Abstract
Beta-hydroxybutyric acid (BHBA), as one of the main metabolic ketones in the rumen epithelium, plays critical roles in cellular growth and metabolism. The ketogenic capacity is associated with the maturation of rumen in young ruminants, and the exogenous BHBA in diet may promote the rumen development. However, the effects of exogenous BHBA on rumen remain unknown. This is the first study to investigate the mechanisms of BHBA on gene expression and metabolism of rumen epithelium using young goats as a model through multi-omics techniques. Thirty-two young goats were divided into control, low dose, middle dose, and high dose groups by supplementation of BHBA in starter (0, 3, 6, and 9 g/day, respectively). Results demonstrated the dietary of BHBA promoted the growth performance of young goats and increased width and length of the rumen papilla (P < 0.05). Hub genes in host transcriptome that were positively related to rumen characteristics and BHBA concentration were identified. Several upregulated hub genes including NDUFC1, NDUFB4, NDUFB10, NDUFA11 and NDUFA1 were enriched in the gene ontology (GO) pathway of nicotinamide adenine dinucleotide (NADH) dehydrogenase (ubiquinone) activity, while ATP5ME, ATP5PO and ATP5PF were associated with ATP synthesis. RT-PCR revealed the expression of genes (HMGCS2, BDH1, SLC16A3, etc.) associated with lipolysis increased significantly by BHBA supplementation (P < 0.05). Metabolomics indicated that some metabolites such as glucose, palmitic acid, cortisol and capric acid were also increased (P < 0.05). This study revealed that BHBA promoted rumen development through altering NADH balance and accelerating lipid metabolism, which provides a theoretical guidance for the strategies of gastrointestinal health and development of young ruminants.
Keywords: Rumen, Beta-hydroxybutyrate acid, Lipid metabolism, Transcriptome, Metabolomics, Goat
1. Introduction
The rumen is a vital organ modulating the absorption and utilization of nutrients in ruminants. The effective nutrient transfer, absorption, and metabolism ruminants depend on the rumen transition from a pre-ruminant to a ruminant state. The rumen epithelium is the core tissue for ruminant feed conversion. Previous studies reported that feeding fermentable carbohydrates to young ruminants increased rumen volatile fatty acid (VFA) production (e.g., butyrate, acetate, and propionate) and stimulated the development of rumen epithelium (Diao et al., 2019). After its absorption by rumen epithelial cells, VFA is oxidized into ketones which facilitate energy transport, signals for gene transcription, and the regulation of metabolism. Therefore, the ketogenic capacity might be correlated with the maturation of rumen function and development (Lin et al., 2020; Lv et al., 2019; Zhuang et al., 2020).
One of the main metabolic ketones of VFA in rumen epithelium is β-hydroxybutyric acid (BHBA) which is considered as an indicator of rumen maturation and its VFA utilization ability (Deelen et al., 2016). Beyond its role as an energy carrier, BHBA also maintains functions involved in cellular signaling to regulate cell growth by activating or inhibiting various signal pathways (Han et al., 2020). Cheng et al. (2019) showed that BHBA boosted notch signaling to bolster intestinal stem cells self-renewing capabilities, including proliferation and function. BHBA can directly facilitate the differentiation and migration of colon cancer cells through active oxidative phosphorylation (Shakery et al., 2018). BHBA associated with metabolism regulation was also reported in previous studies (Deng et al., 2015; Fu et al., 2015; Han et al., 2020). Hence, it is important to investigate the mechanisms of BHBA during the process of rumen development.
Several studies have investigated the functions of endogenous BHBA (Rahman et al., 2014; Yokoo et al., 2003) and found the intake of exogenous BHBA may be beneficial to the body when the ketogenic capacity is weak in young animals. Ang et al. (2020) showed that the dietary synthetic ketone esters (mainly composed of BHBA) modulated the gut microbiota and subsequently reduced the intestinal pro-inflammatory Th17 cell growth in mice and humans. Wang's team reported that the addition of BHBA in the intestinal tract of mice induced the increase of 3-hydroxy-3-methylglutaryl-CoA synthase isoform 2 (HMGCS2) expression, which aided to maintain the homeostasis of the intestinal environment and regulate small intestinal cell differentiation and growth (Wang et al., 2017). Benito et al. (2020) found that BHBA in small keratinocytes which was activated by lipopolysaccharide significantly reduced the ratio of nicotinamide adenine dinucleotide (NADH) to NAD+ in the cytoplasm, the uptake of glucose, and the export of lactic acid. The injection of BHBA into the isolated cerebral cortex could reduce the ratio of NADH to NAD+, enhance the respiration of mitochondria, and delay cellular senescence by reducing the production of free radicals (Shah et al., 2011).
Many studies have initially started to investigate the impacts of BHBA, but these studies focused on monogastric animals or humans, and the important issues remain unaddressed for ruminants. For instance, whether the exogenous dietary supplementation of BHBA promotes the development of rumen epithelium? If so, which genes are regulated by BHBA in the process of rumen development, and how BHBA affects host metabolism and growth performance? In our study, we hypothesized that the exogenous dietary supplementation of BHBA could promote the development of rumen epithelium by regulating the genes associated with the rumen development and influencing the host metabolism and growth performance. The objectives of this study were to determine the impact of BHBA on the development and metabolism in the rumen epithelium. Multi-omics techniques including transcriptomics and metabolomics were used to quantitatively analyze rumen epithelial genes and metabolites. In addition, hub genes and metabolic which is connected with the rumen traits and BHBA treatment were identified. This work prompts us to better comprehend the mechanisms how BHBA affects gene expression and metabolism patterns in rumen epithelium.
2. Materials and methods
2.1. Animal ethics statement
The study was conducted in accordance with the guidance of Animal Ethics Committee of the Chinese Academy of Agricultural Sciences (AEC-CAAS-20200605; Approval date: 6 November, 2020). The experimental protocol was approved by Institute of Feed Research, Chinese Academy of Agricultural Sciences, and all methods were performed in accordance with the relevant guidelines and regulations. We also confirmed that the experiment complied with the Animal Research Reporting of in Vivo Experiments (ARRIVE) guidelines.
