–87c | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 12 | 4 | 0 | 0 | 0 | 7 | 2 | 3 | 8 | 2 | 3 | 0 | 20 | 0 | 63 |
| 1994–95 | 33 | 13 | 3 | 21 | 4 | 2 | 2 | 2 | 1 | 0 | 2 | 3 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 7 | 100 |
| Serotype | |||||||||||||||||||||||
| I | 9 | 2 | 3 | 2 | 9 | 1 | 26 | ||||||||||||||||
| II | 33 | 13 | 3 | 21 | 5 | 3 | 1 | 1 | 13 | 4 | 2 | 3 | 3 | 8 | 5 | 3 | 121 | ||||||
| Others | 1 | 1 | 2 | 3 | 6 | 3 | 16 | ||||||||||||||||
Pulsotype contained more than one serotype.
There was a single isolate in each of these pulsotypes.
Mann-Whitney U test, P = 0.0001.
TABLE 2.
Distribution of S. sonnei isolates in different pulsotypes at Dice coefficient of 50%
| Collection period | No. of isolates in PT50 pulsotype:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | Total | |
| 1986–87a | 15 | 4 | 0 | 0 | 1 | 0 | 4 | 1 | 2 | 3 | 3 | 1 | 34 |
| 1994–95 | 29 | 3 | 2 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 38 |
Mann-Whitney U test, P = 0.0007.
The relative genetic distances of isolates from the two collection periods were calculated as described above and were compared. For S. flexneri isolates, the mean relative genetic distance for those collected in 1986 and 1987 was 0.016 and the 90th percentile was ≤0.089. The corresponding figures for isolates collected in 1994 and 1995 were 0.013 and 0.045 (P < 0.0001), respectively. Because of the large number of isolates from 1994 and 1995 in PT75s 1, 2, and 4, the analysis was also performed with the 67 isolates in these three groups excluded. The significant difference between the two collections remained (P < 0.0001). For S. sonnei, the mean relative genetic distance for isolates from 1986 and 1987 was 0.046, and the 90th percentile was ≤0.078. The corresponding figures for isolates collected in 1994 and 1995 were 0.030 and 0.044 (P < 0.001), respectively. For both species, the relative genetic distances between isolates of the earlier collection period were significantly larger and therefore more diverse than those of the later period.
DISCUSSION
For the estimation of chromosomal genotypic diversity and relationships, methods commonly used by population geneticists include multilocus enzyme electrophoresis to examine protein polymorphisms or nucleotide sequencing of selected genes to examine DNA polymorphisms (9). These techniques are not readily performable in a typical clinical laboratory like ours which nonetheless has collected and typed a substantial number of human pathogens over the years for the investigation of outbreaks of infections. For these investigations, PFGE is now accepted as an effective tool (13). With the introduction of bench top PFGE apparatuses and computer programs to handle large numbers of gel images, it may be possible for typical clinical laboratories, particularly in collaboration with each other, to consider what Tibayrenc labeled long-term epidemiology of the species (country- or continent-wide investigations lasting months or years) (20). Such investigations may contribute to studies of pathogenicity, resistance to drugs, adaptative significance, immunological patterns, and vector and host specificity of microbial genetic diversity (20). It will be interesting to see whether digital networking can further enhance surveillance in different locations.
During an outbreak of infection, the aim of bacterial typing is to provide laboratory evidence that epidemiologically related isolates are also genetically related. For this purpose, PFGE should be applied to small sets of isolates (typically ≤30) that are epidemiologically related (19). In this study, we investigated the relatedness of a large number of isolates collected 10 years apart and used PFGE to distinguish strains at the coefficient of similarity of <50% (showing a difference of seven to eight bands or more and representing three or more independent genetic events) (19). The isolates were obtained from symptomatic patients over a period of 24 months by the same laboratories serving the same population. Detailed epidemiological information normally collected in the investigation of individual outbreaks of infection is not relevant to an investigation such as ours, which looks at the long-term epidemiology.
