Abstract
There is a clear need for the development of a rapid and reliable test for the identification of Candida dubliniensis and for the discrimination of this species from Candida albicans. In the present study we have investigated the potential use of C. dubliniensis-specific antigens as a basis for its identification. We produced an anti-C. dubliniensis serum which, after adsorption with C. albicans blastospores, was found to differentially label C. dubliniensis isolates in an indirect immunofluorescence test. In this test, the antiserum reacted with blastospores and germ tubes of C. dubliniensis and with blastospores of Candida krusei and Rhodotorula rubra but did not react with blastospores of several other Candida species including C. albicans. The antiserum also reacted with C. albicans germ tubes. The anti-C. dubliniensis adsorbed serum reacted with specific components of 25, 28, 37, 40, 52, and 62 kDa in the C. dubliniensis extract and with a variety of antigens from other yeast species. The antigens from non-C. dubliniensis yeasts showing reactivity with the anti-C. dubliniensis adsorbed serum are mostly expressed within the cell walls of these yeast species, and this reactivity does not interfere with the use of the anti-C. dubliniensis adsorbed serum in an indirect immunofluorescence test for the rapid identification of C. dubliniensis.
Oral candidiasis is one of the most prevalent clinical presentations of Candida infections, particularly in human immunodeficiency virus (HIV)-infected individuals. Although Candida albicans remains the most common cause of oral candidiasis, the incidence of disease caused by other species of Candida, including Candida krusei, Candida tropicalis, and Candida glabrata, has been increasing steadily (4, 5). The reason(s) for this epidemiological change is not clear; however, it has been suggested that the reduced susceptibility of these species to commonly used antifungal agents, such as fluconazole, may have led to their selection. Furthermore, different groups of researchers have reported the recovery of atypical C. albicans isolates from HIV-infected patients (2, 10, 20, 22). Some of these organisms have been shown to belong to a recently described species, C. dubliniensis, which appears to be mainly associated with oral disease in HIV-infected individuals (4, 23, 25). First identified as a new species in Ireland in 1995, this organism has since been identified in laboratories around the world (14, 23, 24). Although the majority of isolates so far examined have been associated with oral candidiasis and HIV infection, isolates have also been recovered from other anatomical sites and from healthy individuals (4, 11, 24).
In order to investigate the significance and incidence of C. dubliniensis in clinical disease and to determine the reasons for its recent emergence, an in-depth epidemiological analysis of this species must be performed. However, before this can occur a rapid and simple means of identification of C. dubliniensis must be made available. The development of such a technique has been hampered by the very close phenotypic and genotypic relationships between C. dubliniensis and C. albicans (24, 25). Indeed, the close similarity between these species has led to the misidentification of isolates of C. dubliniensis as C. albicans (4). At present, the most accurate means of differentiating between isolates of the two species requires the use of molecular biology-based techniques such as DNA fingerprinting with repetitive sequence-containing DNA probes, PCR, or pulsed-field gel electrophoresis (3, 4, 25). However, these techniques are not readily applicable for use with the large numbers of isolates regularly encountered in clinical mycology laboratories. Several phenotype-based methods for the identification of C. dubliniensis isolates have been described. The diagnostic characteristics used in these methods include colonial coloration on differential media, such as CHROMagar Candida, atypical carbohydrate assimilation profiles with commercially available kits, such as the API ID 32C system, and a lack of β-glucosidase activity (3, 4, 24). However, there are drawbacks with many of these techniques, since they can be unreliable and/or time-consuming (14, 21, 24). Recently, it was shown that C. dubliniensis can be readily differentiated from C. albicans on the basis of its inability to grow at 45°C (14). Although easy to perform, this test requires that isolates be incubated for 24 to 48 h before the isolates can be discriminated.
In order to facilitate the development of a rapid and reliable test for the identification of C. dubliniensis from clinical specimens, the presence of antigenic differences between C. dubliniensis and C. albicans was investigated. In this study, a rabbit polyclonal anti-C. dubliniensis antiserum was used to identify cell wall antigens specific for C. dubliniensis. The antigenic composition of C. dubliniensis was found to be very similar to that of C. albicans. However, the use of the anti-C. dubliniensis antiserum adsorbed with C. albicans blastospores allowed a clear-cut differentiation between C. dubliniensis and C. albicans by indirect immunofluorescence.
MATERIALS AND METHODS
Yeast strains and clinical isolates.
The reference strains used in this study are listed in Table 1, and the clinical isolates of C. dubliniensis and C. albicans used in this study are listed in Table 2.
TABLE 1.