2.2. Animals trial and sample collection
In this experiment, a total of 32 healthy female Haimen young goats (30 days of age) were selected in a goat farm (Haimen, Jiangsu, China), and randomly divided into four groups based on BHBA supplementation. From 30 to 90 days of age, goats in four groups fed starter diet supplemented with 0, 3, 6, or 9 g/day per goat of BHBA (named as CON, LOW, MIDDLE, and HIGH, respectively). Prior to the study, young goats lived with their dams and consumed milk as their only feed source. From 30 to 60 days, young goats were weaned and fed milk replacer (Table S1) and starter (Table S2). Young goats were provided only starter from 60 to 90 days. During the trial, young goats were offered with 300 mL/time per goat of milk replacer (the ratio of milk replacer to water is 1:5) and fed 3 times a day. Young goats also had ad libitum access to water and starter. The basic diet (milk replacer and starter) did not contain BHBA. Ketone salt was selected as the addition form of exogenous BHBA (99% purity), which was white power with no irritating odor. BHBA was fully mixed with 50 g starter to feed the young goats every morning, which ensured that BHBA could be completely consumed by young goats.
Six young goats were selected from each group and slaughtered at the end of the experiment (90 days of age). After the rumen was collected and washed with phosphate buffered saline (PBS), and 3 pieces of rumen tissue from the ventral sac were collected and stored for analysis of morphology, transcriptomics, and metabolomics.
2.3. Analysis of nutrient composition of starter
The samples of starter were ground to pass through a 1-mm sieve and dried in an oven at 135 °C for 2 h (method 930.15; AOAC, 1990) to measure the dry matter (DM) content. The ash content, nitrogen, neutral detergent fiber (NDF), acid detergent fiber (ADF), calcium, and total phosphorus was measured according to methods described by previous studies (Marshall et al., 1978; Shiqin et al., 2019; Van Soest et al., 1991). Crude protein (CP) was calculated as 6.25 × nitrogen.
2.4. Morphology measurement
A 2 cm × 2 cm section of rumen epithelium tissue was collected from each goat kid and immediately washed with physiological saline and fixed in a 250-mL jar containing 10% neutral formalin solution after slaughter. The samples were dehydrated by different concentrations of ethanol, embedded in paraffin sections, and cut into 6 μm sections. The rumen papilla structure was observed under a light microscope at 40× magnification (Olympus BX-51; Olympus Corporation, Tokyo, Japan) after staining with hematoxylin and eosin (H&E). The image-pro express image analysis processing system (Image-Pro Plus 6.0, Media Cybernetics, Silver Spring, MD, USA) was used to observe and measure the rumen papilla length, papilla width, lamina propria thickness, and epithelial thickness.
2.5. Transcriptomic sequencing and analysis of rumen epithelium
Twenty-four rumen epithelium samples were used for transcriptomic analysis. RNA from the rumen epithelial tissue was extracted using the TRIzol reagent according to manufacturer's protocol (Invitrogen, CA, USA). In order to ensure the RNA concentration and quality, we measured the absorbed optical density ratio of total RNA (OD260/280 between 1.80 and 2.10) using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Madison, WI) and confirmed the RNA integrity by running 1.4% agarose-formaldehyde gel.
DNase I was used to eliminate double-stranded, and single-stranded DNA contaminants in all RNA samples and mRNA molecules were purified from total RNA using oligo(dT)-attached magnetic beads. After fragmenting the mRNA of samples into small pieces using fragmentation reagent, the first-strand cDNA was generated using random hexamer-primed reverse transcription, followed by second-strand cDNA synthesis. The synthesized cDNA was subjected to end-repair and then was 3′ adenylated. Adaptors were ligated to the ends of these 3′ adenylated cDNA fragments. The ligated products were amplified by PCR with specific primers. The PCR product was thermally denatured into a single strand, and then the single-strand DNA was cyclized with a single segment bridge primer to obtain a single strand cyclic DNA library initially on a BGISEQ-500 platform (Beijing Genomics Institute, Shenzhen, China). The quality inspection of the constructed library was carried out and the sequencing was carried out after the library was qualified. The RNA library was sequenced at Beijing Genomics Institute (BGI) Co., Ltd. The Pacbio RS II sequencer (Pacific Biosciences, CA, USA) was used to obtain 150 bp paired end reads according to the manufacturer's instructions.
After quality control of raw reads, clean reads were obtained, and HISAT2 (https://ccb.jhu.edu/software/hisat2/index.shtml) was used to align the clean reads to the host. StringTie (version 1.3.4d) was utilized to map reads for describing the expressions of gene transcripts (Pertea et al., 2016). Then, fragments per kilobase of transcript per million fragments mapped (FPKM) methods had been used to calculate the expression level of genes (Trapnell et al., 2010). The differentially expressed genes (DEGs) were identified by the DESeq method, with threshold values being folded change ≥ 1.37 and Q < 0.05. Only the detected genes (FPKM > 0.1 in each sample) were considered to conduct further analysis. Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were carried out by KOBAS (version 3.0, http://kobas.cbi.pku.edu.cn/kobas3).
2.6. Gene expression quantified by RT-PCR
Based on the discoveries of the analysis of host transcriptomics, the genes related to lipid metabolism were quantified using RT-PCR. RNA extraction was identical to the method of transcriptomic measurement. The RT-PCR was performed in a 20 μL reaction mixture including 10 μL 2× qPCR Master Mix (SinoGene, Beijing, China) as a fluorescent dye, 0.5 μL of each primer (10 μM), 8.0 μL ddH2O and 1 μL DNA templates (10 ng). RT-PCR cycling process was as follows: initial denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 20 s, annealing at 60 °C for 30 s, extension at 72 °C for 30 s and a final extension at 72 °C for 10 min. The primer sequencings designed for this study are listed in Table S3.
2.7. Metabolites identification and quantification of rumen tissues
The metabolites in rumen epithelium tissue, with 24 samples, were measured using metabolomics technology. Each sample was weighed to 100 mg and ground with a SCIENTZ-48 tissue grinder (SCIENTZ, Ningbo, China). Then, the rumen power was transferred into a 5-mL tube mixed with 2 mL tissue extract fluids (75% methanol and 25% ddH2O) and 3 steel beads. After ultrasound at room temperature for 30 min and chilling on ice for 1 h, a refrigerated centrifuge was used to separate the supernatant of the mixture at 6,000 × g for 20 min at 4 °C. The supernatant was removed into 2-mL centrifuge tubes and concentrated to dryness in a vacuum. The resulting sample was dissolved with 200 μL of 50% acetonitrile solution containing 2-chlorobenzalanine, and the supernatant was filtered using a 0.22-μm membrane. By mixing equal volumes (20 μL) from each sample, quality control (QC) samples were made to calibrate the error for the analysis. The remaining sample was prepared for LC-MS detection using Thermo Q Exactive Plus (Thermo Fisher Scientific, Waltham, MA) and Thermo Vanquish (Thermo Fisher Scientific, Waltham, MA) to detect metabolites.
