Summary
Early growth response 1 (EGR1) mediates transcriptional programs that are indispensable for cell division, differentiation and apoptosis in numerous physiologies and pathophysiologies. Whole-body EGR1 knockouts in mice (Egr1KO) have advanced our understanding of EGR1 function in an in vivo context. To extend the utility of the mouse to investigate EGR1 responses in a tissue- and/or cell-type-specific manner, we generated a mouse model in which exon 2 of the mouse Egr1 gene is floxed by CRISPR/Cas9 engineering. The floxed Egr1 alleles (Egr1f/f) are designed to enable spatiotemporal control of Cre-mediated EGR1 ablation in the mouse. To confirm that the Egr1f/f alleles can be abrogated using a Cre driver, we crossed the Egr1f/f mouse with a global Cre driver to generate the Egr1 conditional knockout (Egr1d/d) mouse in which EGR1 expression is ablated in all tissues. Genetic and protein analysis confirmed the absence of exon 2 and loss of EGR1 expression in the Egr1d/d mouse respectively. Moreover, the Egr1d/d female exhibits overt reproductive phenotypes previously reported for the Egr1KO mouse. Therefore, studies described in this short technical report underscore the potential utility of the murine Egr1 floxed allele to further resolve EGR1 function at a tissue- and/or cell-type-specific level.
Keywords: Mouse, early growth response-1, CRISPR/Cas9, floxed, pituitary, ovary, uterus
1 ∣. INTRODUCTION
Acting as an immediate early response transcription factor for a broad spectrum of signaling cues, early growth response 1 (EGR1) mediates transcriptional programs that are essential for cellular replication, differentiation, and apoptosis in normal biology and pathobiology (Sukhatme, 1991; Thiel & Cibelli, 2002). Also known as NGFI-A, ZIF 268, KROX 24 or TIS8 (Thiel & Cibelli, 2002), EGR1 is a modular domain nuclear protein and a founding member of the evolutionary conserved EGR family of Cys2-His2 zinc finger transcription factors, which includes EGR2, EGR3, and EGR4 (Gashler & Sukhatme, 1995). Through binding cognate GC-rich (5’-GCG(T/G)GGGCG-3’) DNA motifs via its three zinc finger DNA binding domains (Swirnoff & Milbrandt, 1995), EGR1 directly activates or represses transcription of a diverse range of target genes, which in turn control a myriad of cellular processes, including (but not limited to) cell division, migration, adhesion, matrix deposition, and thrombosis (Adamson & Mercola, 2002; Bhattacharyya et al., 2011; Blaschke, Bruemmer, & Law, 2004; DeLigio & Zorio, 2009; Du, Wu, Ji, Liang, & Li, 2020; Maurya et al., 2022).
The Milbrandt and Charnay groups independently described the generation and characterization of global null mice for EGR1 (Egr1ko) (Lee et al., 1996; Lee, Tourtellotte, Wesselschmidt, & Milbrandt, 1995; Topilko et al., 1998), which have been invaluable in investigating EGR1 function in an in vivo context (Kim et al., 2018; Krones-Herzig, Adamson, & Mercola, 2003; Renaudineau, Poucet, Laroche, Davis, & Save, 2009; Thierion et al., 2017). Both Egr1ko models exhibit infertility defects in the female, which is primarily caused by a block in expression of the luteinizing hormone β (LH-β) subunit in the pituitary gonadotrope that leads to impairment in ovarian function. Charnay’s group also described a male reproductive defect in the Egr1ko mouse with an unclassified cellular and molecular etiology (Topilko et al., 1998); 16% of male mice heterozygous for the Egr1ko also displayed the male infertility phenotype. A male Egr1ko reproductive phenotype was not reported by the Milbrandt group (Lee et al., 1996).
Although the Egr1ko models have been indispensable in expanding our understanding of EGR1 function in a vivo context, the engineering of mouse models to ablate EGR1 in a tissue- or cell-type selective fashion has not been described. Given EGR1’s pleiotropic functions in multiple tissues, such a conditional knockout mouse would be useful in investigating EGR1 responses in select tissues or cell types that is free from confounding secondary phenotypic effects of EGR1 loss. Therefore, this short technical report comprehensively describes the generation of a mouse model in which exon 2 of the murine Egr1 gene is flanked (or floxed (Egr1f/f)) by loxP sites using a Clustered Regularly Interspersed Short Palindromic Repeats (CRISPR)/CRISPR associated (Cas) 9 (CRISPR/Cas9) editing approach, a genome modification strategy that we have successfully used in the recent past (Hai, Szwarc, Lanza, Heaney, & Lydon, 2019). To confirm that the Egr1f/f alleles operate as designed, we crossed the CRISPR-generated Egr1f/f mouse with an established global cre driver mouse. We demonstrate that the resultant EGR1 knockout (Egr1d/d) mouse neither contains exon 2 of the Egr1 gene nor expresses EGR1 protein. First line phenotypic characterization of Egr1d/d females demonstrates that these mice are infertile due to the absence of LH β subunit expression in the pituitary gland, which in turn causes an anovulatory defect, a hypoplastic uterus and infertility. The anovulatory impairment also causes a persistent pre-pubescent mammary gland phenotype in the adult Egr1d/d female mouse. Having confirmed the functionality of the floxed Egr1 allele in this short technical report, we believe that this new Egr1f/f mouse will significantly extend the utility of the mouse as an experimental model to elucidate EGR1 function in a tissue and cell-type specific manner.