Reference yeast strains and their reactivities by indirect immunofluorescence with the rabbit hyperimmune serum raised against C. dubliniensis NCPF 3949, the serum adsorbed with C. albicansa blastospores, and the anti-C. dubliniensis NCPF 3949 cell wall surface-specific serum
| Species | Strain nameb | Reactivity
|
||
|---|---|---|---|---|
| Unadsorbed antiserum | Adsorbed antiserum | Surface-specific antiserum | ||
| Candida dubliniensis | NCPF 3949, CD57c | + | + | + |
| Candida albicans | NCPF 3153, NCPF 3156, NCPF 3116, NCPF 3119, NCPF 3155, NCPF 3254, ATCC 60458, ATCC 64548, ATCC 76615, ATCC 90029 | + | − | − |
| Candida stellatoidea type I | ATCC 11006 | + | − | − |
| Candida stellatoidea type II | ATCC 20408d | + | − | − |
| Candida glabrata | NCPF 3203, ATCC 90030, UPV94387, UPV94308 | + | − | − |
| Candida guilliermondii | NCPF 3099, ATCC 22995, ATCC 58070 | + | − | − |
| Candida krusei | NCPF 3100, UPV94823, UPV94813, UPV95773 | + | +/− | +/− |
| Candida parapsilosis | NCPF 3103, 3104, ATCC 90018 | + | − | − |
| Candida tropicalis | NCPF 3111, UPV94800, UPV94840, UPV96080 | + | − | − |
| Rhodotorula rubra | UPV94921, UPV95093, UPV95487 | + | + | + |
| Trichosporon beigelii | UPV97398 | + | − | − |
| Saccharomyces cerevisiae | NCPF 3191, CECT1478, CECT1678 | + | − | − |
| Cryptococcus neoformans | ATCC 90112, UPV94448, UPV94450 | + | − | − |
C. albicans NCPF 3153.
The reference strains used in this study were obtained from the British National Collection of Pathogenic Fungi, Bristol, United Kingdom (NCPF); the American Type Culture Collection, Rockville, Md. (ATCC); the Colección Española de Cultivos Tipo, Valencia, Spain (CECT); the Universidad del País Vasco strain collection, Bilbao, Spain (UPV); and the Dublin University strain collection (CD).
CD57 is a vaginal isolate of C. dubliniensis (11).
ATCC 20408 is listed in the American Type Culture Collection catalog under C. albicans strains.
TABLE 2.
C. dubliniensis and C. albicans clinical isolates
| Species and country of isolation | No. of isolatesa | Specimen | HIV status of subjects sampled | Reference or source |
|---|---|---|---|---|
| C. dubliniensis | ||||
| Argentina | 1 | Oral cavity | + | 23 |
| Australia | 5 | Oral cavity | + | 10, 25 |
| Belgium | 5 | Oral cavity | + | 14 |
| Canada | 6 | Oral cavity | + | 14 |
| Finland | 1 | Oral cavity | + | 14 |
| Germany | 4 | Oral cavity | + | 14 |
| Greece | 1 | Oral cavity | + | 14 |
| Ireland | 23 | Oral cavity | + | 4, 25 |
| Ireland | 4 | Oral cavity | − | 4 |
| Ireland | 4 | Vaginal | − | 11, 14 |
| Spain | 10 | Oral cavity | + | 14, this study |
| Switzerland | 6 | Oral cavity | + | 2, 23 |
| United Kingdom | 2 | Oral cavity | + | 14, 23 |
| United Kingdom | 7 | Oral cavity | − | 14 |
| United Kingdom | 4 | Miscellaneousb | − | 14, 23 |
| Total | 83 | |||
| C. albicans | ||||
| Australia | 1 | Oral cavity | + | 25 |
| Hong Kong | 6 | Oral cavity | + | 14 |
| Ireland | 12 | Oral cavity | + | 14 |
| 16 | Oral cavity | − | 14 | |
| United Kingdom | 9 | Oral cavity | − | 14 |
| Total | 44 |
Each isolate was recovered from a separate individual.
These included one isolate each from a blood culture and fecal, sputum, and postmortem lung specimens.
Culture conditions and preparation of antigens.
C. dubliniensis and C. albicans strains were routinely grown in medium 199 (Sigma Chemical Co., St. Louis, Mo.) as described previously (17). Briefly, 48-h-old blastospores grown at 24°C on glucose-yeast extract-agar plates were transferred to Erlenmeyer flasks containing medium 199 at 5 × 107 blastospores/ml, and the flasks were incubated at 24°C for 18 h in a rotary shaker set at 100 rpm. Following incubation, the blastospores were harvested by centrifugation, inoculated into fresh medium, and incubated with shaking as before at 24°C for 24 h to obtain blastospores. The same conditions were used to obtain blastospores of the other yeast species studied. C. albicans germ tubes were induced by incubation of similar cultures in medium 199 at 37°C for 4 h. Since C. dubliniensis failed to produce germ tubes under these conditions, they were obtained by incubation in horse serum (9). The cell walls of C. dubliniensis and the other yeast species studied were extracted in the presence of dithiothreitol (DTT; Sigma) as reported previously (15). To obtain formalin-killed blastospores, the cells of the different Candida species were resuspended in a 4% (vol/vol) formaldehyde solution in phosphate-buffered saline (PBS) and were incubated at 4°C for 18 h. Assessment of the viability of the cells was determined by incubating the suspension on Sabouraud agar plates (Difco, Detroit, Mich.).
