Abstract
Background
Mucoepidermoid carcinoma is a malignant salivary gland tumor which, in most cases, is composed of variable proportions of mucous, epidermoid, and intermediate cells.
Methods
We report a case of parapharyngeal mucoepidermoid carcinoma with highly unusual (“monomorphic”) light microscopic features as well as atypical immunohistochemical properties. Molecular analysis was performed using the TruSight RNA fusion panel.
Results
The tumor featured heretofore undescribed histopathological features: sheets and nests composed of monomorphic neoplastic (plump spindle to epithelioid) cells with no mucous, intermediate, glandular/columnar, or any other cell type identified. The neoplastic cells displayed variable clear cell change and only expressed cytokeratin 7. Despite this non-classical morphology, the presence of the classical CRTC1::MAML2 fusion was demonstrated.
Conclusions
Mucoepidermoid carcinoma featuring a uniform (“monomorphic”) population of neoplastic cells is a novel observation. A confident diagnosis of mucoepidermoid carcinoma can be made upon detection of the CRTC1/3::MAML2 fusion. Our case increases the spectrum of histopathological appearances that mucoepidermoid carcinoma may display.
Keywords: Mucoepidermoid carcinoma, Salivary gland, Spindle cell, Clear cell
Introduction
Mucoepidermoid carcinoma (MEC) is a malignant salivary gland tumor typically comprising variable proportions of mucous, epidermoid, and intermediate cells. Classical MEC is in most instances an easily recognizable tumor [1]. The resultant “multi-morphic” nature of the tumor is diagnostically distinctive. In contrast, non-classical variants of MEC demonstrate a significant component (more than 80%) of non-mucous histology [1]. In the absence of an easily recognizable mucous cell population, the diagnosis is challenging, often requiring ancillary studies such as special (mucin) stains, immunohistochemistry, and detection of classical MEC-associated chromosomal translocations.
We report a case of parapharyngeal MEC with no discernible mucous, intermediate, glandular or epidermoid cell populations. The tumor comprised a fairly monomorphic neoplastic cell population arranged in solid organoid nests and sheets, suggestive of a perivascular epithelioid cell tumor (PEComa). The tumour cells were epithelioid to “plump-spindly” with variable clear cell change. Despite this non-classical, and hitherto unreported, histomorphology, the identification of the classical CRTC1/3::MAML2 fusion was instrumental in the diagnosis of this uncommon histopathologic manifestation of MEC.
Materials and Methods
Our case was identified prospectively in our routine files. Following an extensive work-up (including mucin stains and immunohistochemical studies), the initial incisional biopsy was first interpreted as a carcinoma with unusual features. A salivary gland carcinoma was suspected given the tumor site and histopathological features. In view of this suspicion, the tumor was sent for gene fusion testing.
RNA was isolated from formalin-fixed paraffin-embedded (FFPE) tissue sections using RNeasy FFPE Kit of Qiagen (Hilden, Germany) and quantified spectrophotometrically using NanoDrop-1000 (Waltham, United States). Molecular analysis was performed using the TruSight RNA fusion panel (Illumina, Inc., San Diego, CA, USA) with 500 ng RNA as input according to the manufacturer’s protocol. Libraries were sequenced on a MiSeq (Illumina) with more than 3 million reads per case, and sequences were analyzed using the RNA-Seq Alignment workflow, version 2.0.1 (Illumina). The Integrative Genomics Viewer (IGV), version 2.2.13 (Broad Institute, REF) was used for data visualization. To analyze the mutational status of common cancer-related genes, DNA was isolated from FFPE tissue sections using the Maxwell 16LEV Blood DNA kit (Promega, Madison, USA) and submitted for hybrid-capture enrichment-based sequencing analysis using the TruSight Tumor 170 (TST170) gene panel (Illumina) according to the manufacturer’s protocol. Libraries were sequenced on a Next Seq550 (Illumina) and analyzed for single nucleotide mutations, insertions, deletions and copy number variations using the TruSight Tumor 170 software (BaseSpace Sequence Hub, Illumina) with human genome hg19 as reference.
