Abstract
We have developed the AMPLICOR CMV Test, which is rapid and sensitive for the detection of cytomegalovirus (CMV) in plasma and cerebrospinal fluid (CSF) specimens. The test incorporated an internal control in the reaction mixture to monitor the amplification efficiency and the presence of inhibitors. The AMPLICOR CMV Test was very specific in detecting 12 clinical CMV isolates and four laboratory CMV strains tested. Cross-reactivity with 26 non-CMV pathogens was not observed. The AMPLICOR CMV Test requires only 50 μl of specimen (plasma or CSF) for processing. The performance of the AMPLICOR CMV Test was compared to those of the CMV antigenemia assay and the conventional tube culture method. Among 112 plasma specimens from 43 human immunodeficiency virus-infected patients, CMV was detected in 20 (18%) of the specimens by the AMPLICOR CMV Test, 21 (19%) of the specimens by the CMV antigenemia assay, and 10 (9%) of the specimens by culture. In CSF specimens from AIDS patients, CMV was detected in 10 of 58 (17%) specimens tested by the AMPLICOR CMV Test, 5 of 28 (18%) specimens tested by the antigen assay, and none of the 25 specimens tested by culture. While the performance of the AMPLICOR CMV Test in this study was comparable to that of the CMV antigen assay, processing of specimens by the AMPLICOR CMV Test was much simpler than that by the antigen assay; in addition, the antigen assay requires greater than 105 leukocytes from blood or 1 ml of CSF to perform the assay. Our study suggested that the AMPLICOR CMV Test could provide a rapid and sensitive assay for the detection of CMV in plasma and CSF specimens.
Cytomegalovirus (CMV) is an opportunistic pathogen in immunocompromised individuals, such as AIDS patients and transplant recipients, as well as in neonates. Diseases that are associated with CMV infections in immunocompromised hosts include retinitis, gastrointestinal disease, central nervous system diseases, interstitial pneumonia, and other visceral diseases (2).
Diagnosis of disseminated CMV infection and active visceral disease usually involves detection of the virus in peripheral blood leukocytes (PBLs) by the conventional tube culture method or the shell vial procedure. Both techniques require maintenance of tissue culture cell lines, timely processing of specimens, and labor-intensive preparation of PBLs by dextran sedimentation. CMV disease in AIDS patients and transplant recipients can be difficult to diagnose because cultures are often negative, despite the development of visceral syndromes.
Direct detection of CMV antigens in blood provides a more rapid diagnosis of CMV infection than tissue culture (7, 14). However, it requires timely specimen processing, PBL preparation, and scanning of slides with a fluorescence microscope, making analysis of large numbers of specimens difficult. Direct detection of viral nucleic acid in PBL cells by various methods can be sensitive, but it requires leukocyte preparations and may detect latent CMV rather than active infection (5). Direct detection of CMV DNA in plasma or sera by PCR correlates well with PBL culture positivity and is a sensitive method for the detection of CMV viremia and systemic CMV disease (3, 17, 22).
The availability of anti-CMV therapy delays the development of disease and increases the rate of patient survival (18, 19). In AIDS patients, a longer survival time is also associated with increasing incidences of CMV disease in the central nervous system that are extremely difficult to diagnose by conventional culture of cerebrospinal fluid (CSF). In autopsy studies, CMV-related neurological disorders occur in >20% of AIDS patients (16). The prognosis for patients with CMV-related neurological disorders is poor. Thus, the availability of diagnostic methods that are more sensitive than tissue culture for the detection of diseases of the central nervous system would benefit patient management. The branched DNA (bDNA) assay, the CMV antigen assay, and PCR-based assays have all been shown to be more sensitive and rapid than culture in detecting CMV in CSF (6, 15, 21). However, both CMV antigen and bDNA assays require a large volume of CSF (1 ml) and a centrifugation step. Such a large volume of CSF is not always available.