2.8. Statistical analyses
One-way ANOVA in SPSS software (version 25, SPSS Inc., Chicago, IL) was applied to compare the difference of the indicators including rumen epithelial section parameters, DGEs among four groups. The results with P < 0.05 were adjudicated as statistical significance.
The weighted gene co-expression network analysis (WGCNA) algorithm (Langfelder and Horvath, 2008) in the R package was used to access the modules of genes and metabolites related to the characterization of the rumen epithelium development and BHBA concentration. The network of genes was constructed by Cytoscape (version 3.7.1, Bethesda, MD, USA) for visualization. Hub genes in the network were selected using the MCODE plugin in Cytoscape (version 2.0, http://apps.cytoscape.org/apps/mcode) with programmed parameters (degree cutoff: 2; K-Core: 2; and max depth: 100).
The functional enrichment analysis of metabolites was performed using MetaboAnalyst 5.0 platform (www.metaboanalyst.ca). The Spearman's analysis was performed to calculate the correlations between hub genes and major metabolites with ‘psych’ package in R. The result was visualized using the heatmap. The connections were considered significant if P < 0.05 and |R| > 0.5.
3. Results
3.1. Growth performance and rumen epithelial morphology
In our study, the results showed that the dietary supplementation of BHBA could promote the growth performance of young goats from 30 to 90 days old. Compared with the CON group, the dry matter intake (DMI) of starter were higher in the LOW and HIGH groups significantly (P = 0.016; Table S4). Similarly, we also observed the higher average daily gain (ADG) in the LOW and HIGH groups (P = 0.001). While feed conversion ratio showed no significant difference among these four groups (P > 0.05). As we know, the growth of young goats relied on the healthy and rapid development of rumen. Hence, we further investigated the characterization and molecular mechanism of rumen development of young goats in each group.
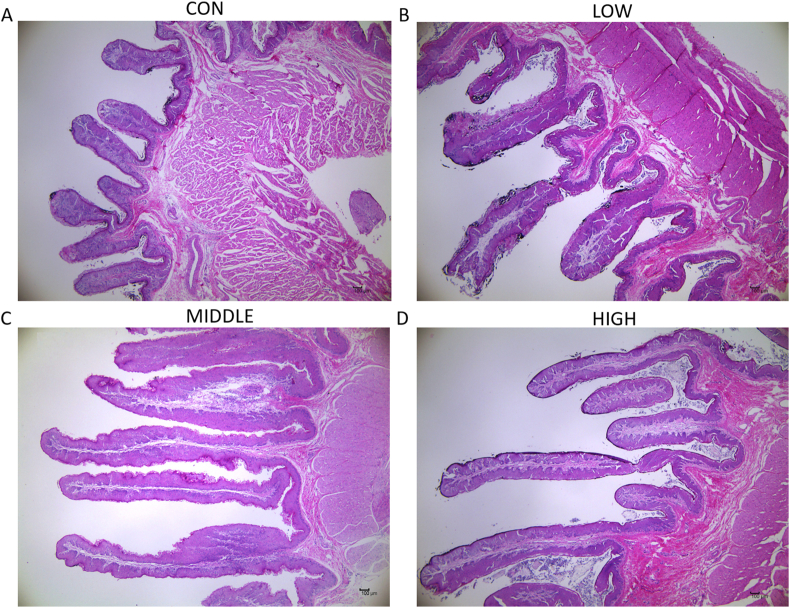
The morphological changes of rumen epithelium influenced by dietary BHBA are shown in Fig. 1 and Table 1. Compared with the control (CON) group, the development of rumen papilla was promoted by BHBA. LOW and HIGH groups' lengths of the rumen papilla were significantly greater (P = 0.023) than the CON group. The ruminal papillae width was increased in response to the supplementation of low and medium doses of BHBA (P = 0.030). In addition, lamina propria thickness and epithelial thickness were not affected by BHBA supplementation (P > 0.05).
Fig. 1.
The effect of BHBA addition to the solid diet on the morphology of the rumen epithelium in young goats. Representative rumen epithelial micrograph in the groups of (A) CON, (B) LOW, (C) MIDDLE and (D) HIGH. Images were obtained through a light micrograph of rumen tissue (40× magnification). CON = starter supplemented with BHBA at 0 g/day per goat; LOW = starter supplemented with BHBA at 3 g/day per goat; MIDDLE = starter supplemented with BHBA at 6 g/day per goat; HIGH = starter supplemented with BHBA at 9 g/day per goat. BHBA = β-hydroxybutyric acid.
Table 1.
Effects of BHBA addition on rumen epithelium parameters in the different groups.
| Epithelial morphology | Groups1 |
SEM | P-value | |||
|---|---|---|---|---|---|---|
| CON | LOW | MIDDLE | HIGH | |||
| Papilla length, μm | 958.60b | 1638.02a | 1263.01ab | 1531.98a | 91.705 | 0.023 |
| Papilla width, μm | 323.71b | 431.43a | 412.48a | 352.94ab | 15.471 | 0.030 |
| Lamina propria thickness, μm | 410.19 | 434.22 | 347.86 | 381.70 | 32.830 | 0.842 |
| Epithelial thickness, μm | 65.61 | 82.73 | 79.78 | 82.18 | 3.717 | 0.312 |
BHBA = β-hydroxybutyric acid.
a,b Values in a row with no common superscripts differ significantly (P < 0.05).
CON = starter supplemented with BHBA at 0 g/day per goat; LOW = starter supplemented with BHBA at 3 g/day per goat; MIDDLE = starter supplemented with BHBA at 6 g/day per goat; HIGH = starter supplemented with BHBA at 9 g/day per goat.