Results
The patient was a 51-year-old Chinese male with no significant past medical history. He presented with per-oral bleeding of two days’ duration. Clinical examination revealed a mass at the left parapharyngeal wall. Magnetic resonance imaging (MRI) (Fig. 1) confirmed the presence of a mass in the left parapharyngeal space, measuring 5.1 × 4.7 × 4.2 cm in size. The mass had filled the vallecula and grown medially, with resultant displacement and severe narrowing of the oropharynx. Inferiorly, the mass extended into the posterior third of the tongue and floor of the mouth. The mass had infiltrated the myohyoid muscle and the left aryepiglottic fold. A fluorodeoxyglucose positron emission tomography/computed tomography (FDG PET/CT) scan performed from the vertex of the skull to the upper thighs revealed the left parapharyngeal space mass to be FDG-avid. There was no evidence of FDG-avid tumor elsewhere in the body.
Fig. 1.
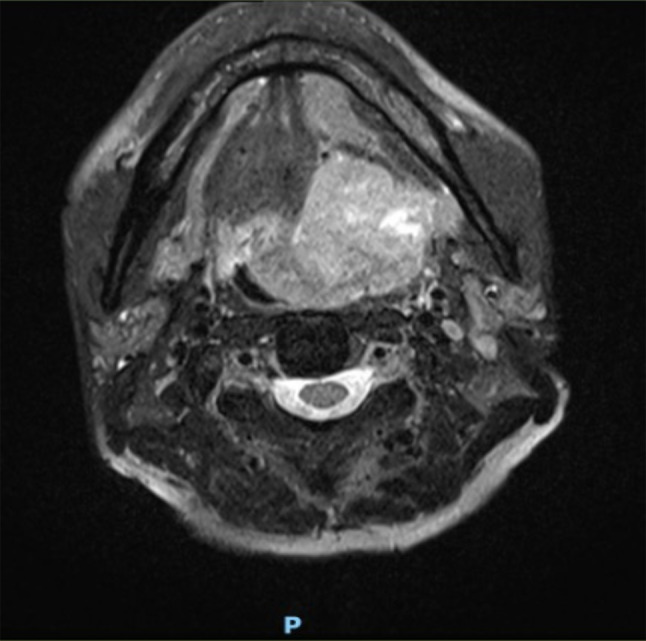
T2-weighted MRI image showing a tumor involving the left lateral oropharyngeal wall and crossing the midline
The initial incisional biopsy was interpreted as carcinoma with unusual features. A repeat biopsy showed identical histopathological features. A salivary gland carcinoma was suspected based on the tumor site and histopathological features. In view of this suspicion, the tumor tissue was sent for gene fusion testing. While awaiting the gene fusion test result, the tumor was resected (sub-total glossectomy with supra-glottic laryngectomy and bilateral cervical lymph node dissection) with clear surgical margins. The tumor measured 5.2 × 4.8 × 2.5 cm in size on gross examination. Cut sections of the tumor showed a soft tan-white solid appearance with small foci of hemorrhage. The overlying mucosa was ulcerated (by the tumor). The tumor was sampled with eight blocks.
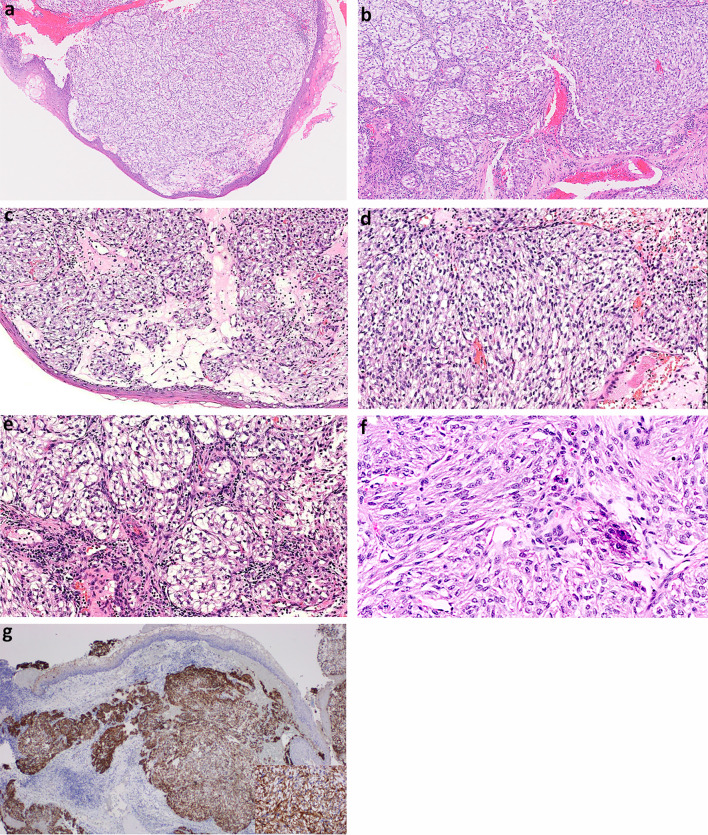
Both the biopsies and the resection specimen showed an unusual tumor with identical histopathological features. The infiltrative tumor was present mainly in the submucosa with overlying mucosal ulceration. The tumor demonstrated solid sheet-like and nested architecture comprising epithelioid to plump spindle cells with variable (cytoplasmic) clear cell change (Fig. 2a, b, c, d). The neoplastic cells featured central ovoid irregular nuclei with small nucleoli and often ample cytoplasm. Areas with epithelioid and plump spindle cells merged imperceptibly into each other [Fig. 2e]. Nuclear pleomorphism was absent. Mitotic activity was up to 3 per 10 high-powered fields. Necrosis was not identified. Epidermoid, glandular and mucous cells were absent. No intra- or extra-cellular mucin was detected on mucicarmine and diastase periodic acid-Schiff (DPAS) stains.