In this report, we describe the development of a simple and rapid AMPLICOR CMV Test for the detection of CMV DNA in 50-μl plasma and CSF specimens from human immunodeficiency virus (HIV)-infected patients and report a comparison of the performance of the AMPLICOR CMV Test with those of the antigen assay and tissue culture.
MATERIALS AND METHODS
Patients and specimens.
One hundred twelve plasma specimens were obtained from 43 HIV-infected patients who were participating in trials of anti-CMV drugs. Fifty-eight CSF samples were obtained from HIV-infected patients who were suspected of having CMV or other neurological diseases.
Viral and bacterial strains.
All viral stocks were obtained from the American Type Culture Collection unless indicated otherwise. Additional isolates were provided by R. Whitely (University of Alabama). Toledo virus was provided by E. S. Mocarski (Stanford University). Hepatitis C virus was from a clinical specimen that was provided by S. Tsang at Roche Molecular Systems, Inc. (RMS). HIV type 1 and type 2 (cloned provirus) were provided by C. Christopherson at RMS. The Candida albicans DNA, Mycobacterium sp. DNA, and DNAs from other bacterial strains in this study were supplied by K. Greisen, K. Young, and D. Leong (all at RMS), respectively (8). Viral DNA preparation was based on a detergent and proteinase K digestion method (1). Viral RNA preparation was based on a guanidine thiocyanate precipitation method (13). The DNA or RNA concentration at an optical density at 260 nm (OD260) and molecular weight were used to determine the copy number. If the concentration of nucleic acid was too low, an endpoint dilution was used to estimate the copy number.
Specimen collection and processing.
Blood specimens were drawn into tubes containing EDTA (0.1 ml EDTA per 10 ml of blood). CSF specimens from patients were obtained from various hospitals and were stored frozen at −40°C.
Dextran sedimentation procedure for collection of leukocytes.
A dextran sedimentation procedure was used to collect PBLs for the direct quantification of CMV DNA. Specimens of whole blood, which had been held at room temperature for up to 8 h, were transferred to 15-ml centrifuge tubes. After the addition of 2 ml of 6% dextran (high-molecular-weight dextran [molecular weight, 76,000]; Sigma Chemical Co., St. Louis, Mo.) in phosphate-buffered saline, the tubes were mixed gently by inversion and were then incubated without additional mixing at 37°C for 15 to 30 min to allow the blood to settle. The leukocyte-rich supernatant (top layer) was collected, with care being taken not to disturb the erythrocyte layer, and was transferred to a new 15-ml centrifuge tube. Puck’s saline without Ca2+ (Gibco BRL, Grand Island, N.Y.) was then added to bring the volume to 15 ml. The leukocyte suspension was centrifuged at 400 to 450 × g for 15 min, and the supernatant was decanted. The pellet was resuspended in 5 ml of sterile distilled H2O with light vortexing to lyse the remaining erythrocytes, and Puck’s saline was added to bring the volume to 15 ml. After centrifugation at 400 to 450 × g for 15 min, the supernatant was decanted and the pellet was resuspended to 2 ml with culture medium.
Conventional tube culture.
A 1-ml aliquot of prepared leukocytes and 0.5 to 1 ml of CSF samples were inoculated into human diploid fibroblast cultures and were examined daily for cytopathic effect. Suspected isolates were confirmed by immunofluorescence by using a monoclonal antibody to CMV antigen.
CMV antigen assay.
The CMV antigen assay was performed by using the CMV-vue (FITC) immunofluorescence kit (INCSTAR, Stillwater, Minn.). This assay detects the lower matrix protein (pp65) of CMV in peripheral blood leukocytes by use of immunocytochemical methods. The dextran sedimentation procedure was used to prepare leukocytes. Twenty-five microliters of cell suspension was spotted onto slides, air dried, and fixed in an FITC-vue fixative. The fixed cells were then incubated with a CMV FITC-vue monoclonal antibody directed against pp65, followed by immunofluorescence staining with fluorescein isothiocyanate (FITC)-labeled anti-mouse immunoglobulin G (IgG). Positively stained cells were then visualized by light microscopy.