We used Upholt’s formula to obtain an approximation of the fraction of nucleotide difference between isolates and MEGA to obtain relative genetic distances so that a quantitative comparison of the relationships among a large body of isolates could be made. The p values obtained should not be seen as precise quantitative estimates of genetic distances. As pointed out by El-Adhami et al., one of the important assumptions inherent in this type of analysis is that the changes seen in the fragment sizes and numbers result from simple point mutations which change endonuclease recognition sites. Insertions, deletions, or transpositions are not theoretically accommodated in the calculations (2). The relatedness of the bacteria may be further ascertained by using several restriction enzymes in PFGE, as has been demonstrated through the analysis of Pseudomonas species (4).
The careful control that should be exercised during PFGE cannot be overemphasized. We standardized reagents and methods of preparation and randomized isolates for the species and the year. We used five to six lanes of lambda ladder on each gel as reference tracks. Ideally, DNA plugs of a suitable control strain, prepared as a single batch, should also be included in each gel (19).
Of the 100 isolates of S. flexneri collected in 1994 and 1995 67 (65%) were found in three PT75s which shared a coefficient of similarity at ≥68% and included 82% of the 44 isolates with the same pattern of resistance to seven antimicrobials (1). This suggests that they may have belonged to the same clone(s). A small number of the 1994 and 1995 isolates (10%) shared the same PT75s (pulsotypes 5, 6, 9, and 14) with some of the 1986 and 1987 isolates (Table 1). Further work, however, is necessary to confirm such parentages. It should also be borne in mind that over time there is an opportunity for isolates of the same parentage to exhibit differences in phenotypic or actual genotypic variations.
With two exceptions, individual PT75s contained only one serotype of S. flexneri (Table 1). Comparison of serotypes and ribotypes showed that different ribotypes belonged to the same serotype but also that similar ribotypes might belong to different serotypes (3). PFGE has also been used successfully to identify genetic subtypes among small numbers of both epidemiologically related and unrelated S. sonnei and S. flexneri isolates (12, 18). Other genetic methods have been used, and the extent of genetic variability varied with the molecular method used (3, 5, 10, 11, 14, 16).
Our results demonstrate that it is possible to analyze a large amount of PFGE information about Shigella isolates when it is obtained under controlled conditions. It remains to be seen whether intra- or interlaboratory results of PFGE from typical clinical laboratories could become comparable through the use of carefully standardized procedures. Such data would enhance surveillance and give indications for further work on various aspects of bacterial pathogenicity as suggested by Tibayrenc (20).
ACKNOWLEDGMENT
This project was supported by grant 2040496 from the University Research Grants Council, Hong Kong.
REFERENCES
- 1.Chu Y-W, Houang E T S, Lyon D J, Ling J M, Ng T-K, Cheng A F B. Antimicrobial resistance in Shigella flexneri and Shigella sonnei in Hong Kong, 1986 to 1995. Antimicrob Agents Chemother. 1998;42:440–443. doi: 10.1128/aac.42.2.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Adhami W, Pobers L, Vickery A, Inglis B, Gibbs A, Stewart P R. Epidemiological analysis of a methicillin-resistant Staphylococcus aureus outbreak using restriction fragment length polymorphisms of genomic DNA. J Gen Microbiol. 1991;137:2713–2720. doi: 10.1099/00221287-137-12-2713. [DOI] [PubMed] [Google Scholar]