Antisera.
Two separate female New Zealand White rabbits with an initial weight of from 2 to 2.5 kg were subcutaneously inoculated with 5 × 108 formalin-killed cells of C. dubliniensis NCPF 3949 (oral isolate and type strain) (25) and C. dubliniensis CD57 (vaginal isolate) (11), respectively, which had been grown at 37°C and resuspended in 0.5 ml of saline and 0.5 ml of Freund’s adjuvant. Immunizations were repeated at weekly intervals. Preimmune sera were obtained from each rabbit before the immunization, and hyperimmune sera were obtained 5 weeks after the start of the immunization.
Aliquots of the hyperimmune anti-C. dubliniensis serum obtained from each rabbit were each adsorbed with C. albicans NCPF 3153 blastospores by previously described methods (17). Briefly, immune sera were mixed and incubated with an equal volume of a formalin-killed blastospore suspension in saline (1010 blastospores/ml) for 2 h at room temperature. After incubation, the suspension was centrifuged and the supernatant was recovered. This process was repeated up to three times. Additionally, antibodies directed against C. dubliniensis cell wall surface components were obtained by a modification of a previously described method (15). Briefly, 0.5 ml of the anti-C. dubliniensis NCPF 3949 serum adsorbed three times with C. albicans blastospores was incubated with 1011 formalin-killed C. dubliniensis NCPF 3949 blastospores for 2 h at room temperature in PBS buffer. Unattached serum proteins were removed by three washes in PBS, and the attached antibodies were then eluted by treatment with 2.5 M NaI in PBS for 1 h at room temperature. Blastospores were harvested by centrifugation, and the supernatant was dialyzed against PBS for 24 h at 4°C.
Immunofluorescence.
Indirect immunofluorescence assay (IFA) was carried out as described previously (1). Briefly, the blastospores from the different yeast species studied were grown on Sabouraud agar plates for 48 h at 24 or 37°C, resuspended in PBS at a cell density of 106 cells/ml, and placed on Teflon-coated immunofluorescence slides. The slides were incubated with the anti-C. dubliniensis serum diluted 1:5 in PBS supplemented with Evans blue (0.05% [wt/vol]) and Tween 20 (0.05% [vol/vol]) and washed, and the reacting antibodies were revealed by incubation with biotin-conjugated anti-rabbit immunoglobulin G (Sigma), followed by incubation with fluorescein isothiocyanate-conjugated ExtrAvidin (Sigma). Slides were mounted with carbonate-glycerol mounting fluid and examined with an epifluorescence microscope.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of yeast cell wall extracts prepared in the presence of DTT was performed by the method of Laemmli (8) in a minigel system (Bio-Rad Laboratories, Richmond, Calif.). Electrophoresis was carried out in 10% (wt/vol) gels at 200 V. Subsequently, the proteins contained in the gels were electrophoretically transferred to a nitrocellulose membrane (Bio-Rad) with a fast blot system (Biometra, Göttingen, Germany). After the transfer, the nitrocellulose membranes were blocked in 10% (wt/vol) nonfat dry milk in Tris-buffered saline, incubated with the anti-C. dubliniensis sera, and finally incubated again with peroxidase-conjugated anti-rabbit immunoglobulin G antibodies (Sigma). Immunoreactive bands were visualized with 4-chloro-1-naphthol by standard procedures.
RESULTS
Reactivities of the anti-C. dubliniensis sera by indirect immunofluorescence.