One milliliter of a CSF specimen was centrifuged in a microcentrifuge at 14,000 × g for 5 min to pellet the cells. The cells were suspended in 40 μl of medium. Next, 15 μl of the cell suspension was spotted onto slides, air dried, and stained by following the protocol used for PBLs.
The results were reported qualitatively as either positive or negative; quantitation of the number of positive cells was not attempted.
Construction of CMV controls.
The CMV internal control (pSYC31) was constructed to include the same primer-binding sequence used in the AMPLICOR CMV Test but had a different probe sequence that allowed the differentiation of the internal control from the CMV target. The amplified product (amplicon) from the internal control was the same size as the amplicon from the CMV target and had a G+C content similar to that of the amplicon from the CMV target, ensuring the same efficiency of the amplification between the internal control and the target. The CMV-positive control (pCMA1) was constructed to include the entire amplicon region. It contains the same primer- and probe-binding sites as the CMV target DNA. The copy number of the control was determined by Poisson analysis.
AMPLICOR CMV Test.
The primers and probe were selected from the 5′ end of the CMV DNA polymerase gene based on the sequence of strain AD169 (10). Both primers were biotinylated at their 5′ ends. The size of the amplicon was 365 bp.
CMV DNA was isolated from 50 μl of a plasma or a CSF specimen by lysing the viral particles in 500 μl of CMV extraction reagent at 100°C for 30 min, and then 50 μl of the processed specimen was added to a PCR tube that contained 50 μl of the CMV master mix and a CMV internal control, yielding ∼5 μl of plasma or CSF equivalent per PCR mixture. Amplification was conducted in a GeneAmp PCR system 9600 (Perkin-Elmer, Norwalk, Conn.). To eliminate any pre-PCR carryover contamination, the amplification reaction was run at 50°C for 10 min to activate the AmpErase (uracil-N-glycosylase) included in the master mix. This was followed by 40 cycles at 94 and 65°C, each for 30 s. The final extension step was 72°C for 10 min.
Detection procedures and reagents are the same as discussed previously (11) for the AMPLICOR PCR Diagnostics Kit for Chlamydia trachomatis except that a CMV-specific capture plate and a control plate were used. The colorimetric results on the microwell plate were read on a spectrophotometric reader (Molecular Devices Corporation, Menlo Park, Calif.) at 450 nm. An OD of 0.35 was the cutoff for the assay, with values of <0.35 interpreted as negative and those of ≥0.3 interpreted as positive. The cutoff was determined by amplification of 220 plasma specimens from healthy individuals: 119 were CMV IgG seronegative specimens (mean OD = 0.091, minimum OD = 0.060, maximum OD = 0.154, standard deviation = 0.017) and 101 were CMV IgG seropositive specimens (mean OD = 0.088, minimum OD = 0.064, maximum OD = 0.146, standard deviation = 0.017) (20).
RESULTS
Inclusivity and exclusivity testing.
The specificity of the AMPLICOR CMV Test was examined with a panel of nucleic acids from CMV isolates and non-CMV pathogens. Twelve clinical CMV isolates and four laboratory CMV strains (strains Davis, Towne, Toledo, and AD169) were included in the CMV panel used for examination of specificity. All clinical isolates and laboratory strains of CMV were detectable by the AMPLICOR CMV Test with a target input of less than 100 copies of DNA.