2 ∣. RESULTS AND DISCUSSION
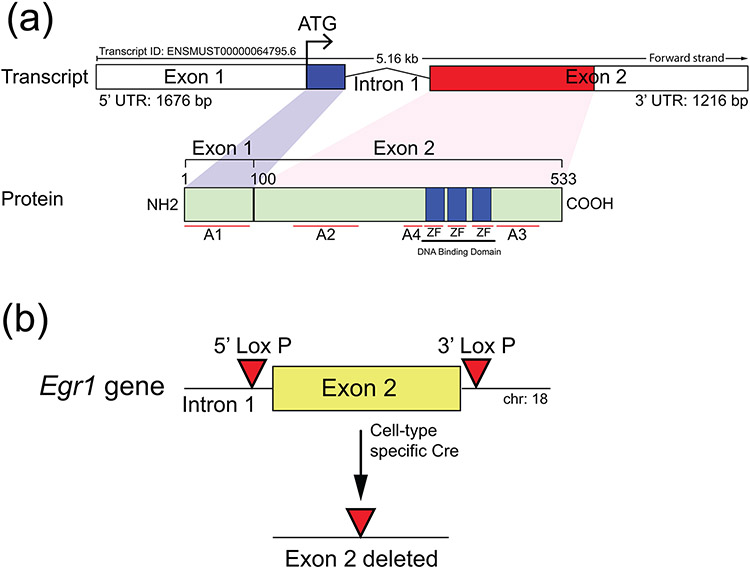
The murine EGR1 protein is 533 amino acids (aa) in length and encoded by two exons that comprise the coding region of the Egr1 gene on chromosome 18 (Figure 1a). In addition to a long 5’ untranslated region (UTR), the sequence of exon 1 contains the initiating ATG and encodes the first 99 aa of the EGR1 protein, which also harbors the activation 1 domain (A1) (Figure 1a). Exon 2 encodes the remainder of the EGR1 protein, which carries A2-4 along with a DNA binding domain consisting of three (C2H2) zinc finger (ZF) motifs; the EGR1 protein also contains inhibitory domains (Russo, Sevetson, & Milbrandt, 1995) that are not shown. Using a CRISPR/Cas9 engineering strategy that we recently used to flox a murine gene encoding another zinc finger transcription factor (Hai et al., 2019), the objective here was to target the insertion of the LoxP sequence to 5’ and 3’ locations relative to exon 2 of the murine Egr1 gene (Figure 1b). Such a LoxP targeting strategy would enable subsequent ablation of exon 2 of the murine Egr1 gene in the presence of a suitable cre driver (Figure 1b). It should be noted that in the generation of the previous Egr1ko mouse by homologous recombination in mouse embryonic stem (ES) cells, the Milbrandt group disrupted exon 2 of the murine Egr1 (NGFIA) gene by inserting a neomycin resistance selection cassette in the middle of the exon, which also resulted in an in-frame stop codon immediately downstream (Lee et al., 1995). In contrast, the Charnay group targeted a selection cassette (including a LacZ reporter gene) to the 5’ UTR of the murine Egr1 (Krox24) gene (Topilko et al., 1998), the insertion site was located 50 base pairs (bp) upstream from the initiating ATG.
FIGURE 1.
General targeting strategy to flox exon 2 of the murine Egr1 gene. (a) The schematic displays the overall structural organization of the mouse Egr1 gene and EGR1 protein (http://useast.ensembl.org and https://www.uniprot.org/uniprotkb/Q544D6/entry). Located on mouse chromosome 18, the Egr1 gene is comprised of two exons, in which exon 1 encodes ~30% of the protein that includes the initiating ATG and the activation 1 (A1) domain. Exon 2 encodes the remainder of the protein, which contains activation domains 2-4 (A2-4) as well as the DNA binding domain with its three zinc finger (ZF) motifs. (b) For effective ablation of EGR1 protein expression by cre-mediated excision in the mouse, exon 2 was chosen to be floxed by 5’ and 3’ LoxP sites using a CRISPR/Cas9 mediated targeting strategy; note: exon 2 of the Egr1 gene in the Egr1KO mouse was ablated by the Milbrandt group (Lee et al., 1996; Lee et al., 1995).
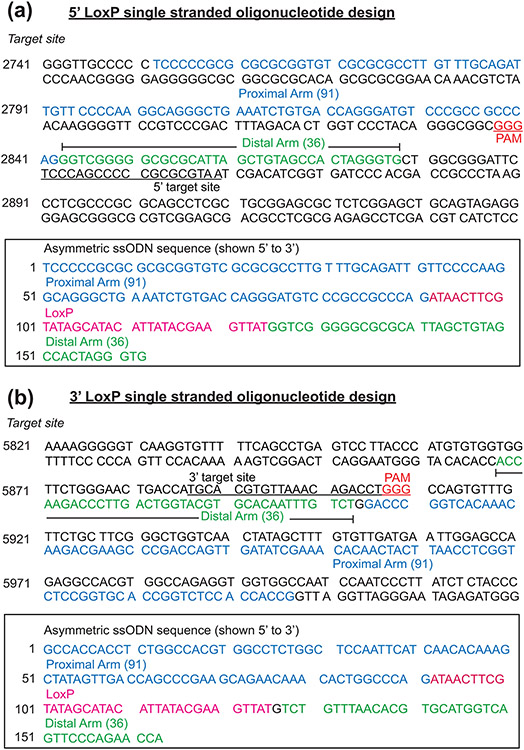
To target LoxP sequences that flox exon 2 of the murine Egr1 gene, Cas9 mRNA, two separate (and prospectively validated) single stranded oligonucleotide (ssODN) donors, each containing the 34 bp LoxP consensus sequence flanked asymmetrically by homology arms (127bp) to each of the 5’ and 3’ insertion sites (Figure 2) were co-injected into approximately 200 single-cell C57BL/6NJ pronuclear-stage zygotes, which were then transferred into ICR recipient pseudopregnant females (Hai et al., 2019). To reduce the chances of off-target events, sgRNAs predicted to have off-target sites with three mismatches or more were only used to target Cas9 endonuclease activity to the intronic sequences on each side of exon 2 (Figure 3a). The sequence location of the 5’ and 3’ sgRNAs in relation to exon 2 of the mouse Egr1 gene is shown in Supporting Information Figure S1.
FIGURE 2.
The design and position of the 5’ and 3’ ssODNs that were used to create the mouse Egr1 floxed allele. (a) Shown is the sequence of the 5’ target site in intron 1 of the mouse Egr1 allele. Sequences in blue (91 base pairs (bp)) and green (36 bp) represent the asymmetric homology arms that are proximal and distal respectively relative to the location of the 3’ protospacer adjacent motif (PAM (red)); the ssODN sequence is complementary to the non-targeted strand. The sequence of the 5’ target site is underlined whereas the 3’ PAM (GGG) site is highlighted in red. The box contains the sequence (in the 5’ to 3’ direction) of the ssODNs that were used for CRISPR/Cas9 mediated homology directed repair. The LoxP sequence is highlighted in red, the underlined sequence shows the target sequence disrupted by the LoxP sequence in the ssODN donor. The proximal and distal homology arm sequences are highlighted in blue and green respectively. (b) The sequence of the 3’ LoxP target site located 3’ to the mouse Egr1 gene. The sequences of the proximal and distal asymmetric homology arms are displayed blue and green respectively. The location and sequence of the 3’ target site is underlined with the 3’ PAM shown in red (GGG). The box encloses the sequence of the ssODN donor that was used to insert the LoxP site in the 3’ location relative to the mouse Egr1 gene. The underlined target sequence is disrupted by the LoxP sequence (highlighted in red), which is flanked by the proximal and distal homology arms that are highlighted in blue and green respectively.