- 3.Faruque S M, Haider K, Rahman M M, Alim A R M A, Ahmad Q S, Albert M J, Sack R B. Differentiation of Shigella flexneri strains by rRNA gene restriction patterns. J Clin Microbiol. 1992;30:2996–2999. doi: 10.1128/jcm.30.11.2996-2999.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grothues D, Tummler B. New approaches in genome analysis by pulsed-field gel electrophoresis: application to the analysis of Pseudomonas species. Mol Microbiol. 1991;5:2763–2776. doi: 10.1111/j.1365-2958.1991.tb01985.x. [DOI] [PubMed] [Google Scholar]
- 5.Hinojosa-Ahumada M, Swaminathan B, Hunter S B, Cameron D N, Kiehlbauch J A, Wachsmuth I K, Strockbine N A. Restriction fragment length polymorphisms in rRNA operons for subtyping Shigella sonnei. J Clin Microbiol. 1991;29:2380–2384. doi: 10.1128/jcm.29.11.2380-2384.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jorgensen M, Givney R, Pegler M, Vickery A, Funnell G. Typing multidrug-resistant Staphylococcus aureus: conflicting epidemiological data produced by genotypic and phenotypic methods clarified by phylogenetic analysis. J Clin Microbiol. 1996;34:398–403. doi: 10.1128/jcm.34.2.398-403.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufmann M E, Pitt T L. Methods in practical laboratory bacteriology. Boca Raton, Fla: CRC Press; 1994. Pulsed-field gel electrophoresis of bacterial DNA; pp. 83–92. [Google Scholar]
- 8.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis, version 1.01. University Park, Penn: The Pennsylvania State University; 1993. [Google Scholar]
- 9.Li J, Smith N H, Nelson K, Crichton P B, Old D C, Whittam T S, Selander R K. Evolutionary origin and radiation of the avian-adapted non-motile salmonellae. J Med Microbiol. 1993;38:129–139. doi: 10.1099/00222615-38-2-129. [DOI] [PubMed] [Google Scholar]
- 10.Ling J M, Shaw P C, Kam K M, Cheng A F, French G L. Molecular studies of plasmids of multiply-resistant Shigella spp. in Hong Kong. Epidemiol Infect. 1993;110:437–446. doi: 10.1017/s095026880005086x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Litwin C M, Storm A L, Chipowsky S, Ryan K J. Molecular epidemiology of Shigella infections: plasmid profiles, serotype correlation, and restriction endonuclease analysis. J Clin Microbiol. 1991;29:104–108. doi: 10.1128/jcm.29.1.104-108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu P Y-F, Lau Y-J, Hu B-S, Shyr J-M, Shi Z-Y, Tsai W-S, Lin Y-H, Tseng C-Y. Analysis of clonal relationships among isolates of Shigella sonnei by different molecular typing methods. J Clin Microbiol. 1995;33:1779–1783. doi: 10.1128/jcm.33.7.1779-1783.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maslow J, Mullingan M E. Epidemiologic typing systems. Infect Control Hosp Epidemiol. 1996;17:595–604. doi: 10.1086/647395. [DOI] [PubMed] [Google Scholar]
- 14.Nastasi A, Pignato S, Mammina C, Giammanco G. rRNA gene restriction patterns and biotypes of Shigella sonnei. Epidemiol Infect. 1993;110:23–30. doi: 10.1017/s0950268800050640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nei M, Li W H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Preston M A, Borczyk A A. Genetic variability and molecular typing of Shigella sonnei strains isolated in Canada. J Clin Microbiol. 1994;32:1427–1430. doi: 10.1128/jcm.32.6.1427-1430.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 18.Soldati L, Piffaretti J C. Molecular typing of Shigella strains using pulsed field gel electrophoresis and genome hybridization with insertion sequences. Res Microbiol. 1991;142:489–498. doi: 10.1016/0923-2508(91)90182-a. [DOI] [PubMed] [Google Scholar]
- 19.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tibayrenc M. Towards a unified evolutionary genetics of micro-organisms. Annu Rev Microbiol. 1996;50:401–429. doi: 10.1146/annurev.micro.50.1.401. [DOI] [PubMed] [Google Scholar]
- 21.Upholt W B. Estimation of DNA sequence divergence from comparison of restriction endonuclease digests. Nucleic Acids Res. 1977;4:1257–1265. doi: 10.1093/nar/4.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]