3.2. Genes in rumen epithelium influenced by adding BHBA
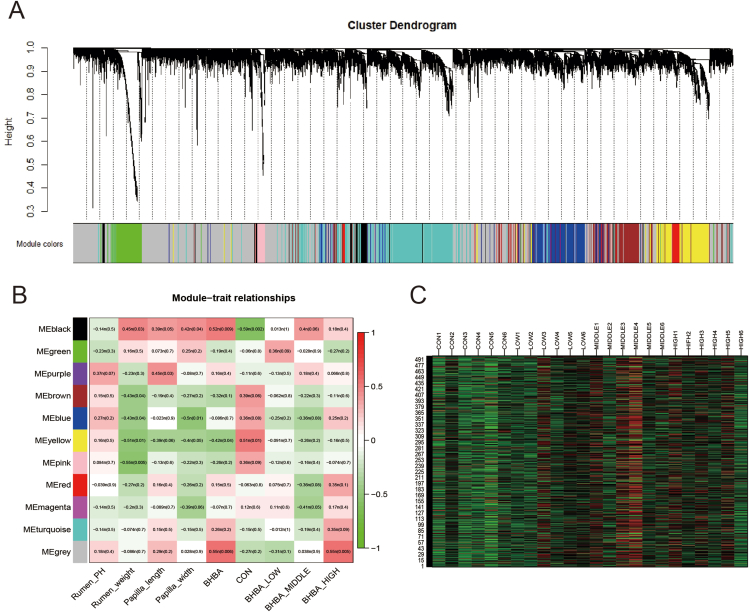
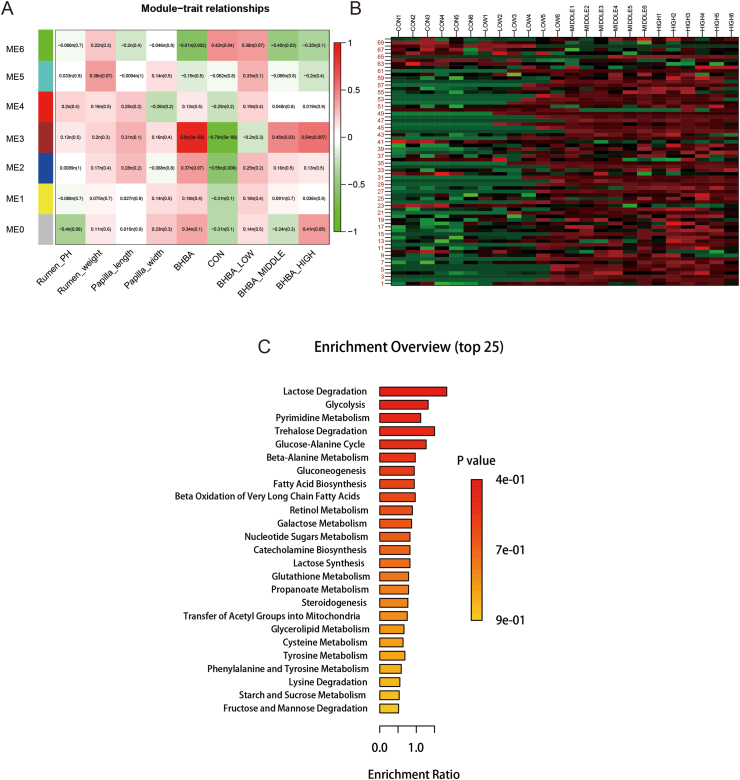
We next sought to investigate the effects of BHBA addition on gene expression in the rumen epithelium. The rumen epithelial transcriptome produced 117.00 GB of clean data. Each sample scored an average of 4.87 GB. The expression profiles of 12,143 genes were determined in all rumen epithelial tissue samples. Compared to the CON group, we identified 58 DEGs (42 upregulated and 16 downregulated) in the LOW group, 44 DEGs (41 upregulated and 3 downregulated) in the MIDDLE group, and 250 DEGs (150 upregulated and 100 downregulated) in the HIGH group, respectively (Fig. S1). To determine comprehensive gene expression influenced by factors including rumen epithelial character and BHBA effects, WGCNA was performed to screen modules of genes. As a result, 11 modules (named colors, MEblack–MEgrey) of co-expressed genes were identified (Fig. 2A and B). Since the MEblack module had the strongest correlation with the indexes (rumen weight, the length of rumen papillae, the width of rumen papillae, dietary factors, and concentration gradient), its major genes are shown in the heat map (Fig. 2C). The MEblack module (496 genes; 4.08% of total genes) was also closely associated with diet, negatively related to low concentrations of BHBA, and positively related to high concentrations of BHBA. Moreover, the expression in MEblack had the highest abundances in the MIDDLE group.
Fig. 2.
Gene co-expression network and its relationship with phenotypes of goats. (A) The dendrogram derived from the gene co-expression network of samples in the four groups (CON, LOW, MIDDLE, and HIGH). (B) The relationship between rumen parameters, dietary factors, concentration gradient and the 11 modules of genes (MEblack to MEgrey). Green colors represent negative correlations, whereas red colors represent positive correlations. Numerical values within a square represent Pearson correlation r values and P values. P values are in the behind parentheses. (C) Heat map to show gene expression of the MEblack module. The color of cells from green to red corresponds to the relative expression of genes from low to high. The numbers on the left side of the figure represent the gene IDs, while group names plus sample ID were labeled over the figure.
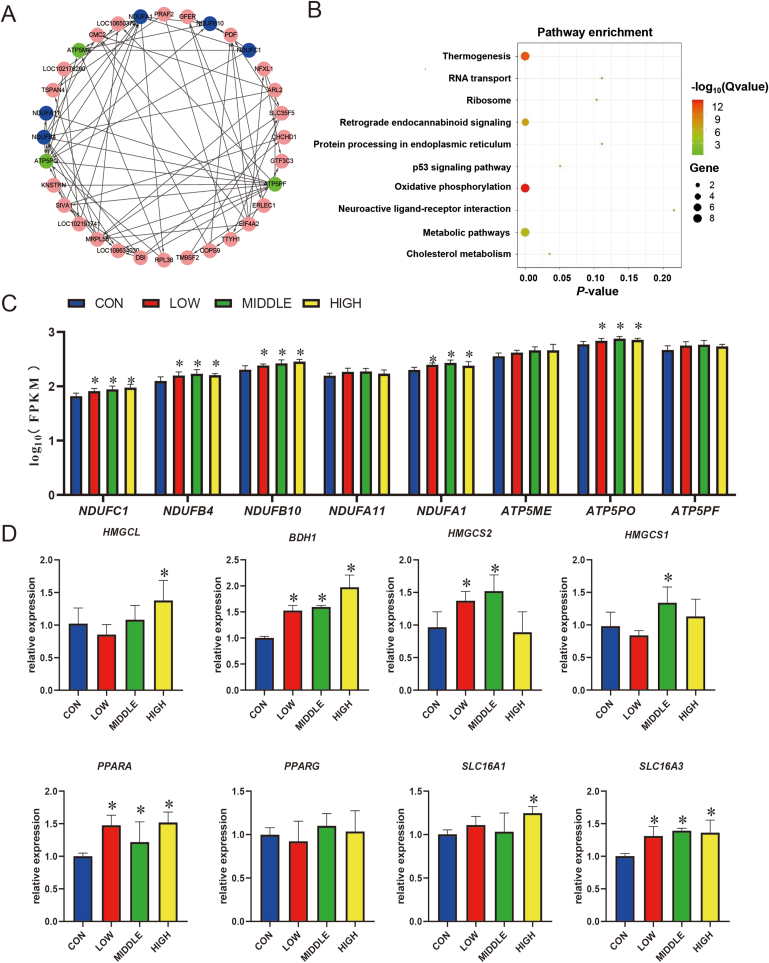
Using the MCODE plugin in Cytoscape, 32 hub genes in the MEblack module were found, and their correlations with other genes were shown in the network plot (Fig. 3A). A total of 88 GO terms targeting these hub genes were induced in the biological processes (P < 0.05) (Table S5). Main functions were related to oxidative metabolism, including mitochondrial respiratory chain complex I (15.63%), NADH dehydrogenase (ubiquinone) activity (12.50%), ATP biosynthetic process (9.38%), mitochondrial ATP synthesis coupled proton transport (9.38%), and long-chain fatty acyl-CoA binding (3.13%). The genes annotated in the above functions mainly covered NADH:ubiquinone oxidoreductase subunit C1 (NDUFC1), NADH:ubiquinone oxidoreductase subunit B4 (NDUFB4), NADH:ubiquinone oxidoreductase subunit B10 (NDUFB10), NADH:ubiquinone oxidoreductase subunit A11 (NDUFA11), NADH:ubiquinone oxidoreductase subunit A1 (NDUFA1), ATP synthase membrane subunit e (ATP5ME), ATP synthase peripheral stalk subunit OSCP (ATP5PO), and ATP synthase peripheral stalk subunit F6 (ATP5PF). Therefore, we compared the relative expression of these 8 genes in each group. As expected, 5 genes (NDUFC1, NDUFB4, NDUFB10, NDUFA1, and ATP5PO) increased significantly (P < 0.05) due to the addition of BHBA (Fig. 3C). In addition, several GO terms belonging to gene expression, such as 5S class rRNA transcription by RNA polymerase III (3.13%), tRNA transcription by RNA polymerase III (3.13%) and