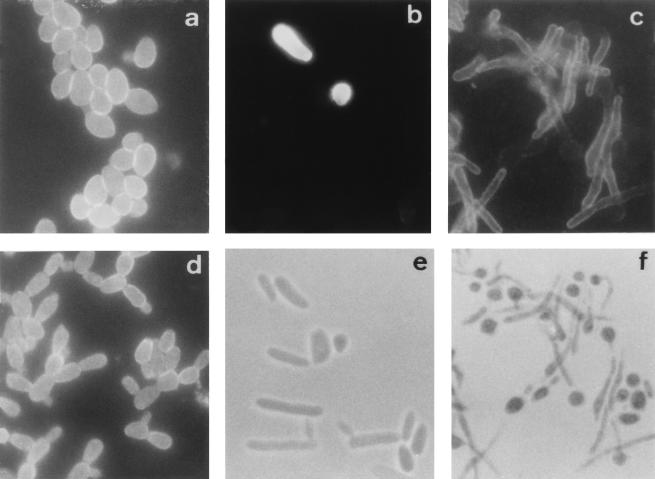
The reactivities of the preimmune sera and the hyperimmune antisera from rabbits immunized with C. dubliniensis NCPF 3949 and C. dubliniensis CD57 were first studied by IFA with reference strains of different yeast species, including several Candida species, Trichosporon beigelii, Saccharomyces cerevisiae, Cryptococcus neoformans, and Rhodotorula rubra (Table 1). The reactivities of the preimmune sera with the Candida species studied were negative. In preliminary experiments, both hyperimmune antisera gave identical results and the antiserum against C. dubliniensis NCPF 3949 was selected for further studies. The unadsorbed antiserum reacted with all the yeast species studied, showing a homogeneous reactivity. The reactivity of the anti-C. dubliniensis serum adsorbed with formalin-killed C. albicans NCPF 3153 blastospores was initially studied with cells grown at 37°C on Sabouraud agar. With this culture medium, all species studied grew as blastospores and the antiserum reacted only with C. dubliniensis, C. krusei, and R. rubra. The expression of antigens reacting with the anti-C. dubliniensis adsorbed serum in the C. dubliniensis and R. rubra strains studied (Table 1) was homogeneous, and most cells showed fluorescence (Fig. 1). However, the reactivity of the adsorbed antiserum with the C. krusei strains studied (Table 1) was very heterogeneous, with only 10% of the cells showing reactivity (Fig. 1).
FIG. 1.
Immunofluorescence photographs of C. dubliniensis NCPF 3949 blastospores (a) and germ tubes (d) stained with the anti-C. dubliniensis adsorbed serum. Immunofluorescence (b and c) and phase-contrast (e and f) photographs, respectively, of the same microscopic fields, show C. krusei NCPF 3100 blastospores (b and e) and C. albicans NCPF 3153 germ tubes (c and f) stained with the anti-C. dubliniensis adsorbed serum. Magnification, ×1,000.
When the yeast species studied (Table 1) were grown at 37°C in medium 199, all species except C. albicans grew as blastospores. C. albicans strains produced a mixture of blastospores and germ tubes. Since the C. dubliniensis strains examined did not produce germ tubes in medium 199, these were obtained by incubation in horse serum. The adsorbed antiserum reacted with blastospores of C. dubliniensis, C. krusei, and R. rubra as well as with C. dubliniensis and C. albicans germ tubes (Fig. 1). Despite the reactivity of C. albicans germ tubes with the anti-C. dubliniensis adsorbed serum, the blastospores attached to them remained negative. Adsorption of the anti-C. dubliniensis antiserum with C. albicans germ tubes resulted in a loss of its reactivity with C. dubliniensis. The anti-C. dubliniensis cell wall surface-specific serum showed a reactivity by IFA identical to that shown by the anti-C. dubliniensis adsorbed serum (Table 1).
The ability of the anti-C. dubliniensis adsorbed serum to differentiate clinical isolates of C. dubliniensis from clinical isolates of C. albicans by IFA was also confirmed with a group of 83 C. dubliniensis and 44 C. albicans isolates (Table 2). These isolates were recovered mainly from the oral cavities of patients from disparate geographical locations and had been identified by both phenotypic and genotypic methods (23, 25). A number of isolates from nonoral sources were also studied (Table 2). The antiserum allowed the rapid and precise discrimination of the isolates, since all of the C. dubliniensis isolates were positive by immunofluorescence assays with the antiserum, while all of the C. albicans isolates were negative.
Identification of C. dubliniensis antigens reactive with the anti-C. dubliniensis serum.
By immunoblotting, the unadsorbed anti-C. dubliniensis serum reacted with antigens in DTT extracts from both C. dubliniensis NCPF 3949 and C. albicans NCPF 3153 strains spanning a wide range of molecular weights (Fig. 2A). Most antigenic components present in the extracts from both species had similar molecular masses, although differences in banding intensity were observed for some components. Despite the common reactivity observed between C. dubliniensis and C. albicans, the antiserum reacted with three components of 25, 35, and 50 kDa in the C. albicans extract which were not observed in the C. dubliniensis extract, while the C. dubliniensis extract contained 52- and 62-kDa components which were not observed in the C. albicans extract. The unadsorbed antiserum also reacted with a variety of components in cell wall extracts from the rest of the yeast species studied (data not shown).