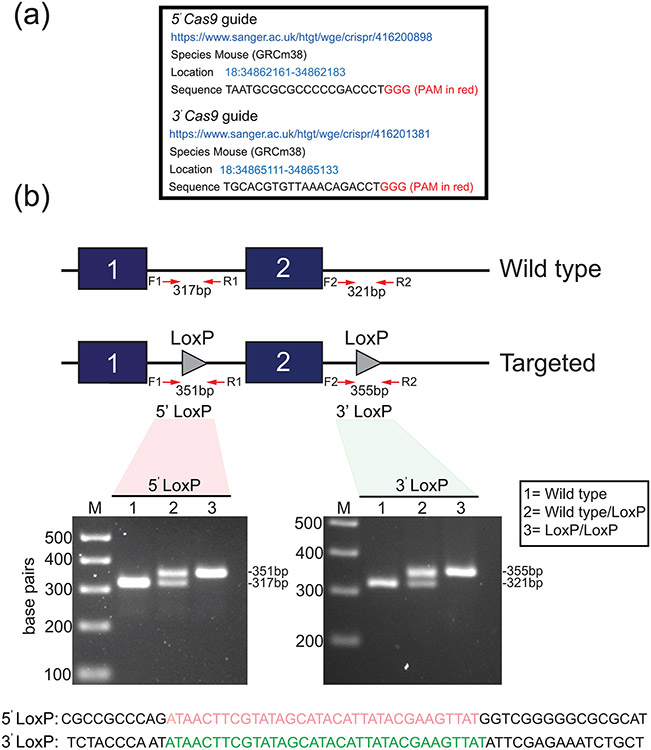
FIGURE 3.
Generation of the murine Egr1 floxed allele. (a) The location information of the 5’ and 3’ Cas9 guide sequences (sense strand) that were used to generate the murine Egr1 floxed allele by CRISPR/Cas9 genome editing is shown. Note: in vitro transcribed sgRNAs were microinjected into C57BL/6NJ zygotes. (b) Schematic of the CRISPR/Cas9-mediated targeting strategy to insert the LoxP sites (grey triangles) into intron 1 of and 3’ from the Egr1 gene to flox exon 2. The location of the forward and reverse PCR primers to amplify the 5’ and 3’ LoxP sites is indicated. Using forward and reverse PCR primers (F1 and R1 primers to amplify the 5’ LoxP site (351 bp); F2 and R2 primers to amplify the 3’ LoxP site (355 bp)), the gel shows a typical genotype result for wild type (lane 1 (no LoxP sites (317/321 bp)), wild type/LoxP (lane 2 (heterozygote (Egr1f/+ (317/321 and 351/355 bp)), and LoxP/LoxP (lane 3 (homozygous (Egr1f/f) for the 5’ and 3’ LoxP insertion (351/355 bp). The PCR genotyping result shown was performed on tail biopsy genomic DNA from F2 generation mice. The sequences for the 5’ (red) and 3’ (green) LoxP sites that were carried by these mice are shown.
Fifty-seven pups were delivered by seven ICR dams and were PCR screened for the insertion of the 5’ and 3’ LoxP insertion into the Egr1 allele. Table 1 lists the PCR primers used to genotype each LoxP insertion site. One PCR primer in the primer-pair is located outside the sequences of the homology arms in each of the ssODNs; the position of these primers in relation to exon 2 is shown in Figure 3b and detailed in Supporting Information Figure S2. From fifty-seven pups screened, two potential male founders ((F0) #G17775 and #G17779) carried the 3’ LoxP insertion site and one potential male founder carried the 5’ LoxP site ((F0) #G177739); five mice in this screen were shown to contain indels. Unlike our recently published CRISPR-engineered mouse model (Hai et al., 2019), potential F0 mice carrying both 5’ and 3’ LoxP site insertions in cis (or on the same Egr1 allele) were not detected. Therefore, the male F0 mouse (#G177739) carrying the 5’ LoxP site was breed for germline transmission of the LoxP insertion. Progeny (F1 and F2), which were positive for the 5’ LoxP insertion, were bred to generate embryos that were homozygous for the 5’ LoxP insertion. Embryos, which were homozygous for the 5’ LoxP insertion, were used for CRISPR/Cas 9-mediated insertion of the 3’ LoxP site. From twelve potential founders, which were born from transferring sixty embryos to three pseudopregnant ICR females, one potential F0 male (#G20079) was genotyped as carrying the 5’ and 3’ LoxP insertions on the same Egr1 allele. Subsequent breeding from this F0 male confirmed germline transmission of the 5’ and 3’ LoxP insertions (Figure 3b), and that these insertions were carried on the same Egr1 allele. Subsequent progeny heterozygous or homozygous for the floxed Egr1 allele were termed Egr1f/+ or Egr1f/f respectively. These studies also confirmed that Egr1f/f mice were phenocopies of their wild type siblings. Sanger sequencing of the 5’ and 3’ LoxP insertions using genomic DNA isolated from progeny of F2 mice confirmed that each of the LoxP sites is intact (Supporting Information Figure S2). Importantly, off-target Cas9 cleavage activity was not detected through analysis of the top five potential off-target sites as scored by the CRISPR Finder tool ((https://www.sanger.ac.uk/htgt/wge/) and Supporting Information Figure S3).
TABLE 1.
List of Primers used for genotyping
| F1 primer | 5’-GGCAGGACGTTTGTTTTGGA-3’ |
| R1 primer | 5’-CCATTTCAACAGCTGACGCA-3’ |
| F2 primer | 5’-GTGGTTCTGGGAACTGACCA-3’ |
| R2 primer | 5’-TCCTGGATTCTTGGCTGGAC-3’ |
| F3 primer | 5’-CGCCTTGTTTGCAGATTGTT-3’ |
| R3 primer | 5’-GATGAAGAGGTCGGAGGATTG-3’ |
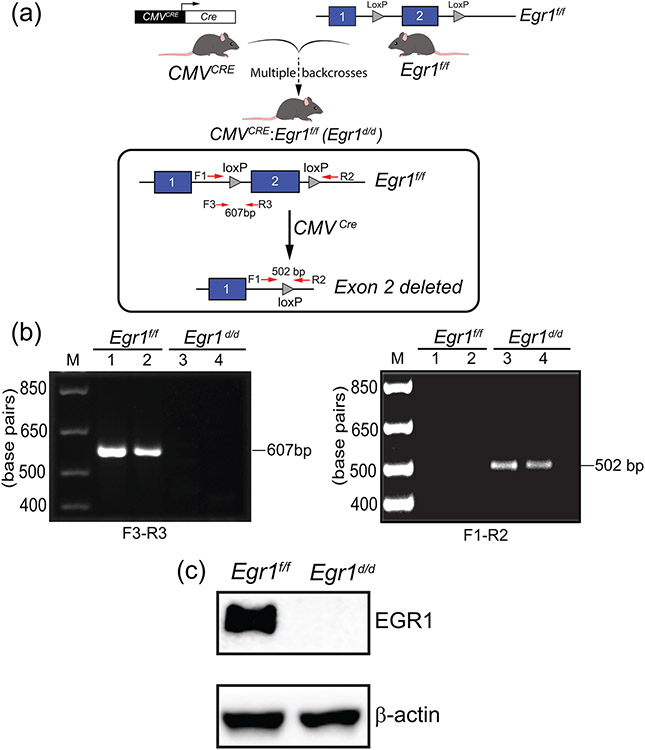
To assess that the Egr1f/f alleles can be abrogated by cre-mediated excision, the Egr1f/f mouse line was crossed with a global cre-driver transgenic mouse in which the cytomegalovirus (CMV) promoter drives cre recombinase (CMVcre) expression in both somatic and germ cells (Schwenk, Baron, & Rajewsky, 1995), to generate the CMVcre: Egr1f/+ heterozygote, which is then intercrossed to generate CMVcre: Egr1f/f bigenic mouse (abbreviated Egr1d/d). The resulting bigenic mouse (CMVcre: Egr1f/f) is termed the Egr1d/d mouse from hereon (Figure 4a). Genotyping with the PCR primer-pairs: F1/R2, and F3/R3 (Table 1 and Supporting Information Figure S2), confirmed the absence of exon 2 in the Egr1d/d mouse (Figure 4b). Moreover, western blot analysis showed the absence of EGR1 protein expression in whole brain tissue from the Egr1d/d mouse (Figure 4c). Adult Egr1d/d mice of both sexes were of normal weight and size, which agrees with observations made from studies on the Egr1KO model described by the Milbrandt group (Lee et al., 1996; Lee et al., 1995); note: Charnay’s group reported a small but significant difference in body weight between their Egr1KO and control siblings (Topilko et al., 1998).