The panel used for examination of exclusivity consisted of nucleic acids isolated from 5 non-CMV herpesviruses and 21 nonherpesvirus pathogens. At 5 × 103 copies of DNA or RNA input molecules or more, these pathogens showed no cross-reactivity with the CMV-specific primers and probe, and the internal control in each test was amplified and detected. A 10-copy sensitivity was demonstrated with the CMV-positive control and with purified DNA of CMV AD169. The five non-CMV herpesviruses were herpes simplex virus type 1, herpes simplex virus type 2, Epstein-Barr virus, varicella-zoster virus, and human herpesvirus 6. The 21 non-herpesvirus pathogens included two fungi (C. albicans and Aspergillus niger), seven viruses (HIV types 1 and 2, human hepatitis C virus, human hepatitis B virus, and human papillomavirus types 6, 16, and 18), and 12 bacteria (Mycobacterium tuberculosis, Mycobacterium avium, Mycobacterium intracellulare, Klebsiella pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus pneumoniae, Propionibacterium acnes, Neisseria meningitidis, Haemophilus influenzae, Listeria monocytogenes, and Escherichia coli).
Analysis of plasma specimens by AMPLICOR CMV Test.
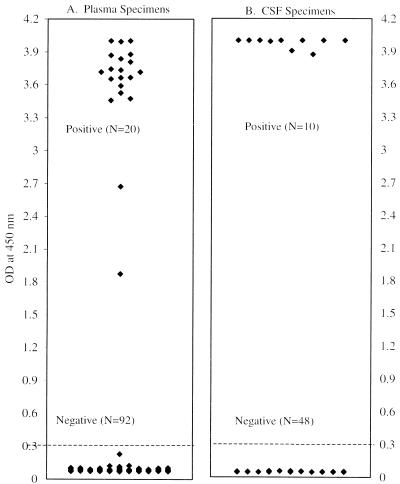
One hundred twelve plasma specimens from 43 HIV-infected patients who were enrolled in anti-CMV drug studies were tested by the AMPLICOR CMV Test. Matched leukocyte specimens from these patients were also tested by a CMV antigenemia assay and the conventional tube culture method. Among the 112 plasma specimens, 20 (18%) were positive by the PCR assay. The CMV antigen assay detected 21 (19%) CMV-positive specimens, and the conventional tube culture method detected 10 (9%) CMV-positive specimens. The 20 AMPLICOR CMV Test-positive specimens were from eight patients. A scattergram of AMPLICOR CMV Test results (Fig. 1A) demonstrated a clear-cut separation between CMV-positive and CMV-negative plasma specimens.
FIG. 1.
Detection of CMV from plasma and CSF specimens by AMPLICOR CMV Test. An OD of 0.35 was the cutoff for the assay, with values of <0.35 being interpreted as negative and those of ≥0.35 being interpreted as positive. Among 112 plasma specimens tested by the AMPLICOR CMV Test, 20 were positive and 92 were negative. Among 58 CSF specimens tested, 10 were positive and 48 were negative.
In order to compare the three tests, a consensus analysis was performed as follows. If two of the three assays detected CMV, the specimens were designated consensus positive, and if two of the three assays were negative, the specimens were designated consensus negative. The consensus analysis showed that 93 specimens were consensus negative and that 19 specimens were consensus positive. The results of the three assays relative to the consensus results are shown in Table 1. The AMPLICOR CMV Test had a sensitivity of 95% and a specificity of 98%. The sensitivity and specificity of the antigen assay were 100 and 98%, respectively, and those of the conventional tube culture method were 42 and 98%, respectively. Compared to the consensus results, three specimens had discordant results by the AMPLICOR CMV Test. One specimen was PCR negative but consensus positive, and two specimens were consensus negative but PCR positive.
TABLE 1.
Consensus comparison of AMPLICOR CMV Test, antigen assay, and conventional culture assay for plasma specimens from HIV-infected patients
| Assay and result | Consensus result (no. of specimens)a
|
Sensi- tivity (%) | Speci- ficity (%) | PPVb (%) | NPVc (%) | |
|---|---|---|---|---|---|---|
| Positive | Negative | |||||
| AMPLICOR CMV Test | ||||||
| Positive | 18 | 2 | 95 | 98 | 90 | 99 |
| Negative | 1 | 91 | ||||
| Antigen assay | ||||||
| Positive | 19 | 2 | 100 | 98 | 90 | 100 |
| Negative | 0 | 91 | ||||
| Culture | ||||||
| Positive | 8 | 2 | 42 | 98 | 80 | 89 |
| Negative | 11 | 91 | ||||
If two of the three assays were positive, the specimen was designated consensus positive. If two of the three assays were negative, the specimen was designated consensus negative.