transcription by RNA polymerase III (3.13%) were induced. Notably, L-glutamate transmembrane transport (3.13%) and regulation of steroid biosynthetic process (3.13%) were also enriched (Table S5). These results indicated that BHBA may manipulate cell growth by regulating the mitochondrial energy supply system. Then, the hub genes were casted to KEGG pathways, which were grouped in 10 functions (Fig. 3B). The most active enrichments were oxidative phosphorylation, thermogenesis, retrograde endocannabinoid signaling and metabolic pathway. In addition, cholesterol metabolism, RNA transport and protein processing in endoplasmic reticulum were also enriched. Consequently, similar to the results presented by GO, the hub genes were mainly associated with energy production, lipid metabolism, RNA transcription, and protein synthesis.
Fig. 3.
Network plot, functional description, and relative expressions of the hub genes in the M10 module. (A) Network analysis of the hub genes. The circle shape represents the hub gene. The 5 genes in the blue color were enriched in the respiratory electron-transport chain, the 3 genes in the green color were enriched in ATP synthesis, and other genes labeled with the pink color were related to other functions. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the hub genes. (C) The relative expressions of 8 genes (including NDUFC1, NDUFB4, NDUFB10, NDUFA11, NDUFA1, ATP5ME, ATP5PO and ATP5PF) based on transcriptomics are shown by histograms. (D) The relative expressions of these 8 genes related to lipid metabolism were quantified by RT-PCR. Four groups were differentiated by color (blue, red, green, and yellow). CON = starter supplemented with BHBA at 0 g/day per goat; LOW = starter supplemented with BHBA at 3 g/day per goat; MIDDLE = starter supplemented with BHBA at 6 g/day per goat; HIGH = starter supplemented with BHBA at 9 g/day per goat. FPKM = fragments per kilobase of transcript per million fragments mapped; NDUFC1 = NADH:ubiquinone oxidoreductase subunit C1; NDUFB4 = NADH:ubiquinone oxidoreductase subunit B4; NDUFB10 = NADH:ubiquinone oxidoreductase subunit B10; NDUFA11 = NADH:ubiquinone oxidoreductase subunit A11; NDUFA1 = NADH:ubiquinone oxidoreductase subunit A1; ATP5ME = ATP synthase membrane subunit e; ATP5PO = ATP synthase peripheral stalk subunit OSCP; ATP5PF = ATP synthase peripheral stalk subunit F6; HMGCL = 3-hydroxy-3-methylglutaryl-CoA lyase; BDH1 = β-hydroxybutyrate dehydrogenase-1; HMGCS2 = 3-hydroxy-3-methylglutaryl-CoA synthase isoform 2; HMGCS1 = 3-hydroxy-3-methylglutaryl-CoA synthase isoform 1; PPARA = peroxisome proliferator-activated receptor alpha; PPARG = peroxisome proliferator-activated receptor gamma; SLC16A1 = solute carrier family 16 member 1; SLC16A3 = solute carrier family 16 member 3; NADH = nicotinamide adenine dinucleotide. The asterisks over bars represent these groups had significantly higher (P < 0.05) values than the CON group.
Regarding fatty acids as the most important substrate for energy metabolism in the rumen and the active performance of mitochondrial energy synthesis revealed by transcriptomics, the genes associated with lipid metabolism (including 3-hydroxy-3-methylglutaryl-CoA lyase [HMGCL], β-hydroxybutyrate dehydrogenase-1 [BDH1], HMGCS2, 3-hydroxy-3-methylglutaryl-CoA synthase isoform 1 [HMGCS1], peroxisome proliferator-activated receptor alpha [PPARA], peroxisome proliferator-activated receptor gamma [PPARG], solute carrier family 16 member 1 [SLC16A1], and solute carrier family 16 member 3 [SLC16A3]) were further verified by RT-PCR. Intriguingly, the relative abundances of BDH1, PPARA, and SLC16A3 genes demonstrated similar trends with the hub genes enriched in energy metabolism (P < 0.05; Fig. 3D). Additionally, the expressions of HMGCL, HMGCS1 and SLC16A1 were increased in the MIDDLE or HIGH groups (P < 0.05).