FIGURE 4.
Generation of the Egr1d/d bigenic mouse. (a) The schematic summarizes the breeding scheme to generate the Egr1d/d bigenic mouse. The CMVcre transgenic mouse was crossed with Egr1f/f mice to generate the CMVcre: Egr1f/+ heterozygote, which is then intercrossed to generate CMVcre: Egr1f/f bigenic mouse (abbreviated Egr1d/d). The location of the F1, R2, F3, and R3 PCR primers to detect loss of the Egr1 exon 2 in the Egr1d/d bigenic mouse is shown. (b) Using the F1 and F3 forward and R2 and R3 reverse primers, the PCR genotyping result confirms the absence of exon 2 in the Egr1d/d bigenic mouse. (c) Western analysis using brain tissue protein isolates confirms that EGR1 protein is not produced in the Egr1d/d mouse. Lanes 1 and 2 represent protein isolated from Egr1f/f and Egr1d/d brain tissue respectively. Note the absence of EGR1 protein in the Egr1d/d lane. The experiment was performed in triplicate and β-actin served as a loading control.
For this short technical report, the female Egr1d/d mouse was the focus of the initial phenotype analysis to confirm that the Egr1f/f alleles operate as designed. Similar to the Egr1KO models reported by the Milbrandt and Charnay groups (Lee et al., 1996; Lee et al., 1995; Topilko et al., 1998), Egr1d/d female mice are infertile. Using a standard six-month breeding trial using proven stud wild type males, Egr1d/d females (n=5) failed to produce litters unlike their control siblings (n=6). Immunohistological studies revealed that EGR1 protein is not expressed in the female Egr1d/d pituitary gland, unlike control siblings (Figure 5c-h). In addition, expression of the LH β-subunit was absent in the adenohypophysis of the female Egr1d/d pituitary gland (Figure 5i-j), which was also reported for the Egr1KO models described by the Milbrandt and Charnay groups (Lee et al., 1996; Lee et al., 1995; Topilko et al., 1998). Of note, a number of molecular studies have demonstrated that EGR1 regulates the expression of the pituitary LH β-subunit (Halvorson, Ito, Jameson, & Chin, 1998), which in turn is required for a number of reproductive functions in the female, including ovulation (Lee et al., 1996; Topilko et al., 1998).
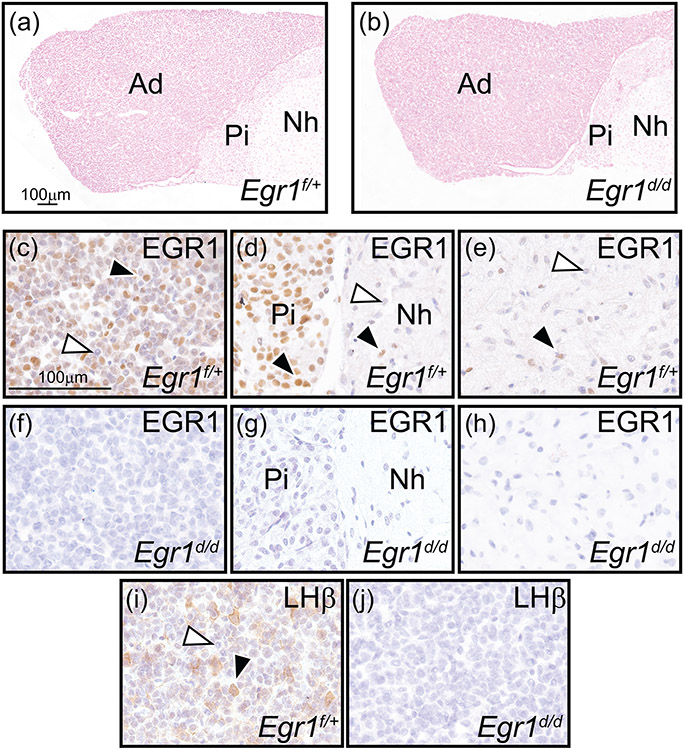
FIGURE 5.
Absence of EGR1 and LH-β subunit expression in the female Egr1d/d pituitary gland. (a) and (b) show representative nuclear-fast red stained sections of Egr1f/+ and Egr1d/d pituitary gland tissue respectively; representative of four adult (nine weeks old) mice per genotype. Pituitary gland size and histomorphology were equivalent for both genotypes (adenohypophysis (or the anterior pituitary gland or pars distalis), pars intermedia (or intermediate gland), and neurohypophysis (or posterior pituitary gland or pars nervosa) are indicated by Ad, Pi, and Nh respectively). Scale bar in (a) also applies to (b). (c-e) Immunohistochemical analysis of EGR1 expression in the Ad, Pi, and Nh of the Egr1f/+ pituitary gland respectively. Note in (c) the presence of numerous EGR1 positive cells in the Ad (black arrowhead); a subgroup of Ad cells is EGR1 negative (white arrowhead). (d) The majority of cells in the Pi region is immunopositive for EGR1 (black arrowhead), with few cells EGR1 positive in the Nh region ((d) and (e) (black arrowhead)); EGR1 negative cells in the Nh are indicated by a white arrowhead ((d) and (e)). (f-h) Expression of EGR1 is absent in all regions of the Egr1d/d pituitary gland. (i) Immunohistochemical detection of the LH β subunit in the Ad region of the Egr1f/+ pituitary gland. Note a subset of Ad cells is positive for LH β expression (black arrowhead); the white arrowhead indicates an Ad cell that is LH β negative. (j) Expression of the LH β subunit is absent in the Ad region of the Egr1d/d pituitary gland. Scale bar in (c) applies to (d-j).