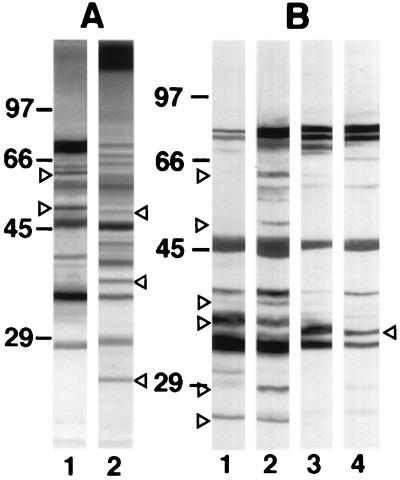
FIG. 2.
Western blots of 10% slab gels loaded with extracts from C. dubliniensis NCPF 3949 blastospores grown at 37°C (lanes A1 and B2) or 24°C (lane B1) and C. albicans NCPF 3153 germ tubes (lanes A2 and B4) or blastospores (lane B3) stained with the anti-C. dubliniensis serum (A) or with the anti-C. dubliniensis adsorbed serum (B). The molecular masses of standard proteins (in kilodaltons) are listed to the left of the gels. Relevant antigenic bands are indicated by arrowheads.
After the adsorption with C. albicans blastospores, the anti-C. dubliniensis serum showed a decrease in reactivity with components in the C. albicans extract but not with components in the C. dubliniensis extract (Fig. 2B). The adsorbed antiserum showed a higher reactivity with extracts from both C. dubliniensis NCPF 3949 and C. albicans NCPF 3153 cells grown at 37°C than with extracts from the cells grown at 24°C. Furthermore, the adsorbed antiserum stained several components of 25, 28, 37, 40, 52, and 62 kDa in the C. dubliniensis extracts which were not stained in the C. albicans extracts. In contrast, a component of 35 kDa present in the C. albicans extracts was not observed in the extracts from C. dubliniensis cells grown at both 24 and 37°C. Similar results were observed when extracts from C. dubliniensis CD57 and C. albicans ATCC 60458 were examined (data not shown).
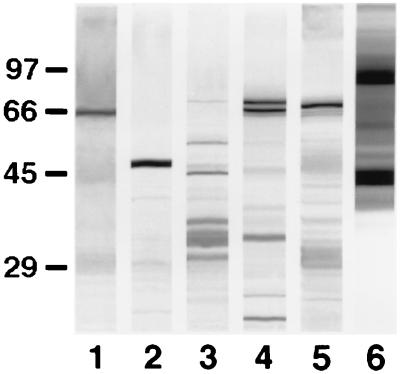
The anti-C. dubliniensis adsorbed serum also reacted by immunoblotting with antigens from other yeast species (Fig. 3). Major antigens comprised a 66-kDa component in the C. krusei NCPF 3100 extract, a 47-kDa component in the C. tropicalis NCPF 3111 extract, several components ranging between 29 and 68 kDa in the Candida parapsilosis NCPF 3104 extract, two components of 66 and 68 kDa in the Candida guilliermondii NCPF 3099 extract, a component of 67 kDa in the C. glabrata NCPF 3203 extract, and several components with molecular masses of >40 kDa in the R. rubra UPV94921 extract.
FIG. 3.
Western blots of 10% slab gels loaded with extracts from C. krusei NCPF 3100 (lane 1), C. tropicalis NCPF 3111 (lane 2), C. parapsilosis NCPF 3104 (lane 3), C. guilliermondii NCPF 3099 (lane 4), C. glabrata NCPF 3203 (lane 5), and R. rubra UPV94921 (lane 6) stained with the anti-C. dubliniensis adsorbed serum. The molecular masses of standard proteins (in kilodaltons) are listed to the left of the gel.
Identification of C. dubliniensis antigens reactive with the anti-C. dubliniensis cell wall surface-specific serum.
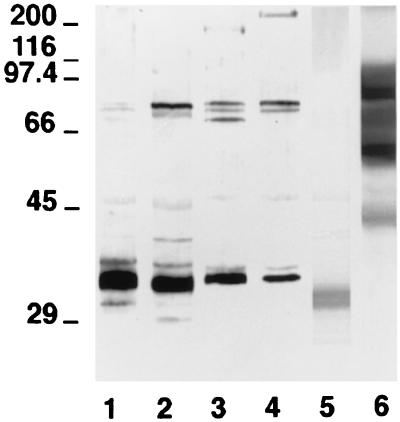
In order to identify the antigens from the cell wall surface responsible for the fluorescence observed on the C. dubliniensis cells, the reactivity of an anti-C. dubliniensis cell wall surface-specific serum was studied by immunoblotting with cell wall extracts from C. dubliniensis, C. albicans, C. krusei, and R. rubra. The anti-C. dubliniensis cell wall surface-specific serum reacted with a component of 33 kDa in the extract from C. dubliniensis cells grown at 24°C and with components of 32 and 77 kDa in the extract from C. dubliniensis cells grown at 37°C (Fig. 4). This antiserum also stained four components of 33, 70, 74, 77, and 180 kDa in the C. albicans blastospore extract and with components of 33, 74, 77, and >200 kDa in the C. albicans germ tube extract. The anti-C. dubliniensis cell wall surface-specific serum also stained a component of 30 kDa in the C. krusei NCPF 3100 extract and several components ranging from 43 to 100 kDa in the R. rubra UPV94921 extract. In one experiment, the anti-C. dubliniensis cell wall surface-specific serum was diluted in order to identify the most reactive components present in the extracts. Under these conditions, only the component of 33 kDa present in the C. dubliniensis NCPF 3949 extract and the components ranging from 60 to 100 kDa present in the R. rubra UPV94921 extract were stained (data not shown).