Fig. 2.
a Solid sheets of tumor present mainly in the submucosa. Hematoxylin & Eosin stain, 2X magnification. b Nested architecture (left half) transitions into solid sheet-like architecture (right half). Hematoxylin & Eosin stain, 4X magnification. c Areas of tumor with nested architecture. Hematoxylin & Eosin stain, 10X magnification. d Areas of tumor with solid sheet-like architecture. Hematoxylin & Eosin stain, 10X magnification. e In most areas, the tumor cells are epithelioid with ample clear cytoplasm and central ovoid irregular nuclei. Hematoxylin & Eosin stain, 20X magnification. f In other areas, the tumor cells have a plump spindled appearance with eosinophilic cytoplasm and absence of clear cell change. Hematoxylin & Eosin stain, 10X magnification. g The tumor is strongly positive for CK7. Immunohistochemistry, 4X magnification and 40X magnification (inset)
On immunohistochemistry, the tumor cells were diffusely positive for CK7 (Fig. 2f). The neoplastic cells were negative for CK5/6 and p63. No expression of NUT1, S100 protein, SOX10, HMB45, MelanA, GFAP, desmin, SMA, synaptophysin, PAX8, TTF1 and TFE3 was detected. The neoplastic cells showed retained nuclear INI1 expression.
The tumor was completely resected. One hundred and thirty-eight lymph nodes were sampled in the bilateral neck dissections, and no metastatic tumor was identified.
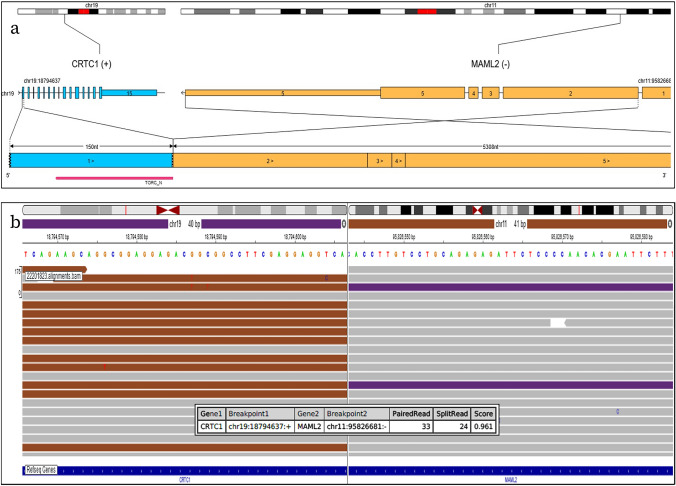
TruSight RNA fusion panel testing using the Illumina panel revealed the presence of a CRTC1::MAML2 fusion (Fig. 3a , b). Exon 1 of CRTC1 replaced Exon 1 of MAML2 with retention of the TORC_N domain. The breakpoints are chr19:18794637:+ and chr11:95826681:-.
Fig. 3.
a Visualization of the detected CRTC1::MAML2 fusion. Exon 1 of CRTC1 replaced Exon 1 of MAML2 with retention of the TORC_N domain. Visualization was done using the Oviz-Bio software package (https://bio.oviz.org/demo-project/analyses/FusionTransJunc). b Integrated Genome Viewer (IGV) split-screen view of read alignments of the identified CRTC1::MAML2 fusion event. Shown are mate pairs mapped to the fusion reads in the CRTC1 (purple color) and MAML2 (brown color) loci, respectively
Following tumor resection, the patient underwent adjuvant radiation therapy.