As reported by the Milbrandt and Charnay groups (Lee et al., 1996; Topilko et al., 1998), the reproductive tract is conspicuously atrophied in the adult Egr1d/d female compared with controls (Figure 6a-e). The diameter of the uterine horn is significantly smaller in the Egr1d/d mouse (Figure 6c (right panel)), and immunohistochemical analysis again confirms the absence of EGR1 expression in the Egr1d/d uterus (Figure 6d, e). Compared with ovaries from control siblings, the Egr1d/d ovary exhibits a significant size reduction, a corpora lutea deficit, and the presence of overt parenchymal angiectasis (Figure 7a-f), which are hallmarks of an atrophied ovary (Davis, Dixon, & Herbert, 1999). The above ovarian phenotypes were also reported in whole or in part by the Milbrandt and Charnay groups (Lee et al., 1996; Topilko et al., 1998). Because of the anovulatory phenotype by the Egr1d/d mouse, the mammary gland fails to undergo post-pubertal ductal epithelial expansion that drives secondary and tertiary branching to the periphery of the fat pad (Figure 8a-d). The presence of a pre-pubertal mammary morphology in the adult Egr1d/d mouse is likely due to inadequate ovarian-derived estrogen and progesterone stimulation (Bocchinfuso & Korach, 1997; Fernandez-Valdivia et al., 2005; Hai et al., 2018). Again, immunohistochemical analysis confirmed the absence of EGR1 expression in both the epithelial and stromal compartments of the Egr1d/d mammary gland (Figure 8e-h).
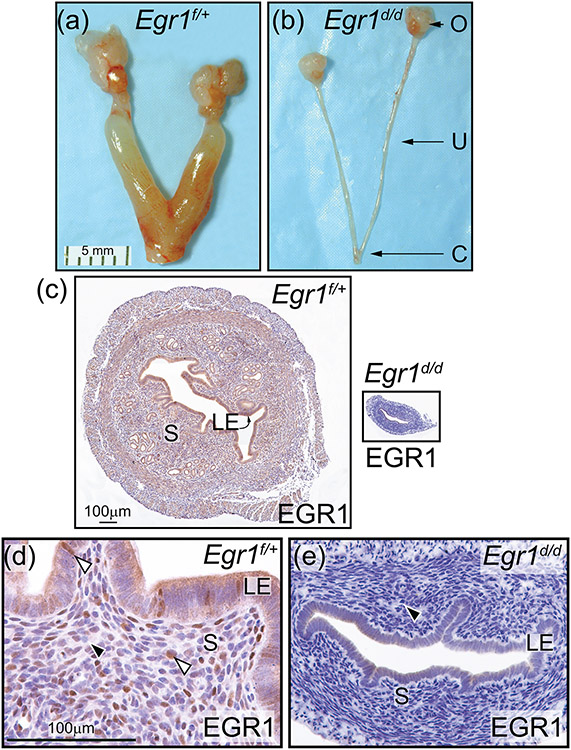
Figure 6.
Abrogation of EGR1 results in a hypoplastic uterus in the Egr1d/d mouse. (a) and (b) show the gross morphology of the Egr1f/+ and Egr1d/d female reproductive tract respectively; representative of five adult mice per genotype. Compared with the Egr1f/+ uterus (a), note the thin Egr1d/d uterine horn (b); ovary, uterus, and cervix are denoted by O, U, and C respectively. Scale bar in (a) applies to (b). (c) Transverse section of the mid-region of the Egr1f/+ and Egr1d/d uterine horn (left and right panels respectively) is shown; luminal epithelium and stroma are indicated by LE and S respectively. Both tissue sections were immunohistochemically stained for EGR1 expression. Note EGR1 expression in S and LE compartments of the Egr1f/+ uterus whereas EGR1 expression is not detected in the Egr1d/d uterus. Scale bar in the left panel applies to right panel. (d) and (e) are higher magnification images of the micrographs shown in the left and right panels in (c) respectively. (d) Expression of EGR1 is detected in subsets of cells in the S and LE compartment of the Egr1f/+ uterus (white arrowhead). (e) Expression of EGR1 is absent in the Egr1d/d uterus (black arrowhead). Scale bar in (d) applies to (e).
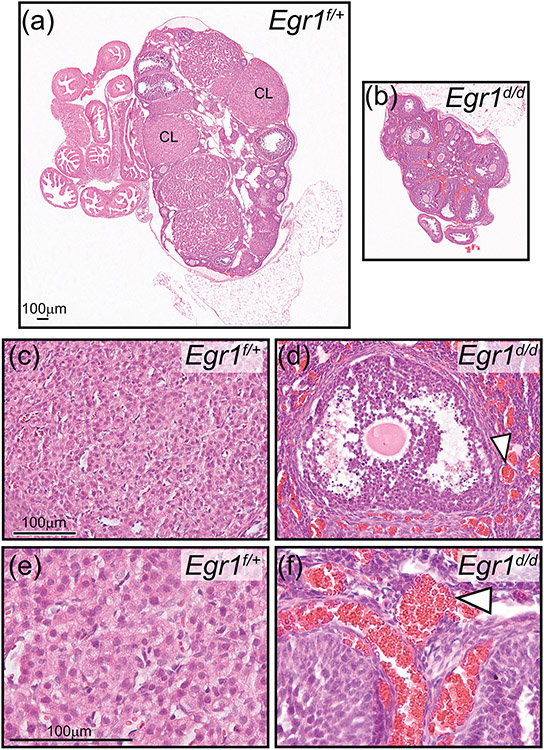
Figure 7.
Absence of corpora lutea in the atrophied Egr1d/d ovary. (a) Hematoxylin and eosin stained section of the Egr1f/+ ovary; a corpus luteum is indicated by CL. (b) Similarly stained Egr1d/d ovarian tissue section; note the absence of CLs. Also note the diminished size of the Egr1d/d ovary compared with the Egr1f/+ ovary shown in (a). Scale bar in (a) applies to (b). The histological data are representative of five adult mice per genotype. (c) Field of view shows luteal cells within the CL of an Egr1f/+ ovary. (d) Image shows an antral follicle within the Egr1d/d ovary; note the juxtaposed hemorrhagic area indicated by the white arrowhead. Scale bar in (c) applies to (d). (e) and (f) represent higher magnification images of regions in (c) and (d) respectively. Again, note the hemorrhagic areas in the Egr1d/d ovary (white arrowhead). Scale bar in (e) applies to (f).
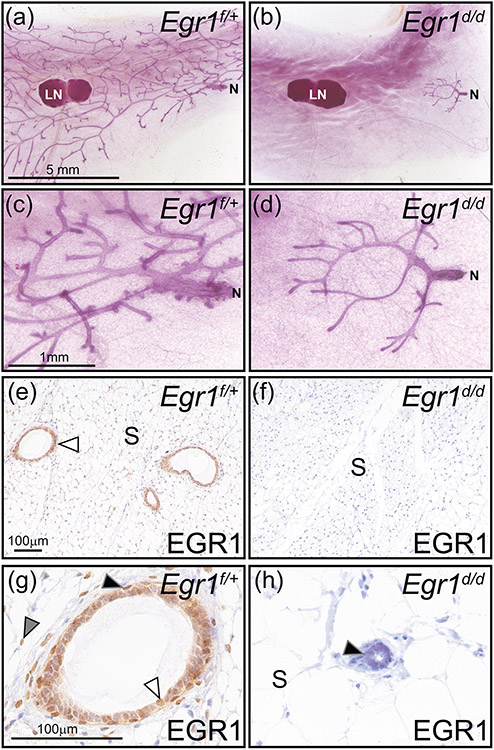
Figure 8.