3.3. The metabolites in rumen epithelium influenced by BHBA addition
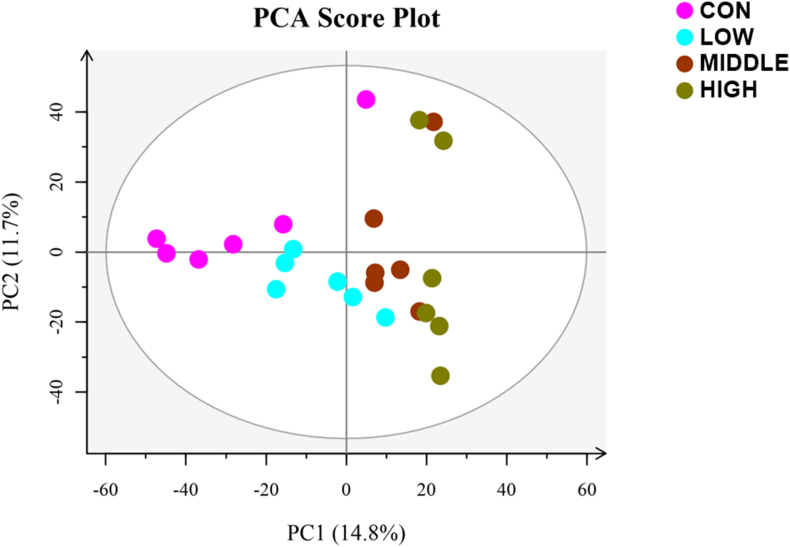
Metabolomics were performed to detect the metabolites in rumen epithelium in response to BHBA supplementation. Across the 24 samples, a total of 657 metabolites were detected by LC-MS instrumentation. The principal component analysis (PCA) showed the remarkable difference of metabolite composition between CON and BHBA groups (Fig. 4). Furthermore, WGCNA was performed to cluster metabolites and identify modules associated with rumen epithelium features and the BHBA. As a result, 7 modules (named ME0 to ME6) were classified (Fig. 5). Although none of the modules showed significant correlations with the traits of the rumen, the ME3 module of metabolomics (70 metabolites; 10.65% of total metabolites) demonstrated strong, positive correlations with the concentration gradient of BHBA supplementation (Fig. 5A), consistent with the results of the MEblack module in the transcriptome. The concentration of metabolites in ME3 were increased as the dose of BHBA supplementation increased (Fig. 5B). In the ME3 module, BHBA increased glycation end products such as D-glucose, β-D-glucosamine, etc. Metabolites related to lipid metabolism, including palmitic acid, 25-hydroxycholesterol, cortisol, and capric acid, also increased with BHBA. Of note, products of nucleotide metabolism were also stimulated, such as cytidine, nicotinamide riboside, hypoxanthine (Table S6).
Fig. 4.
The principal component analysis (PCA) of metabolite composition in different groups (CON, LOW, MIDDLE, and HIGH). Each point represents a unique sample, and different colors represent different groups. CON = starter supplemented with BHBA at 0 g/day per goat; LOW = starter supplemented with BHBA at 3 g/day per goat; MIDDLE = starter supplemented with BHBA at 6 g/day per goat; HIGH = starter supplemented with BHBA at 9 g/day per goat.
Fig. 5.
The functions of key metabolites associated with BHBA addition. (A) The relationship between rumen parameters, dietary factors, concentration gradient and the 7 modules (ME0 to ME6) of metabolites calculated by WGCNA. (B) Heat map of metabolites in the ME3 module. The color of cells from green to red corresponds to the relative expression of metabolites from low to high. The numbers on left side of the figure represent the metabolites IDs, while group names plus sample ID were labeled over figure. (C) Enrichment analysis of the metabolites in the ME3 module. BHBA = β-hydroxybutyric acid.
Then, the pathway of 70 metabolites in the ME3 module was determined using enrichment analysis (Fig. 5C). The subjected metabolic pathways were associated with lactose degradation, glycolysis, pyrimidine metabolism, fatty acid biosynthesis, and beta oxidation of very long chain fatty acids. Moreover, amino acid metabolism, including glutathione metabolism, cysteine metabolism, tyrosine metabolism, phenylalanine and tyrosine metabolism, and lysine degradation were enriched.
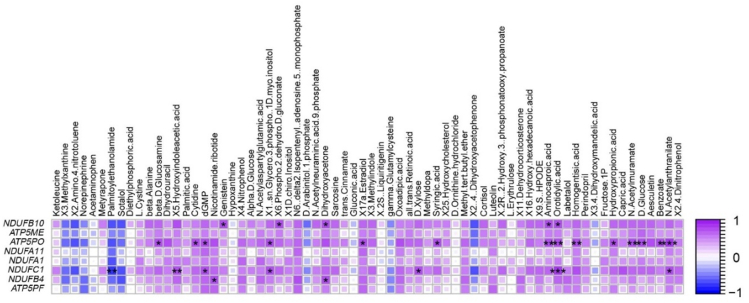
3.4. Correlation between hub genes of MEblack module and major metabolites of ME3 module
The correlation between hub genes of the MEblack module and major metabolites of the ME3 module was calculated (Fig. 6). The hub genes, including NDUFB10, ATP5PO, NDUFC1, and NDUFB4, were strongly correlated (P < 0.05) with some metabolites. For example, ATP5PO was strongly correlated with D-glucose, beta-D-glucosamine, homogentisic acid, orotidylic acid, and dGMP. NDUFC1 was positively correlated (P < 0.05) with palmitoylethanolamide (PEA), dGMP, and D-xylose. Gene NDUFB10 showed a strong correlation (P < 0.05) with genistein, dihydroxyacetone (DHA), and orotidylic acid.
Fig. 6.
Spearman's correlation between the hub genes related to oxidative phosphorylation and metabolites in the ME3 module. The correlation coefficients between the two factors are indicated by the intensity of the color. The colors of the squares range from blue to purple to indicate negative (closer to −1) to positive (closer to 1) correlations. NDUFB10 = NADH:ubiquinone oxidoreductase subunit B10; ATP5ME = ATP synthase membrane subunit e; ATP5PO = ATP synthase peripheral stalk subunit OSCP; NDUFA11 = NADH:ubiquinone oxidoreductase subunit A11; NDUFA1 = NADH:ubiquinone oxidoreductase subunit A1; NDUFC1 = NADH:ubiquinone oxidoreductase subunit C1; NDUFB4 = NADH:ubiquinone oxidoreductase subunit B4; ATP5PF = ATP synthase peripheral stalk subunit F6; NADH = nicotinamide adenine dinucleotide. The degree of significance is indicated by the asterisk symbol, ∗ 0.01 < P < 0.05, ∗∗ 0.001< P < 0.01, ∗∗∗P < 0.001.