The patient is currently six months post-surgical resection of tumor and remains disease-free.
Discussion
The diagnostically challenging case presented here shows hitherto unreported histopathological features of mucoepidermoid carcinoma and the presence of a classical CRTC1::MAML2 fusion. The historical/classical definition of MEC states that it is a malignant salivary gland tumor composed of three different neoplastic cell types (epidermoid, intermediate, and mucous cells) in variable proportions [1]. Thus, classical MEC comprises easily discernible mucous cells with associated populations of epidermoid cells and/or intermediate cells, resulting in a “bi-” or “tri-morphic” appearance that is generally apparent even on low to medium-power light microscopic examination [1]. When mucous cells are lacking, a MEC is considered to demonstrate “non-classical” or “variant” morphology. While variant MEC can be challenging to diagnose, there are often interspersed or adjacent areas of classical MEC that provide important morphological clues for tumor recognition. Our case was diagnostically challenging in that no areas of classical MEC were detected.
A particularly novel feature of our tumor is its “monomorphic” nature combined with the tumor cell morphology. No mucous, intermediate, columnar or epidermoid cells were present. The tumor comprised exclusively of epithelioid to plump neoplastic cells arranged in nests and sheets. MEC mainly composed of spindle cells and oncocytic cells are rare, but have previously been described [1–8]. However, to the best of our knowledge, there has not been a case of MEC comprising exclusively epithelioid to plump spindle cells, devoid of mucous and epidermoid cells.
The morphological features of our case were reminiscent of a PEComa. However, diffuse cytokeratin expression eliminates this differential diagnosis [9].
Of note, the tumor presented herein lacked expression of p63 and CK5/6. Epidermoid differentiation in MEC is sometimes not readily apparent on light microscopy, necessitating immunohistochemical assessment with squamous markers such as p63 and CK5/6 [7, 10]. The absence of p63 and CK5/6 expression was misleading in this case, and these immunohistochemical stains should not be taken as an infallible screening panel for MEC.
Our case of MEC was negative for SOX10, consistent with the literature. SOX10 is usually negative in MEC, including clear cell MEC [11, 12]. An unusual SOX10-positive subtype of MEC featuring polygonal cells with pale to eosinophilic cytoplasm, forming solid nests with occasional glandular structures filled with colloid-like dense eosinophilic secretions, in addition to conventional MEC elements (epidermoid cells, mucous cells and intermediate cells), was described by Hsieh et al. [12].
The presence of a prominent spindle cell component in our case, while unusual in MEC, should not be mistaken for sarcomatoid de-differentiation or high-grade transformation (HGT). HGT in salivary gland carcinoma is a well-established phenomenon where there is clonal progression of a high-grade neoplasm arising from a low-grade salivary gland carcinoma [13]. HGT is known to occur in MEC [14–16]. Previously reported cases of HGT in MEC described undifferentiated anaplastic/sarcomatoid carcinoma [14, 16]. The absence of nuclear pleomorphism, brisk mitotic activity and tumor necrosis in our tumor argue strongly against progression to a high-grade malignancy. The distinction is important because HGT in salivary gland carcinoma portends far more aggressive clinical behavior with frequent regional and distant metastases [13].
Based on the tumor morphology, in particular the nested appearance with spindle to clear neoplastic cells, important differential diagnoses for this case included PEComa, alveolar soft part sarcoma, paraganglioma and metastatic renal cell carcinoma. However, these differential diagnoses were readily excluded with our immunohistochemical studies and correlation with the radiological staging scans.