A prepubescent mammary gland phenotype in the adult Egr1d/d mouse. (a) Whole mount of an inguinal mammary gland from an adult Egr1f/+ mouse, note the extensive epithelial ductal branching from the nipple (N) to the periphery of the fat pad. The lymph node is indicated by LN, which is used as a structural reference point in the inguinal gland. (b) Absence of epithelial ductal elongation and dichotomous branching in the mammary of the adult Egr1d/d mouse. Note that the majority of the Egr1d/d mammary gland fat pad is devoid of the epithelial compartment. Mammary gland results are representative of five adult mice per genotype. Scale bar in (a) applies to (b). (c) and (d) represent higher magnification images shown in (a) and (b) respectively; scale bar in (c) applies to (d). (e) Immunohistochemical detection of EGR1 expression in a section derived from Egr1f/+ mammary gland tissue. Immunopositivity in the mammary epithelium is indicated by white arrowhead; mammary stroma is denoted as S. (f) Immunopositivity for EGR1 is absent in the mammary gland of the Egr1d/d mouse. As indicated in panel (b), the majority of the mammary gland is devoid of the epithelial compartment in the Egr1d/d mouse; scale bar in (e) applies to (f). (g) Higher magnification image clearly showing EGR1 expression in the basal epithelium (black arrowhead), a subset of luminal epithelial cells (white arrowhead), and stromal cells (gray arrowhead). (h) Immunopositivity for EGR1 is not detected in cells of vestigial epithelial ducts that are localized to the nipple region of the adult Egr1d/d mammary gland; the stromal compartment is also negative for EGR1 immunopositivity. Scale bar in (g) applies to (h).
Collectively, our genetic, protein and phenotypic analysis in this short technical report demonstrates that the Egr1f/f allele operates as designed and therefore will be of significant utility for researchers interested in resolving EGR1 responses at a tissue and/or cell-type specific level. For example, we have recently demonstrated that EGR1 is critical for both normal and abnormal uterine function (Maurya et al., 2022; Park et al., 2021; Szwarc et al., 2019). As EGR1 is expressed in endometrial epithelial and stromal cells (Maurya et al., 2022; Park et al., 2021; Szwarc et al., 2019), selective ablation of the EGR1 in the epithelial or stromal specific cell type will provide an unprecedented opportunity to investigate EGR1’s role in endometrial biology and pathobiology at a cell-type specific level. In closing, it should be noted that the use of an Egr1f/f mouse was recently reported by a research group in China (Lei et al., 2022; Wu et al., 2019; Wu et al., 2018); however, details on how this mouse was generated or whether the mouse will be made available to the scientific community has not been published.
3 ∣. MATERIALS AND METHODS
3.1 ∣. Generation of the Egr1f/f mouse using CRISPR/Cas9 gene editing
Two sgRNAs along with two ssODNs containing LoxP sequences were used to insert a LoxP site into the 5’ and 3’ positions of exon 2 of the mouse Egr1 gene. Exon 2 was selected because it encodes greater than 70% of the murine EGR1 protein (Figure 1). Standard methods were used to co-microinject 100 ng/μl Cas9 mRNA, 20 ng/μl validated sgRNA (each) and 100 ng/μl of ssODNs (each) into the cytoplasm of 200 C57BL/6NJ embryos (Hai et al., 2019; Lanza et al., 2018). Microinjectioned zygotes were transferred into pseudopregnant ICR recipient females (~25–32 zygotes per recipient). The C57BL/6NJ and ICR mouse strains were purchased from the Jackson Laboratory (Bar Harbor, ME). Standard genotyping methods of potential founder mice were used (Hai et al., 2019; Lanza et al., 2018). Sanger sequencing of the 5’ and 3’ LoxP insertions in the mouse genome was performed as previously described (Hai et al., 2019; Lanza et al., 2018).
3.2 ∣. Off-target Cas9 activity analysis
The top five candidate off-target sites for each sgRNA used in the targeting of the 5’ and 3’ LoxP sequences into the murine Egr1 gene were identified using the Wellcome Trust Sanger Institute Genome Editing website (https://wge.stemcell.sanger.ac.uk//). Information on the forward and reverse PCR primers used to amplify 80–180 base pair amplicons are described in Supporting Information Figure 3, which lists the location, sequence, and the number of mismatches for each of the sgRNAs. Genomic DNA from F2 mice was prepared as previously reported (Hai et al., 2019; Lanza et al., 2018). Genomic DNA analysis was conducted with the HighResolution Melt Analysis method, using the MeltDoctor HRM Master Mix (ThermoFisher Scientific Inc., Waltham, MA (#4415440)) and run on the Applied Biosystems QuantStudio 7 Flex Real-Time PCR System (ThermoFisher Scientific Inc.). In addition to the test samples, at least two wild-type samples were concurrently analyzed. For samples with a HRM analysis result that deviated from the wild-type sample (suggesting a mutagenesis event), PCR products were cloned and Sanger sequenced as described (Hai et al., 2019; Lanza et al., 2018).
3.3 ∣. Generation of the conditional knockout Egr1d/d mouse
To generate the Egr1d/d mouse, Egr1f/f mice were crossed with the CMVcre mouse in the C57/BL6 inbred strain (The Jackson Laboratory, Bar Harbor, ME (stock number: 006054; strain name B6. C-Tg(CMV-cre)1 Cgn/J (Schwenk et al., 1995))). With this conditional knockout mouse strategy, the expression of the targeted protein is ablated in all tissues, including germ cells (Hai et al., 2019; Schwenk et al., 1995).
Mice were housed and maintained in an AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care) accredited vivarium, administered by the Center for Comparative Medicine at Baylor College of Medicine. In temperature-controlled animal rooms (22 ± 2°C) with a 12-hr lights-on: 12-hr lights-off photocycle, mice received an irradiated Tekland global soy protein-free extruded rodent diet (Harlan Laboratories, Inc., Indianapolis, IN) and fresh water ad libitum. Mouse experimental protocols were followed in accordance with the guidelines detailed in the Guide for the Care and Use of Laboratory Animals (“The Guide”, 8th ed., 2011)), published by the National Research Council of the National Academies, Washington, DC (www.nap.edu). All animal procedures described in this study were prospectively approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine. As with our previous engineered mouse models, the Egr1f/f mouse will be made freely available to members of the scientific community upon request.