PPV, positive predictive value.
NPV, negative predictive value.
The only specimen that was negative by PCR but positive by the antigen assay and culture was from patient A, from whom a total of six sequential specimens were tested in the study (Table 2). The internal control for this specimen, which was AMPLICOR CMV Test negative but consensus positive, was positive, indicating that the specimen was noninhibitory to the PCR and that the detection procedure worked well. Repeated assay of this specimen showed sporadic positive results, and the specimen was consistently positive if the plasma input was increased to ∼25 μl of plasma equivalent per PCR by a different specimen processing method, suggesting that the plasma had a low viral load (data not shown). The fifth specimen received from patient A was positive by the PCR assay but consensus negative. Even though two specimens from patient A had discordant results, the detection of CMV in the plasma of patient A by the AMPLICOR CMV Test occurred 43 days earlier than the diagnosis of retinitis.
TABLE 2.
Discordant analysis
| Patient and date (mo/day/yr) | Resulta
|
||
|---|---|---|---|
| AMPLICOR CMV Test | Antigen assay | Culture | |
| Patient Ab | |||
| 11/16/1993c | Negd | Pos | Pos |
| 1/11/1994 | Pos | Pos | Pos |
| 2/4/1994 | Pos | Pos | Pos |
| 2/24/1994 | Pos | Pos | Neg |
| 4/19/1994 | Pos | Pos | Pos |
| 6/24/1994c | Posd | Neg | Neg |
| Patient Be | |||
| 10/20/1993 | Neg | Neg | Neg |
| 12/16/1993 | Pos | Pos | Pos |
| 2/8/1994c | Posf | Neg | Neg |
The other consensus-negative but PCR-positive specimen was from patient B, from whom a total of three sequential plasma samples were obtained.
By a prototype quantitative CMV assay (12), the viral loads of the two specimens from patient A with discordant results were <400 copies per ml of plasma and the viral load of the specimen from patient B with discordant results was 2,610 copies per ml of plasma. By the CMV antigen assay two specimens had discordant results: antigen assay positive but consensus negative. Both specimens were weakly positive, with low numbers of positive cells by the antigen assay (1 positive cell among 2 × 105 PBLs).
Analysis of CSF specimens by AMPLICOR CMV Test.
The AMPLICOR CMV Test was used to evaluate 58 CSF specimens from AIDS patients suspected of having an infection of the central nervous system. The antigen assay and tissue culture results were also available for some but not all of the CSF specimens. The AMPLICOR CMV Test detected 17% (10 of 58) CMV-positive CSF specimens (Fig. 1B). The antigen assay detected 18% (5 of 28) positive specimens, and tissue culture detected no CMV-positive specimens among the 25 CSF specimens tested. Results of all three tests were available for only 23 of the 58 specimens. Among the 23 specimens, PCR detected CMV in 1 specimen, and the antigen assay also detected CMV in this specimen. Of the remaining 35 of the 58 specimens, 9 of them were positive by PCR and 4 of them were confirmed to be positive by the CMV antigen assay. Of the other five PCR-positive specimens, four were from patients with polyradiculopathy, and the fifth specimen was from a patient who showed varicella-zoster virus infection and neurological weakness (Table 3). The CMV culture results for the fifth patient were negative. For the 48 negative specimens, the internal controls, which were coamplified in the PCR, were positive, suggesting that there were no detectable inhibitors in these specimens. The clinical symptoms of the patients with PCR-negative results revealed that, except for the specimen from one patient, these specimens were unlikely to be from patients who had CMV infection. The one exception was a specimen from a patient who was diagnosed with polyradiculopathy but who was undergoing treatment.