The presence of CRTC1::MAML2 fusion in our case is strongly supportive of the diagnosis of MEC. In the absence of molecular testing, a confident diagnosis of MEC would have been exceedingly difficult. The importance of molecular testing in the diagnosis of salivary gland tumors have been emphasized by Fonseca et al. and Skalova et al. [17, 18]. The classical underlying molecular mechanism driving MEC is a t(11;19) translocation resulting in a CRTC1::MAML2 translocation. Rarely, there is a variant fusion CRTC3::MAML2 [19]. There are numerous histo-morphological subtypes of MEC, including squamoid (epidermoid), eosinophilic (oncocytoid), clear cell, spindle cell, sclerosing, ciliated, muco-acinar and Warthin-like variants [1, 20–22]. The CRTC1/3::MAML2 translocation is frequently found in classical and variant MEC arising from salivary gland and non-salivary gland tissue [7, 22–25]. Bishop et al. described a series of 10 cases of MEC devoid of squamous cell differentiation, both by morphology and immunohistochemistry [7]. Of note, one tumor from Bishop’s series demonstrated an admixture of spindle and clear cells as well as a CRTC3::MAML2 translocation, features very similar to the case we have described here. In Bishop’s series, three other tumors showed predominant clear cell morphology (CRTC1::MAML2 translocations) while one tumor showed predominant spindle cell morphology (CRTC3::MAML2 translocation). All tumors from Bishop’s series were p63 negative, as was the tumor we have described in this case report.
The CRTC1/3::MAML2 translocation is more frequently found in low- and intermediate-grade MEC, as compared to high-grade MEC [17, 26, 27]. Whether the presence of CRTC1/3::MAML2 translocation in MEC confers better prognosis remains contentious, but a recent large study by Fehr et al. concluded that fusion-negative MECs had worse 5-year progression-free survival [19, 28].
Apart from MEC, CRTC1/3::MAML2 fusions have also been detected in cutaneous hidradenoma and in odontogenic cysts with mucous prosoplasia [29, 30]. However, these entities are clearly not diagnostic considerations in our case.
Our case supports the growing body of opinion that the historical definition of MEC as a carcinoma with epidermoid, intermediate and mucous cells should be revisited. [7]
In summary, we report a case of “monomorphic” mucoepidermoid carcinoma composed of a uniform population of epithelioid to plump spindle cells, and the diagnosis is supported by documentation of a CRTC1::MAML2 fusion. The absence of mucous cells and epidermoid cells is particularly novel for a mucoepidermoid carcinoma. Awareness of this rare histopathological manifestation of mucoepidermoid carcinoma is crucial to direct appropriate molecular testing.
Funding
This study was not supported by any funding. The authors did not receive support from any organization for the submitted work. The authors have no relevant financial or non-financial interests to disclose.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
It is our institutions’ policy not to require formal Institutional Review Board (IRB) ethics approval for retrospective case reports on up to two patients.
Consent for publication
Consent for publication was obtained for every individual person’s data included in the study.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schwarz S, Stiegler C, Müller M, Ettl T, Brockhoff G, Zenk J, et al. Salivary gland mucoepidermoid carcinoma is a clinically, morphologically and genetically heterogeneous entity: a clinicopathological study of 40 cases with emphasis on grading, histological variants and presence of the t(11;19) translocation. Histopathology. 2011;58(4):557–570. doi: 10.1111/j.1365-2559.2011.03777.x. [DOI] [PubMed] [Google Scholar]