Immunoblotting
Protein isolates (20 μg) were resolved on 7.5% sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gels before transfer to polyvinylidene difluoride (PVDF) membranes. After protein transfer, PVDF membranes were blocked for 1 h with 5% non-fat dry milk ((sc-2324 (Blotto)) Santa Cruz Biotechnology Inc.) in Tris-buffered saline with Tween 20 (TBS-T) before incubation overnight at 4°C with the following primary antibodies: anti-EGR1 (#4154, Cell Signaling Technology, Inc.) diluted 1:1000 and anti-β-actin (#A00702, GenScript Biotech, Piscataway, NJ) diluted 1:100,000 in 5% non-fat milk in TBS-T buffer. Following primary antibody incubation, immunoblots were probed with anti-rabbit ((A27036 (1:5000 dilution)) ThermoFisher Scientific Inc.) and anti-mouse IgG secondary antibodies conjugated with horse radish peroxidase (HRP ((#7076 (1:10,000 dilution)) Cell Signaling Technology, Inc.) respectively in 5% non-fat milk in TBS-T buffer for 1 h at room temperature. The resultant chemiluminescence was detected with the SuperSignal West Pico PLUS Chemiluminescent Substrate ((#1863097) ThermoFisher Scientific, Inc.). Immunoreactive bands were digitally imaged using the Azure 300 Chemiluminescent Western Blot Imager (Azure Biosystems, Inc., Dublin, CA).
Immunohistochemistry
For immunohistochemical detection, tissues were fixed overnight in 4% paraformaldehyde in phosphate buffered saline (PBS) before pre-processing, paraffin embedding and sectioning onto glass slides. Immunohistochemical detection of EGR1 was achieved using a primary rabbit monoclonal antibody ((#4153); diluted 1:100; Cell Signaling Technology Inc., Danvers, MA). A primary rabbit polyclonal antibody was used to immunohistochemically detect the luteinizing hormone (LH) beta-subunit (1:200 dilution; #PA5-102674; ThermoFisher Scientific Inc.). Following incubation with a HRP-conjugated goat anti-rabbit secondary antibody ((P-1000); diluted 1:200; Vector Laboratories, Burlingame CA), peroxidase activity was detected with the Vectastain Elite ABC-HRP kit (Vector Laboratories Inc.). Tissue sections were then counterstained with hematoxylin before sequentially applying Permount mounting medium and coverslips. Nuclear fast red staining was performed according to the manufacturer’s instructions (#J61010.AP ThermoFisher Scientific Inc.). Images of immunostained tissue sections were digitally captured using a color chilled AxioCam MRc5 digital camera affixed to a Zeiss AxioImager A1 upright microscope (Carl Zeiss AG, Jena, Germany). Digital images were processed using the latest version of Adobe Photoshop and Illustrator software programs (Adobe Systems Inc. San Jose, CA).
Mammary gland whole mounts
As previously described (Hai et al., 2018), inguinal (or #4) mammary glands were fixed overnight in an ethanol: acetic acid (3:1) solution before rehydration with 70% ethanol and water. Rehydrated tissues were then stained with a carmine aluminum solution for 24 hours at room temperature. The mammary fat pad was cleared in toluene solvent before mounting the cleared tissue on slides with Permount (Fisher Scientific, Pittsburgh, PA) (Hai et al., 2018). Mammary gland whole mounts were digitally imaged using a Zeiss stereo-microscope with an attached AxioCam MRC-5 digital camera (Carl Zeiss AG, Jena, Germany).
Supplementary Material
SUPPORT INFORMATION FIGURE S1 Location of the insertion sites for 5’ and 3’ LoxP sequences that flox exon 2 of the murine Egr1 gene. The exon 2 coding region is highlighted in yellow whereas the 5’ and 3’ guide sequences are indicated by green and blue respectively.
SUPPORT INFORMATION FIGURE S3 The list of the locations, sequence, and the number of mismatches for each of the sgRNAs used in this study.
SUPPORT INFORMATION FIGURE S2 Germline transmission of the 5’ and 3’ LoxP insertions into the murine Egr1 locus. Sequence traces of 5’ and 3’ LoxP sites (red) in genomic DNA from tail biopsy tissue from F2 generation Egr1f/f mice. Sequence of the 5’ and 3’ asymmetric oligonucleotides are highlighted in green. Note: both sequences match the predicted sequences for both LoxP insertions (see: 1, 2, and 3 sequences).
ACKNOWLEDGMENTS
The authors thank Jie Li and Rong Zhao for their outstanding technical expertise. We thank the Genetically Engineered Rodent Models Core at Baylor College of Medicine for assistance with mouse production. Resources accessed through the core were supported by a National Institutes of Health gran (P30CA12512) to the Dan L. Duncan Comprehensive Cancer Center. Finally, this research was supported by the National Institutes of Health/ National Institute of Child and Human Development R01 grant: HD042311 to JPL.
REFERENCES
- Adamson ED, & Mercola D (2002). Egr1 transcription factor: multiple roles in prostate tumor cell growth and survival. Tumour Biol, 23(2), 93–102. doi: 10.1159/000059711 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Wu M, Fang F, Tourtellotte W, Feghali-Bostwick C, & Varga J (2011). Early growth response transcription factors: key mediators of fibrosis and novel targets for anti-fibrotic therapy. Matrix Biol, 30(4), 235–242. doi: 10.1016/j.matbio.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke F, Bruemmer D, & Law RE (2004). Egr-1 is a major vascular pathogenic transcription factor in atherosclerosis and restenosis. Rev Endocr Metab Disord, 5(3), 249–254. doi: 10.1023/B:REMD.0000032413.88756.ee [DOI] [PubMed] [Google Scholar]
- Bocchinfuso WP, & Korach KS (1997). Mammary gland development and tumorigenesis in estrogen receptor knockout mice. J Mammary Gland Biol Neoplasia, 2(4), 323–334. doi: 10.1023/a:1026339111278 [DOI] [PubMed] [Google Scholar]
- Davis BJ, Dixon D, & Herbert RA (1999). Ovary, oviduct, uterus, cervix, and vagina. In Maronpot RR, Boorman GA, & Gaul BW (Eds.), Pathology of the Mouse: Reference and atlas (pp. 409–444). Vienna, IL: Cache River Press. [Google Scholar]
- DeLigio JT, & Zorio DA (2009). Early growth response 1 (EGR1): a gene with as many names as biological functions. Cancer Biol Ther, 8(20), 1889–1892. doi: 10.4161/cbt.8.20.9804 [DOI] [PubMed] [Google Scholar]
- Du K, Wu X, Ji X, Liang N, & Li Z (2020). Early growth response 1 promoted the invasion of glioblastoma multiforme by elevating HMGB1. J Neurosurg Sci. doi: 10.23736/S0390-5616.20.05107-3 [DOI] [PubMed] [Google Scholar]
- Fernandez-Valdivia R, Mukherjee A, Mulac-Jericevic B, Conneely OM, DeMayo FJ, Amato P, & Lydon JP (2005). Revealing progesterone's role in uterine and mammary gland biology: insights from the mouse. Semin Reprod Med, 23(1), 22–37. doi: 10.1055/s-2005-864031 [DOI] [PubMed] [Google Scholar]