TABLE 3.
AMPLICOR CMV Test-positive CSF specimens
| Patient no. | Resulta
|
Diagnosis | ||
|---|---|---|---|---|
| AMPLICOR CMV Test | Antigen assay | Culture | ||
| 1 | Pos | Pos | NDb | PRIc |
| 2 | Pos | Pos | ND | Progressive weakness |
| 3 | Pos | Pos | ND | Progressive weakness |
| 4 | Pos | Pos | Neg | Encephalitis |
| 5 | Pos | Pos | ND | Progressive weakness |
| 6 | Pos | ND | Neg | Clinical CMV infection and PRI |
| 7 | Pos | ND | Neg | PRI |
| 8 | Pos | ND | Neg | PRI |
| 9 | Pos | ND | Neg | Weaknessd |
| 10 | Pos | ND | ND | PRI |
Pos, positive; Neg, negative.
ND, not done.
PRI, polyradiculopathy.
The patient also had varicella-zoster virus infection.
DISCUSSION
The AMPLICOR CMV Test amplifies a portion of the coding region of the CMV DNA polymerase gene. The primers and probe selected for the test are specific for CMV. Twelve clinical CMV isolates and four laboratory CMV strains could be detected by the AMPLICOR CMV Test with a target input of <100 copies. The test had a 10-copy sensitivity, as demonstrated with CMV-positive control DNA and viral DNA. With DNA or RNA inputs of ≥5 × 103 molecules, the CMV-specific primers and probe did not cross-react with the 26 non-CMV pathogens tested. These pathogens include the herpesviruses and blood pathogens, such as HIV type 1, HIV type 2, hepatitis C virus, hepatitis B virus, and C. albicans.
The AMPLICOR CMV Test results for 112 plasma specimens showed that, by comparison with the consensus results, the sensitivity and specificity of the test are 95 and 98%, respectively, and the sensitivity and specificity of the CMV antigen assay are 100 and 98%, respectively. Three specimens had discordant results by the AMPLICOR CMV Test compared to the consensus results. These three specimens were from two patients: patients A and B. The two specimens from patient A had discordant results due to a low viral load, since the viral loads in these two specimens were <400 copies per ml of plasma when plasma specimens were analyzed by a prototype quantitative test for CMV (12). The viral load in the specimen from patient B with discordant results was 2,610 copies per ml of plasma (12). It was unclear why the antigen assay failed to detect CMV in PBLs while there was a moderate viral load in plasma. Regardless of the discordance, the detection of CMV by the AMPLICOR CMV Test in both patients occurred much earlier than the diagnosis of CMV disease. For patient A, PCR of plasma detected CMV 43 days before the diagnosis of retinitis, and for patient B, PCR of plasma detected CMV 41 days before the diagnosis of colitis. Overall, the performance of the AMPLICOR CMV Test is comparable to the performance of the CMV antigen assay. However, the CMV antigen assay with blood specimens requires >105 PBLs. Recently, it has been reported that the bDNA assay can also detect CMV in PBLs, but it requires 2 × 106 to 6 × 106 PBLs for the assay (4). Large numbers of PBLs may be hard to obtain from bone marrow transplant recipients and AIDS patient with low CD4 cell counts. The AMPLICOR CMV Test requires the processing of only 50 μl of plasma. The requirement of such a small volume of plasma also makes the detection of CMV in neonates possible.
The clinical utility of the AMPLICOR CMV Test with plasma specimens has recently been demonstrated in a study with bone marrow transplant recipients (9). That study showed that plasma PCR is more sensitive than the antigen assay at detecting CMV disease.