FIG. 4.
Western blots of 10% (wt/vol) acrylamide slab gels loaded with extracts from C. dubliniensis NCPF 3949 grown at 24°C (lane 1) and 37°C (lane 2), C. albicans NCPF 3153 grown at 24°C (lane 3) and 37°C (lane 4), C. krusei NCPF 3100 (lane 5), and R. rubra UPV94921 (lane 6) stained with the anti-C. dubliniensis cell wall surface-specific serum. The molecular masses of standard proteins (in kilodaltons) are listed to the left of the gel.
DISCUSSION
Because species of Candida other than C. albicans continue to be recovered from patients with infection with increasing frequency, it is clear that routine diagnostic laboratories should be in a position to identify these species rapidly and accurately. Precise identification of these species will allow epidemiological studies to be conducted and may be of benefit in selecting the most effective therapies (12, 13). However, Candida species identification can be time-consuming and inaccurate. This is further complicated by the many anomalies in candidal classification (4), particularly with the recent identification of new Candida species such as C. dubliniensis and C. fermentatii (19, 25). As far as possible, species identification techniques should be easy to perform, rapid, reliable, inexpensive, and applicable to a large volume of isolates. In this study, we attempted to develop such a technique for the identification of clinical C. dubliniensis isolates. Reliable phenotypic identification of C. dubliniensis has proved to be problematic (21, 24). Although C. dubliniensis and C. albicans are readily distinguishable at the genetic level, the techniques capable of detecting the discriminatory differences are not easily applicable to the large numbers of isolates tested routinely in clinical mycology laboratories. Furthermore, the phenotypic similarities of the two species, including the ability to produce germ tubes and chlamydospores, has hampered the development of a rapid phenotypic test that can discriminate between isolates of both species (24).
In this paper, we present evidence, for the first time, of the existence of antigenic differences between C. dubliniensis and C. albicans, thus providing the basis for the development of a rapid identification test. Anti-C. dubliniensis sera were raised in rabbits by inoculation with formalin-killed strains of C. dubliniensis. In order to remove antibodies raised against common antigens present on the cell wall surfaces of C. dubliniensis and C. albicans, the antisera were adsorbed with C. albicans blastospores, a technique originally used by Hasenclever and Mitchell (7) to identify the existence of two serotypes in C. albicans. These adsorbed sera were then investigated for their ability to identify C. dubliniensis in indirect immunofluorescence experiments.
The IFA developed with the anti-C. dubliniensis adsorbed serum takes less than 2 h to perform and correctly identified the C. dubliniensis reference strains NCPF 3949 and CD57 and a further 83 clinical isolates of C. dubliniensis which had been identified previously by both phenotypic and genotypic methods. In all cases, most cells showed fluorescence on the entire cell wall, although differences in reactivity were observed among C. dubliniensis isolates. This antiserum showed no indirect immunofluorescence with blastospores of species closely related to C. dubliniensis, such as C. albicans, Candida stellatoidea type I, C. stellatoidea type II, and C. tropicalis. However, the anti-C. dubliniensis adsorbed serum reacted with R. rubra and C. krusei, the latter showing a heterogeneous reactivity with only 10% of the cells being positive. The reactivity with the anti-C. dubliniensis adsorbed serum shown by C. krusei and R. rubra is not likely to be an important problem when identifying C. dubliniensis by indirect immunofluorescence, since R. rubra has a distinctive colony color on conventional mycological media and C. krusei can be easily differentiated from C. dubliniensis by the conventional identification methods used in the clinical microbiology laboratory such as the germ tube or Krusei color tests (6, 9) as well as by growth on CHROMagar Candida medium (18).