4. Discussion
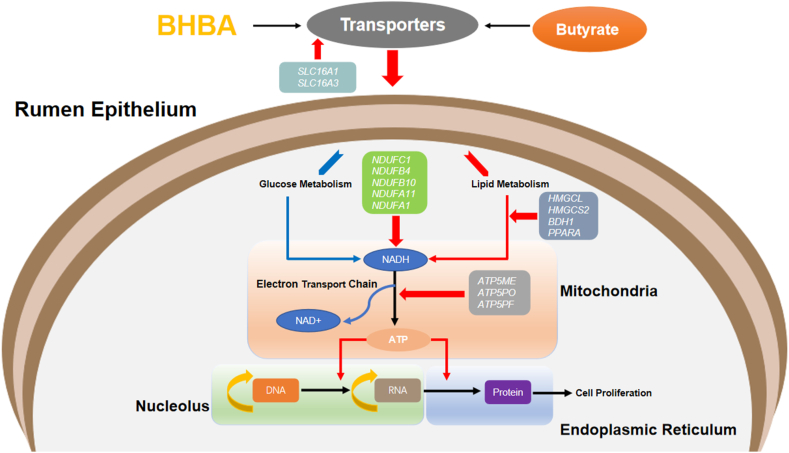
BHBA, as one of the significant physiological ketones produced by the rumen of ruminants, represents an indication of rumen development integrity and plays essential roles in the regulation of cell growth and metabolism. The mature rumen epithelium can convert VFA into ketone bodies (mainly BHBA). However, due to the undeveloped rumen and low ketogenic capacity of epithelial cells in neonatal ruminants, supplementation of exogenous BHBA and its mechanism to improve rumen development remains unknown. This study found that the length and width of papillae were increased when young goats consumed exogenous BHBA. The analysis of genes and metabolites showed that BHBA promoted the occurrence of oxidative phosphorylation to produce ATP for rumen growth and maintain the balance of NADH/NAD+ of cells according to the activation of lipid metabolism and inhibition of glycogen decomposition. In addition, the abundance of metabolites related to amino acids, cholesterol, and nucleotides were increased.
At present, exogenous BHBA forms include ketone salts, medium-chain triglyceride (MCT) oil, and ketone esters (Stubbs et al., 2017). In this study, BHBA salts were chosen based on the economic cost, palatability, and stability of their exogenous form. Moreover, to determine the impacts of BHBA supplementation and its dose on young goats, our preliminary experiment (data not shown) was performed, and the supplementation level of BHBA was established based on a previous human study (Kackley et al., 2020). Then, we found that BHBA addition (6 g/day) could significantly increase the body weights of young goats compared to the control group, butyrate, and γ-aminobutyric acid. Thus, based on the results of preliminary experiment, we chose 6 g/day as the intermediate dose of the additive to conduct this experiment in further.
The papillae of rumen epithelium could expend the contact area of the rumen wall with the digesta and enhance nutrient absorption (Malmuthuge et al., 2019). The length and width of rumen papillae are the most typical indexes for evaluating the state of rumen development (Lesmeister et al., 2004). In the present study, dietary BHBA increased rumen development. BHBA was metabolized from VFA (mainly butyric acid) through oxidative metabolism in the rumen epithelial cell's mitochondria. Similar studies on exogenous butyrate regulating rumen development have been extensive and gradually deepened. Many studies (Cavini et al., 2015; Gorka et al., 2018) have demonstrated that the infusion of sodium butyrate promoted the growth of rumen papillae. This may be due to the large amount of energy released during the conversion of sodium butyrate to BHBA that is absorbed by growing epithelial cells (Allen and Michael, 2014). The metabolic product of BHBA also acts as an energy substrate and regulatory factor to promote the further development of rumen epithelium. In our study, based on our initial view of morphological pictures and corresponding indicators, it was undoubted that BHBA had remarkable effects on rumen development. In addition, in this study, we have also confirmed that different level of BHBA addition could significantly improve the DMI and ADG of young goats and so on (Abdelsattar et al., 2022). On the one hand, the mature rumen improved by BHBA showed higher efficient digestion to consume more solid feed for growth needs. On the other hand, the previous studies have showed the introduction of solid feed and increase of the intake could further promote rumen development. This is because the physical friction of the solid feed against the rumen wall stimulates the growth of the rumen papillae (Lv et al., 2019) and the nutrients from the feed also provide the necessary energy for rumen development (Lin et al., 2019), which indicated that BHBA also could promote growth performance and rumen development of goats by improving feed intake.
Transcriptomics and untargeted metabolomics were applied to further analyze the internal mechanisms of rumen epithelial differences in response to dietary BHBA. BHBA is involved in maintaining NADH/NAD+ equilibrium in cells (Newman and Verdin, 2014). The excessive attrition of NAD+ may accelerate cell apoptosis and the occurrence of metabolic diseases (Stein and Imai, 2012). In this study, the expression of hub genes (NDUFC1, NDUFB4, NDUFB10, NDUFA11, and NDUFA1) belonging to the MEblack module were increased in respiratory complex I (CI, NADH:ubiquinone oxidoreductase) (Abu Dawud et al., 2012). These genes were all an accessory subunit of CI and responsible for the transferring of electrons from NADH to the respiratory chain required for oxidative phosphorylation (reaction equation: NADH + H+ + Q + 4H+ (matrix) → NAD+ + QH2 + 4H (intermembrane space [IMS])) (Wu et al., 2016). Therefore, BHBA replenished the concentration of NAD+ by increasing the activity of CI, thereby regulating cell homeostasis and growth.
On the other hand, nicotinamide ribotide (NMN) is the most direct precursor of NAD+ in mammals. Evidence accumulated in studies of rodents proved that NMN infusion effectively strengthened NAD+ biosynthesis in various tissues and cells. In this study, NMN abundance showed a significant increase in response to BHBA supplementation. This may be another method for BHBA to adjust the balance of NADH/NAD+. However, the signal pathway of NMN induced by BHBA remains unknown, which is another crucial question to address. ATP5ME, ATP5PO, and ATP5PF are all ATP synthase subunits in the mitochondrial membrane (F1F0 ATP synthase or Complex V), which participated in the process of ATP assembly. Under a high proton gradient caused by CI activity, it is unsurprising that their expressions increased significantly. Veech (2004) reported that BHBA supplementation improved the efficiency of ATP production based on the mitochondrial proton gradient and inhibited free radicals. Shakery et al. (2018) also found BHBA addition stimulated oxidative phosphorylation and increased the amount of ATP in colon cells, consistent with the results of this experiment.