- 2.Ide F, Mishima K, Saito I. Mucoepidermoid carcinoma with spindle cell change: a low-grade lesion potentially mistaken for sarcomatoid dedifferentiation. Head Neck Pathol. 2008;2(3):227–230. doi: 10.1007/s12105-008-0067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goh GH, Lim CM, Vanacek T, Michal M, Petersson F. Spindle cell Mucoepidermoid Carcinoma of the Palatine Tonsil with CRTC1-MAML2 Fusion transcript: report of a rare case in a 17-Year-old boy and a review of the literature. Int J Surg Pathol. 2017;25(8):705–710. doi: 10.1177/1066896917714890. [DOI] [PubMed] [Google Scholar]
- 4.Liyanage RL, Wadusinghearachchi NS, Siriwardena BS, Jayasooriya PR, Tilakaratne WM. Pigmented mucoepidermoid carcinoma with spindle cell differentiation. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;117(6):e449–e451. doi: 10.1016/j.oooo.2013.08.031. [DOI] [PubMed] [Google Scholar]
- 5.Love GL, Sarma DP. Spindle cell mucoepidermoid carcinoma of submandibular gland. J Surg Oncol. 1986;31(1):66–68. doi: 10.1002/jso.2930310116. [DOI] [PubMed] [Google Scholar]
- 6.Terada T, Ikeuchi S, Inomoto C, Shimamura K. Mucoepidermoid carcinoma of the palate composed exclusively of clear cells (clear cell variant) Virchows Arch. 2004;445(5):541–543. doi: 10.1007/s00428-004-1086-1. [DOI] [PubMed] [Google Scholar]
- 7.Bishop JA, Thompson LDR, Siegele B, Gagan J, Mansour M, Chernock RD, et al. Mucoepidermoid carcinoma may be devoid of squamoid cells by immunohistochemistry: expanding the histologic and immunohistochemical spectrum of MAML2- rearranged salivary gland tumours. Histopathology. 2023;82(2):305–313. doi: 10.1111/his.14817. [DOI] [PubMed] [Google Scholar]
- 8.Skálová A, Agaimy A, Stanowska O, Baneckova M, Ptáková N, Ardighieri L, et al. Molecular Profiling of Salivary Oncocytic Mucoepidermoid Carcinomas helps to resolve Differential Diagnostic Dilemma with Low-grade oncocytic lesions. Am J Surg Pathol. 2020;44(12):1612–1622. doi: 10.1097/pas.0000000000001590. [DOI] [PubMed] [Google Scholar]
- 9.Folpe AL, Kwiatkowski DJ. Perivascular epithelioid cell neoplasms: pathology and pathogenesis. Hum Pathol. 2010;41(1):1–15. doi: 10.1016/j.humpath.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Sivakumar N, Narwal A, Pandiar D, Devi A, Anand R, Bansal D, et al. Diagnostic utility of p63/p40 in the histologic differentiation of salivary gland tumors: a systematic review. Oral Surg Oral Med Oral Pathol Oral Radiol. 2022;133(2):189–198. doi: 10.1016/j.oooo.2021.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Lee JH, Kang HJ, Yoo CW, Park WS, Ryu JS, Jung YS, et al. PLAG1, SOX10, and myb expression in Benign and malignant salivary gland neoplasms. J Pathol Trans Med. 2019;53(1):23–30. doi: 10.4132/jptm.2018.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh MS, Lee YH, Chang YL. SOX10-positive salivary gland tumors: a growing list, including mammary analogue secretory carcinoma of the salivary gland, sialoblastoma, low-grade salivary duct carcinoma, basal cell adenoma/adenocarcinoma, and a subgroup of mucoepidermoid carcinoma. Hum Pathol. 2016;56:134–142. doi: 10.1016/j.humpath.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 13.Petersson F. High-grade transformation (“dedifferentiation”)—malignant progression of salivary gland neoplasms, including varcinoma ex pleomorphic adenoma: a review. Pathol Case Rev. 2016;20(1):27–37. doi: 10.1097/PCR.0000000000000076. [DOI] [Google Scholar]
- 14.Lee H, Roh JL, Choi YJ, Choi J, Cho KJ. High Grade Transformation in Mucoepidermoid Carcinoma of the minor salivary gland with Polyploidy of the rearranged MAML2 gene. Head Neck Pathol. 2020;14(3):822–827. doi: 10.1007/s12105-019-01064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagao T, Gaffey TA, Kay PA, Unni KK, Nascimento AG, Sebo TJ, et al. Dedifferentiation in low-grade mucoepidermoid carcinoma of the parotid gland. Hum Pathol. 2003;34(10):1068–1072. doi: 10.1053/s0046-8177(03)00418-0. [DOI] [PubMed] [Google Scholar]
- 16.Subramaniam MM, Ng SB, Seah SB, Anuar D, Soong R, Lee VK. Molecular characterization of dedifferentiated mucoepidermoid carcinoma of the trachea using laser microdissection-based TP53 mutation analysis. Histopathology. 2009;55(4):472–475. doi: 10.1111/j.1365-2559.2009.03385.x. [DOI] [PubMed] [Google Scholar]