- Gashler A, & Sukhatme VP (1995). Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog Nucleic Acid Res Mol Biol, 50, 191–224. doi: 10.1016/s0079-6603(08)60815-6 [DOI] [PubMed] [Google Scholar]
- Hai L, Szwarc MM, Lanza DG, Heaney JD, & Lydon JP (2019). Using CRISPR/Cas9 engineering to generate a mouse with a conditional knockout allele for the promyelocytic leukemia zinc finger transcription factor. Genesis, 57(3), e23281. doi: 10.1002/dvg.23281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai L, Szwarc MM, Wetendorf M, Wu SP, Peavey MC, Grimm SL, … Lydon JP (2018). A mouse model engineered to conditionally express the progesterone receptor-B isoform. Genesis, 56(8), e23223. doi: 10.1002/dvg.23223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorson LM, Ito M, Jameson JL, & Chin WW (1998). Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone beta-subunit gene expression. J Biol Chem, 273(24), 14712–14720. doi: 10.1074/jbc.273.24.14712 [DOI] [PubMed] [Google Scholar]
- Kim HR, Kim YS, Yoon JA, Yang SC, Park M, Seol DW, … Song H (2018). Estrogen induces EGR1 to fine-tune its actions on uterine epithelium by controlling PR signaling for successful embryo implantation. FASEB J, 32(3), 1184–1195. doi: 10.1096/fj.201700854RR [DOI] [PubMed] [Google Scholar]
- Krones-Herzig A, Adamson E, & Mercola D (2003). Early growth response 1 protein, an upstream gatekeeper of the p53 tumor suppressor, controls replicative senescence. Proc Natl Acad Sci U S A, 100(6), 3233–3238. doi: 10.1073/pnas.2628034100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza DG, Gaspero A, Lorenzo I, Liao L, Zheng P, Wang Y, … Heaney JD (2018). Comparative analysis of single-stranded DNA donors to generate conditional null mouse alleles. BMC Biol, 16(1), 69. doi: 10.1186/s12915-018-0529-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SL, Sadovsky Y, Swirnoff AH, Polish JA, Goda P, Gavrilina G, & Milbrandt J (1996). Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1). Science, 273(5279), 1219–1221. doi: 10.1126/science.273.5279.1219 [DOI] [PubMed] [Google Scholar]
- Lee SL, Tourtellotte LC, Wesselschmidt RL, & Milbrandt J (1995). Growth and differentiation proceeds normally in cells deficient in the immediate early gene NGFI-A. J Biol Chem, 270(17), 9971–9977. doi: 10.1074/jbc.270.17.9971 [DOI] [PubMed] [Google Scholar]
- Lei X, Xu Q, Li C, Niu B, Ming Y, Li J, … Mao Y (2022). Egr1 confers protection against acetaminophen-induced hepatotoxicity via transcriptional upregulating of Acaa2. Int J Biol Sci, 18(9), 3800–3817. doi: 10.7150/ijbs.71781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya VK, Szwarc MM, Fernandez-Valdivia R, Lonard DM, Yong S, Joshi N, … Lydon JP (2022). Early growth response 1 transcription factor is essential for the pathogenic properties of human endometriotic epithelial cells. Reproduction, 164(2), 41–54. doi: 10.1530/REP-22-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Park SH, Park H, Kim HR, Lim HJ, & Song H (2021). ADAMTS-1: a novel target gene of an estrogen-induced transcription factor, EGR1, critical for embryo implantation in the mouse uterus. Cell Biosci, 11(1), 155. doi: 10.1186/s13578-021-00672-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudineau S, Poucet B, Laroche S, Davis S, & Save E (2009). Impaired long-term stability of CA1 place cell representation in mice lacking the transcription factor zif268/egr1. Proc Natl Acad Sci U S A, 106(28), 11771–11775. doi: 10.1073/pnas.0900484106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo MW, Sevetson BR, & Milbrandt J (1995). Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc Natl Acad Sci U S A, 92(15), 6873–6877. doi: 10.1073/pnas.92.15.6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Baron U, & Rajewsky K (1995). A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res, 23(24), 5080–5081. doi: 10.1093/nar/23.24.5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhatme VP (1991). The Egr family of nuclear signal transducers. Am J Kidney Dis, 17(6), 615–618. doi: 10.1016/s0272-6386(12)80333-7 [DOI] [PubMed] [Google Scholar]
- Swirnoff AH, & Milbrandt J (1995). DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Mol Cell Biol, 15(4), 2275–2287. doi: 10.1128/MCB.15.4.2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwarc MM, Hai L, Gibbons WE, Mo Q, Lanz RB, DeMayo FJ, & Lydon JP (2019). Early growth response 1 transcriptionally primes the human endometrial stromal cell for decidualization. J Steroid Biochem Mol Biol, 189, 283–290. doi: 10.1016/j.jsbmb.2019.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G, & Cibelli G (2002). Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol, 193(3), 287–292. doi: 10.1002/jcp.10178 [DOI] [PubMed] [Google Scholar]
- Thierion E, Le Men J, Collombet S, Hernandez C, Coulpier F, Torbey P, … Gilardi-Hebenstreit P (2017). Krox20 hindbrain regulation incorporates multiple modes of cooperation between cis-acting elements. PLoS Genet, 13(7), e1006903. doi: 10.1371/journal.pgen.1006903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topilko P, Schneider-Maunoury S, Levi G, Trembleau A, Gourdji D, Driancourt MA, … Charnay P (1998). Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol Endocrinol, 12(1), 107–122. doi: 10.1210/mend.12.1.0049 [DOI] [PubMed] [Google Scholar]
- Wu J, Tao W, Bu D, Zhao Y, Zhang T, Chong D, … Li C (2019). Egr-1 transcriptionally activates protein phosphatase PTP1B to facilitate hyperinsulinemia-induced insulin resistance in the liver in type 2 diabetes. FEBS Lett, 593(21), 3054–3063. doi: 10.1002/1873-3468.13537 [DOI] [PubMed] [Google Scholar]
- Wu J, Tao WW, Chong DY, Lai SS, Wang C, Liu Q, … Li CJ (2018). Early growth response-1 negative feedback regulates skeletal muscle postprandial insulin sensitivity via activating Ptp1b transcription. FASEB J, 32(8), 4370–4379. doi: 10.1096/fj.201701340R [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORT INFORMATION FIGURE S1 Location of the insertion sites for 5’ and 3’ LoxP sequences that flox exon 2 of the murine Egr1 gene. The exon 2 coding region is highlighted in yellow whereas the 5’ and 3’ guide sequences are indicated by green and blue respectively.
SUPPORT INFORMATION FIGURE S3 The list of the locations, sequence, and the number of mismatches for each of the sgRNAs used in this study.
SUPPORT INFORMATION FIGURE S2 Germline transmission of the 5’ and 3’ LoxP insertions into the murine Egr1 locus. Sequence traces of 5’ and 3’ LoxP sites (red) in genomic DNA from tail biopsy tissue from F2 generation Egr1f/f mice. Sequence of the 5’ and 3’ asymmetric oligonucleotides are highlighted in green. Note: both sequences match the predicted sequences for both LoxP insertions (see: 1, 2, and 3 sequences).