The anti-C. dubliniensis adsorbed serum was also used to identify the antigens responsible for the fluorescence observed on the C. dubliniensis cell wall and to study the presence of similar antigens in the cell walls of other yeasts. This antiserum reacted with several cell wall components with molecular masses ranging from 25 to 77 kDa, and when compared with the reactivity observed in the C. albicans extract, some of them seemed to be present only in the C. dubliniensis extract. Some of the components stained by the anti-C. dubliniensis adsorbed serum in the C. dubliniensis extract are likely to be located on the C. dubliniensis cell wall surface since they were stained with an anti-C. dubliniensis cell wall surface-specific serum. Among these antigens, the component of 33 kDa seems to be the most reactive with the anti-C. dubliniensis cell wall surface-specific serum, and it may be a good candidate for use in the production of monoclonal antibodies to be used in the identification of C. dubliniensis.
In addition to the reactivity observed with C. dubliniensis, the anti-C. dubliniensis adsorbed serum also reacted with other yeast species. On the basis of their reactivities by both indirect immunofluorescence and Western blotting, we can speculate about the distribution of the antigens in the cell wall. Since C. albicans blastospores did not react by indirect immunofluorescence with either the anti-C. dubliniensis adsorbed serum or the anti-C. dubliniensis cell wall surface-specific serum, the antigenic bands observed by Western blotting in the extract from C. albicans blastospores stained with these antisera must be located in the inner layers of the cell wall. However, the antigen of >200 kDa observed in the C. albicans germ tube extracts is likely to be located on the germ tube cell wall surface since the cell wall of C. albicans germ tubes is stained by indirect immunofluorescence with the anti-C. dubliniensis adsorbed serum and the adsorption of the anti-C. dubliniensis antiserum with C. albicans germ tubes abolishes the reactivity of this antiserum with C. dubliniensis. The distribution of this antigen in the cell wall of C. albicans is in agreement with that shown by the previously described type I antigens, which are specifically expressed on the germ tube cell wall surface (16). The component of 30 kDa and several components ranging from 43 to 100 kDa are likely to be located on the cell wall surface of C. krusei and R. rubra, respectively, while the components from C. tropicalis, C. parapsilosis, C. guilliermondii, and C. glabrata extracts reacting with the anti-C. dubliniensis adsorbed serum are located within the cell wall.
In conclusion, our results indicate that C. dubliniensis and C. albicans share a group of cell wall antigens and that a specific anti-C. dubliniensis serum can be prepared after removal of the antibodies against the common antigens present on the cell wall surfaces of both species. The adsorbed anti-C. dubliniensis serum reacts with several components expressed specifically on the C. dubliniensis cell wall surface. These antigens or other antigens showing cross-reactivity with them are expressed on the cell walls of other Candida species and R. rubra, but this reactivity does not interfere with the use of the anti-C. dubliniensis adsorbed serum in an indirect immunofluorescence test for the rapid identification of C. dubliniensis.
ACKNOWLEDGMENTS
Research performed in Bilbao was supported by grant UPV 093.327-eb131/96 from the Universidad del País Vasco and grant EX97/4 from the Departamento de Educación, Universidades e Investigación del Gobierno Vasco. Research performed in Dublin was supported by Irish Health Research Board grant 41/96 and by the School of Dental Science, Trinity College, Dublin.
REFERENCES
- 1.Barturen B, Bikandi J, San Millán R, Moragues M D, Regúlez P, Quindós G, Pontón J. Variability in expression of antigens responsible for serotype specificity in Candida albicans. Microbiology. 1995;141:1535–1543. doi: 10.1099/13500872-141-7-1535. [DOI] [PubMed] [Google Scholar]
- 2.Boerlin P, Boerlin-Petzold F, Durussel C, Addo M, Pagani J-L, Chave J-P, Bille J. Cluster of atypical Candida isolates from a group of human immunodeficiency virus-positive drug users. J Clin Microbiol. 1995;30:1129–1135. doi: 10.1128/jcm.33.5.1129-1135.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coleman, D., D. Sullivan, K. Haynes, M. Henman, D. Shanley, D. Bennett, G. Moran, C. McCreary, L. O’Neill, and B. Harrington. 1997. Molecular and phenotypic analysis of Candida dubliniensis: a recently identified species linked with oral candidosis in HIV-infected and AIDS patients. Oral Dis. 3(Suppl. 1):S96–S101. [DOI] [PubMed]