- 17.Fonseca FP, Sena Filho M, Altemani A, Speight PM, Vargas PA. Molecular signature of salivary gland tumors: potential use as diagnostic and prognostic marker. J Oral Pathol Med. 2016;45(2):101–110. doi: 10.1111/jop.12329. [DOI] [PubMed] [Google Scholar]
- 18.Skálová A, Stenman G, Simpson RHW, Hellquist H, Slouka D, Svoboda T, et al. The role of Molecular Testing in the Differential diagnosis of salivary gland carcinomas. Am J Surg Pathol. 2018;42(2):e11–e27. doi: 10.1097/pas.0000000000000980. [DOI] [PubMed] [Google Scholar]
- 19.Fehr A, Werenicz S, Trocchi P, Falk M, Friedrich RE, Stammler A, et al. Mucoepidermoid carcinoma of the salivary glands revisited with special reference to histologic grading and CRTC1/3-MAML2 genotyping. Virchows Arch. 2021;479(5):975–985. doi: 10.1007/s00428-021-03146-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bundele M, Weinreb I, Xu B, Chiosea S, Faquin W, Dias-Santagata D, et al. Mucoacinar Carcinoma: a rare variant of Mucoepidermoid Carcinoma. Am J Surg Pathol. 2021;45(8):1028–1037. doi: 10.1097/pas.0000000000001752. [DOI] [PubMed] [Google Scholar]
- 21.Zhang D, Liao X, Tang Y, Meyer RG, Van Dyke DL, Liu X, et al. Warthin-like Mucoepidermoid Carcinoma of the parotid gland: unusual morphology and diagnostic Pitfalls. Anticancer Res. 2019;39(6):3213–3217. doi: 10.21873/anticanres.13461. [DOI] [PubMed] [Google Scholar]
- 22.Bishop JA, Cowan ML, Shum CH, Westra WH. MAML2 rearrangements in variant forms of Mucoepidermoid Carcinoma: Ancillary Diagnostic Testing for the Ciliated and Warthin-like variants. Am J Surg Pathol. 2018;42(1):130–136. doi: 10.1097/pas.0000000000000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lennerz JK, Perry A, Mills JC, Huettner PC, Pfeifer JD. Mucoepidermoid carcinoma of the cervix: another tumor with the t(11;19)-associated CRTC1-MAML2 gene fusion. Am J Surg Pathol. 2009;33(6):835–843. doi: 10.1097/PAS.0b013e318190cf5b. [DOI] [PubMed] [Google Scholar]
- 24.Tirado Y, Williams MD, Hanna EY, Kaye FJ, Batsakis JG, El-Naggar AK. CRTC1/MAML2 fusion transcript in high grade mucoepidermoid carcinomas of salivary and thyroid glands and Warthin’s tumors: implications for histogenesis and biologic behavior. Genes Chromosom Cancer. 2007;46(7):708–715. doi: 10.1002/gcc.20458. [DOI] [PubMed] [Google Scholar]
- 25.Behboudi A, Enlund F, Winnes M, Andrén Y, Nordkvist A, Leivo I, et al. Molecular classification of mucoepidermoid carcinomas-prognostic significance of the MECT1-MAML2 fusion oncogene. Genes Chromosomes Cancer. 2006;45(5):470–481. doi: 10.1002/gcc.20306. [DOI] [PubMed] [Google Scholar]
- 26.Okabe M, Miyabe S, Nagatsuka H, Terada A, Hanai N, Yokoi M, et al. MECT1-MAML2 fusion transcript defines a favorable subset of mucoepidermoid carcinoma. Clin Cancer Res. 2006;12(13):3902–3907. doi: 10.1158/1078-0432.Ccr-05-2376. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama T, Miyabe S, Okabe M, Sakuma H, Ijichi K, Hasegawa Y, et al. Clinicopathological significance of the CRTC3-MAML2 fusion transcript in mucoepidermoid carcinoma. Mod Pathol. 2009;22(12):1575–1581. doi: 10.1038/modpathol.2009.126. [DOI] [PubMed] [Google Scholar]
- 28.Miyabe S, Okabe M, Nagatsuka H, Hasegawa Y, Inagaki A, Ijichi K, et al. Prognostic significance of p27Kip1, Ki-67, and CRTC1-MAML2 fusion transcript in mucoepidermoid carcinoma: a molecular and clinicopathologic study of 101 cases. J Oral Maxillofac Surg. 2009;67(7):1432–1441. doi: 10.1016/j.joms.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 29.Kuma Y, Yamada Y, Yamamoto H, Kohashi K, Ito T, Furue M, et al. A novel fusion gene CRTC3-MAML2 in hidradenoma: histopathological significance. Hum Pathol. 2017;70:55–61. doi: 10.1016/j.humpath.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 30.Argyris PP, Wehrs RN, García JJ, Koutlas IG. Fluorescence in-situ hybridization identifies mastermind-like 2 (MAML2) rearrangement in odontogenic cysts with mucous prosoplasia: a pilot study. Histopathology. 2015;66(6):791–797. doi: 10.1111/his.12526. [DOI] [PubMed] [Google Scholar]