Analysis of CSF specimens demonstrated that both the PCR assay and the antigen assay detected the same CMV-positive specimens. Clinical information suggested that these CMV-positive CSF specimens were collected from patients who had developed CMV disease (Table 3). Detection of CMV in CSF by the conventional tube culture method is insensitive. In fact, the culture technique detected no CMV in 25 CSF specimens tested in our study, and 5 of 25 specimens were positive by the AMPLICOR CMV Test, and one AMPLICOR CMV Test-positive specimen was also positive by the antigen assay (Table 3). PCR, the antigen assay, and the bDNA assay have been demonstrated to be more sensitive than culture for the detection of CMV in CSF specimens (6, 15, 21). However, both the CMV antigen assay and the bDNA assay require at least 1 ml of CSF for the test. In addition, the CMV antigen assay requires centrifugation of CSF in a microcentrifuge for 5 min to pellet the cells, while the bDNA assay requires an ultracentrifugation step of 1 h. Although the CSF specimens tested in this study were limited in number and more work needs to be done in well-designed studies, these results demonstrate the potential for the use of CSF in the AMPLICOR CMV Test.
In this report, we show that the performance of the AMPLICOR CMV Test is similar to the performance of the CMV antigen assay. The advantage of the AMPLICOR CMV Test is that it requires the processing of only 50 μl of plasma or CSF and does not require the preparation of leukocytes. The specimen processing procedure for the AMPLICOR CMV Test involves a single 30-min heating step before the PCR. The test incorporates an internal control that is coamplified with the CMV target in clinical specimens, providing a useful tool for monitoring specimen amplification and specimen inhibition. In this study, the results for plasma and CSF by the AMPLICOR CMV Test indicated a clear separation between the positive and the negative specimens (Fig. 1). The test is easy to perform and requires less than 5 h for specimen processing, amplification, and detection. It is therefore more rapid and technically easier for large-scale screening than the CMV antigen assay.
ACKNOWLEDGMENTS
We thank Sheng-Yung Chang for the construction of the CMV internal control and Beverly Dale for critical reading of the manuscript.
REFERENCES
- 1.Bauer H M, Greer C E, Manos M M. Determination of genital human papillomavirus infection by consensus polymerase chain reaction. In: Herrington C S, McGee J O, editors. Diagnostic molecular pathology: a practical approach. New York, N.Y: IRL Press; 1992. pp. 131–152. [Google Scholar]
- 2.Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, et al., editors. Virology. New York, N.Y: Lippincott-Raven Publishers; 1996. pp. 2439–2533. [Google Scholar]
- 3.Brytting M, Xu W, Warren B, Sundqvist V-A. Cytomegalovirus detection in sera from patients with active CMV infections. J Clin Microbiol. 1992;30:1937–1942. doi: 10.1128/jcm.30.8.1937-1941.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chernoff D N, Miner R C, Hoo B S, Shen L-P, Kelso R J, Jekic-McMullen D, Lalezari J B, Chou S, Drew W L, Kolberg J A. Quantification of cytomegalovirus DNA in peripheral blood leukocytes by a branched-DNA signal amplification assay. J Clin Microbiol. 1997;35:2740–2744. doi: 10.1128/jcm.35.11.2740-2744.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delgado R, Lumbreras C, Alba C, Pedraza M A, Otero J R, Gomez R, Moreno E, Noriega A R, Paya C V. Low predictive value of polymerase chain reaction for diagnosis of cytomegalovirus disease in liver transplant patients. J Clin Microbiol. 1992;30:1876–1878. doi: 10.1128/jcm.30.7.1876-1878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flood J, Drew W L, Miner R, Jekic-McMullen D, Shen L-P, Kolberg J, Garvey J, Follansbee S, Poscher M. Diagnosis of cytomegalovirus polyradiculopathy and documentation of in vivo anti-CMV activity in cerebrospinal fluid by using branched DNA signal amplification and antigen assays. J Infect Dis. 1997;176:348–352. doi: 10.1086/514051. [DOI] [PubMed] [Google Scholar]