- 4.Coleman D C, Sullivan D J, Bennett D E, Moran G P, Barry H J, Shanley D B. Candidiasis: the emergence of a novel species, Candida dubliniensis. AIDS. 1997;11:557–567. doi: 10.1097/00002030-199705000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Delgado W, Aguirre J M. Las micosis orales en la era del sida. Rev Iberoam Micol. 1997;14:14–22. [PubMed] [Google Scholar]
- 6.Freydière A M, Buchaille L, Guinet R, Gille Y. Evaluation of latex reagents for rapid identification of Candida albicans and Candida krusei colonies. J Clin Microbiol. 1997;35:877–880. doi: 10.1128/jcm.35.4.877-880.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasenclever H F, Mitchell W O. Antigenic studies of Candida. I. Observation of two antigenic groups in Candida albicans. J Bacteriol. 1961;82:570–573. doi: 10.1128/jb.82.4.570-573.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 9.Mackenzie D W R. Serum tube identification of Candida albicans. J Clin Pathol. 1962;15:563–565. doi: 10.1136/jcp.15.6.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCullough M J, Ross B C, Dwyer B D, Reade P C. Genotype and phenotype of oral Candida albicans from patient infected with the human immunodeficiency virus. Microbiology. 1994;140:1195–1202. doi: 10.1099/13500872-140-5-1195. [DOI] [PubMed] [Google Scholar]
- 11.Moran G P, Sullivan D J, Henman M C, McCreary C E, Harrington B J, Shanley D B, Coleman D C. Antifungal drug susceptibility of oral Candida dubliniensis isolates from HIV-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob Agents Chemother. 1997;41:617–623. doi: 10.1128/aac.41.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfaller M A. Laboratory aids in the diagnosis of invasive candidiasis. Mycopathologia. 1992;120:65–72. doi: 10.1007/BF00578290. [DOI] [PubMed] [Google Scholar]
- 13.Pfaller M A, Rex J H, Rinaldi M G. Antifungal susceptibility testing: technical advances and potential clinical applications. Clin Infect Dis. 1997;24:776–784. doi: 10.1093/clinids/24.5.776. [DOI] [PubMed] [Google Scholar]
- 14.Pinjon E, Sullivan D, Salkin I, Shanley D, Coleman D. A simple, inexpensive, reliable method for the differentiation of Candida dubliniensis from Candida albicans. J Clin Microbiol. 1998;36:2093–2095. doi: 10.1128/jcm.36.7.2093-2095.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponton J, Jones J M. Identification of two germ tube specific cell wall antigens of Candida albicans. Infect Immun. 1986;54:864–868. doi: 10.1128/iai.54.3.864-868.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pontón J, Marot-Leblond A, Ezkurra P A, Barturen B, Robert R, Senet J M. Characterization of Candida albicans cell wall antigens with monoclonal antibodies. Infect Immun. 1993;61:4842–4847. doi: 10.1128/iai.61.11.4842-4847.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Regúlez P, Arilla M C, Bikandi J, Quindós G, Cisterna R, Pontón J. Identification of antigens reacting with anti-Candida albicans germ tube antibodies. Eur J Epidemiol. 1992;8:356–361. doi: 10.1007/BF00158568. [DOI] [PubMed] [Google Scholar]
- 18.San Millán R, Ribacoba L, Pontón J, Quindós G. Evaluation of a commercial medium for the identification of Candida species. Eur J Clin Microbiol Infect Dis. 1996;15:153–158. doi: 10.1007/BF01591489. [DOI] [PubMed] [Google Scholar]
- 19.San Millán R M, Wu L-C, Salkin I F, Lehmann P F. Clinical isolates of Candida guilliermondii included Candida fermentati. Int J Syst Bacteriol. 1997;47:385–393. doi: 10.1099/00207713-47-2-385. [DOI] [PubMed] [Google Scholar]
- 20.Schmid J, Odds F C, Wiselka M J, Nicholson K G, Soll D R. Genetic similarity and maintenance of Candida albicans strains from a group of AIDS patients, demonstrated by DNA fingerprinting. J Clin Microbiol. 1992;30:935–941. doi: 10.1128/jcm.30.4.935-941.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schoofs A, Odds F C, Colebunders R, Ieven M, Goosens H. Use of specialised isolation media for recognition and identification of Candida dubliniensis isolates from HIV-infected patients. Eur J Clin Infect Dis. 1997;16:296–300. doi: 10.1007/BF01695634. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan D, Bennett D, Henman M, Harwood P, Flint S, Mulcahy F, Shanley D, Coleman D. Oligonucleotide fingerprinting of isolates of Candida species other than C. albicans and of atypical Candida species from human immunodeficiency virus-positive and AIDS patients. J Clin Microbiol. 1993;31:2124–2133. doi: 10.1128/jcm.31.8.2124-2133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan D, Haynes K, Bille J, Boerlin P, Rodero L, Lloyd S, Henman M, Coleman D. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J Clin Microbiol. 1997;35:960–964. doi: 10.1128/jcm.35.4.960-964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan D, Coleman D. Candida dubliniensis: characteristics and identification. J Clin Microbiol. 1998;36:329–334. doi: 10.1128/jcm.36.2.329-334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan D J, Westerneng T J, Haynes K A, Bennett D E, Coleman D C. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]