- 7.Gerna G, Parea M, Percivalle E, Zipeto D, Sillini E, Barbini G, Milanesi G. Human cytomegalovirus viraemia in HIV-1 seropositive patients at various clinical stages of infection. AIDS. 1990;4:1027–1031. doi: 10.1097/00002030-199010000-00014. [DOI] [PubMed] [Google Scholar]
- 8.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiyoshi M, Taqawa S, Takubo T, Tanaka K, Nakao T, Higashihata Y, Yasui Y, Kim T, Hiraoka A, Tatsulmi N. Evaluation of the AMPLICOR CMV Test for direct detection of cytomegalovirus in plasma specimens. J Clin Microbiol. 1997;35:2692–2694. doi: 10.1128/jcm.35.10.2692-2694.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kouzarides T, Banjier A T, Satchwell S C, Weston K, Tomllinson P, Barell B G. Sequence and transcription analysis of the human cytomegalovirus DNA polymerase gene. J Virol. 1987;61:125–133. doi: 10.1128/jvi.61.1.125-133.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeffelholz M J, Lewinski C A, Silver S R, Purohit A P, Herman S A, Buonagurio D A, Dragon E A. Detection of Chlamydia trachomatis in endocervical specimens by polymerase chain reaction. J Clin Microbiol. 1992;30:2847–2851. doi: 10.1128/jcm.30.11.2847-2851.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long, C. M., and S.-Y. Kao. Unpublished data.
- 13.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kowk S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Revello M G, Furione M, Zavattoni M, Gerna G. Human cytomegalovirus infection: diagnosis by antigen and DNA detection. Rev Med Microbiol. 1994;5:265–276. [Google Scholar]
- 15.Revello M G, Perecivalle E, Sarasini A, Baldanti F, Furione M, Gerna G. Diagnosis of human cytomegalovirus infection of the nervous system by pp65 detection in polymorphonuclear leukocytes of cerebrospinal fluid from AIDS patients. J Infect Dis. 1994;170:1275–1279. doi: 10.1093/infdis/170.5.1275. [DOI] [PubMed] [Google Scholar]
- 16.Roullet E, Assuerus V, Conzlan J, Ropert A, Sais G, Baudrilmont M, El Amrani M, Jacomet C, Duvivier C, Gonzales-Canali G, Kirstetter M, Meyohas M C, Picard O, Rozenbaum W. Cytomegalovirus multifocal neuropathy in AIDS: analysis of 15 consecutive cases. Neurology. 1994;44:2174–2182. doi: 10.1212/wnl.44.11.2174. [DOI] [PubMed] [Google Scholar]
- 17.Spector S A, Merrill R, Wolf D, Dankner W M. Detection of human cytomegalovirus in plasma of AIDS patients during acute visceral disease by DNA amplification. J Clin Microbiol. 1992;30:2359–2365. doi: 10.1128/jcm.30.9.2359-2365.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spector S A, McKinley G F, Lalezari J P, Samo T, Andruczk R, Follansbee S, Sparti P D, Havlir D V, Simpson G, Buhles W, Wong R, Stempien M J. Oral ganciclovir for the prevention of cytomegalovirus disease in persons with AIDS. N Engl J Med. 1996;334:1491–1497. doi: 10.1056/NEJM199606063342302. [DOI] [PubMed] [Google Scholar]
- 19.Spector, S. A. 1997. Current therapeutic challenges in the treatment of cytomegalovirus retinitis. J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 14(Suppl. 1):S32–S35. [DOI] [PubMed]
- 20.Tevere, V. (Roche Molecular Systems, Inc.). Personal communication.
- 21.Wolf D, Spector S A. Diagnosis of human cytomegalovirus central nervous disease in AIDS patients by DNA amplification from cerebrospinal fluid. J Infect Dis. 1992;166:1412–1415. doi: 10.1093/infdis/166.6.1412. [DOI] [PubMed] [Google Scholar]
- 22.Wolf D, Spector S A. Early diagnosis of human cytomegalovirus disease in transplant recipients by DNA amplification in plasma. Transplantation. 1993;56:330–334. doi: 10.1097/00007890-199308000-00014. [DOI] [PubMed] [Google Scholar]