NADH is the material basis of oxidative phosphorylation and derives from the processes of biochemical metabolism, such as glycolysis, tricarboxylic acid cycle, lipid oxidation, etc (Wilson, 2017). The results of RT-PCR showed several genes (HMGCS2, BDH1, PPARA, and SLC16A3) associated with VFA metabolism were increased with BHBA supplementation, which implied lipid metabolism was one major pathway to induce mitochondrial ATP production. Additionally, BHBA providing roles as a signal molecule or ligand to regulate mitochondrial enzymes is a broad consensus. HMGCS2 is the rate-limiting enzyme involved in the generation of ketone bodies from lipids in the rumen (Wang et al., 2019). The initiation of its transcription is mainly controlled by two signal molecules, the positive regulation of FOXA2 (Vonmeyenn et al., 2013) and the negative regulation of mTORC (Howell and Manning, 2011). Firstly, histone deacetylases (HDACs) mediated deacetylation leads to the inactivation of FOXs. Many studies proved BHBA was an endogenous inhibitor of class I HDACs (Mikami et al., 2020). Simultaneously, acetyl-coenzyme and NADH generated from BHBA's metabolism increased the acetylation level of mitochondrial proteins, which indirectly led to the transcription of metabolic enzymes. Secondly, mTOR reduces mitochondrial activity by inhibiting PPARA, the dominant regulator of mitochondrial function enzymes (Kersten and Stienstra, 2017). Fatty acids and their derivatives, including BHBA, are highly effective ligands for inducing PPARA. As expected, high-level expression of PPARA in treated groups was our primary focus. Another important note is that recent studies have shown the accumulation of BHBA and the increase of HMGCS2 expression strengthened the ability of intestinal epithelial cells to proliferate and differentiate. Exogenous BHBA supplementation repaired the blockage of intestinal regeneration caused by HMGCS2 deletion, which suggested that BHBA may be a direct promoter of epithelial cell growth (Wang et al., 2017). In addition, the expression of two VFA transporters (SLC16A3 and SLC16A1) (Kuzinski et al., 2012) also increased accordingly, which seemed to verify the active performance of lipid metabolism pathways.
Glucose metabolism is another important source of energy productivity in the cell. Some studies have reported that infusion of BHBA in the heart resulted in glycogen synthesis and the inhibition of glycolysis (Kashiwaya et al., 1994; Lund et al., 2015). The underlying mechanism may be that BHBA reduced pyruvate dehydrogenase complex activity (PDH complex), which triggered the stacking effect of pyruvate, which prevented glycogen degradation via negative feedback loops. Another study involving the nervous system reported that some glycolytic enzymes combined their complexes with subunits of the ATP sensitive potassium (KATP) channels, and the presence of BHBA increased the channel's inhibitory transmitter release and thereby interfered with the normal function of the enzymes (Lund et al., 2015). In the current study, the abundance of glucose (alpha-D-glucose, beta-D-glucosamine, and D-glucose) enriched in glycolysis and gluconeogenesis increased with BHBA supplementation. Accordingly, we assumed that BHBA might inhibit glucose metabolism in rumen epithelial cells. In addition, norepinephrine can activate the biochemical pathway of lipolysis (Schaff et al., 2014), and alterations initiate the stimulation dynamics of the adrenal medulla in nutrient concentration. BHBA, like glucose and short-chain fatty acid, acts as a feedback regulation signal to activate the phospholipase C β (PLCβ)/extracellular signal-regulated kinase (ERK)/synapsin 2b/G-protein coupled receptor 41 (GPR41) pathways in sympathetic neurons, and in turn, regulates hormone release in adrenal medulla (D. Inoue and Takei, 2012; Stricker et al., 1977; Won et al., 2013). In our study, the increased abundance of norepinephrine performed dose dependence of BHBA. Thus, we made a conjecture that BHBA supplement also led the facilitation of lipolysis according to noradrenaline. Notably, with lipolysis, fat synthesis accordingly changes for the balance of fat metabolism. In our study, palmitic acid and capric acid were all enriched in the pathway of fatty acid biosynthesis. We also found that the expression of diazepam-binding inhibitor (DBI) was increased. DBI was regarded as lipogenesis, and dietary enhancers with mice experiment (Pedro et al., 2019). Taken together, we supposed that BHBA promoted the metabolism of lipid and inhibited glycogen breakdown.
On the other hand, in immature organs, BHBA is considered an important precursor for amino acid synthesis (aspartate, glutamate, and glutamine, etc.) and is also necessary for phospholipid bilayer construction cells and repair of damaged cell connections (Müller et al., 1976). Analysis based on the metabolome revealed that several amino acids were increased, such as L-cystine and beta-alanine with BHBA supplementation. Moreover, cholesterol increase (hydroxycholesterol, oxidized cholesterol) and the activation of its metabolic pathway were also observed. HMGCS1, as the rate-limiting enzyme for the conversion of fatty acids to cholesterol (Yao et al., 2020), significantly increased expression in the MIDDLE group compared to the CON group via PCR quantification. Cholesterol is a major component of cell membranes, which maintains the stability and permeability of cells (Elustondo et al., 2017). Due to the sensitivity of mitochondria to cholesterol content, mitochondrial biology is also affected, including oxidative stress, oxidative phosphorylation, etc (Martin et al., 2016). Considering the above, it is supposed that BHBA assisted amino acid synthesis and promoted cellular biofilm construction and mitochondrial activity with the regulation of the cholesterol pathway.
Finally, we found that the abundances of nucleotide-related metabolites (cytidine, orotidylic acid, dGMP, and NMN) were increased with BHBA dietary. However, the exact mechanisms of these metabolites in response to BHBA are unclear so far. Several studies have proved that purines and pyrimidines had effects on modulating cell proliferation and differentiation (Burnstock and Ralevic, 2014; Burnstock and Verkhratsky, 2010). Adenosine mediated by P2Y induced the mRNA expression of genes related to cell cycles, leading to muscle cell proliferation in rats. Nucleotide ligands enhanced DNA synthesis and migration by apamin sensitive small-conductance Ca2+-activated K+ channels in most cells (Yamazaki et al., 2011). In addition, nucleotides are precursors for the synthesis of biological macromolecular-nucleic acids (DNA and RNA), which are essential for DNA replication and transcription.
5. Conclusions
In summary, we demonstrated that BHBA addition in solid feed promoted the growth performance and stimulated rumen epithelial development in young goats. Based on gene quantification and metabolomics, BHBA activated oxidative phosphorylation to maintain NADH/NAD+ equilibrium and enhanced ATP formation via the accelerating of lipid metabolism and inhibition of the glycolysis (Fig. 7). Furthermore, cholesterol and nucleotide metabolomics were also active. These findings in our study highlight the beneficial effects of BHBA on rumen development.
Fig. 7.
Comprehensive response of gene expression and metabolic pathways in rumen epithelial to the introduction of BHBA and the interaction between them. The red arrows represent activation, and the blue arrows represent inhibition. BHBA = β-hydroxybutyric acid; NADH = nicotinamide adenine dinucleotide; SLC16A1 = solute carrier family 16 member 1; SLC16A3 = solute carrier family 16 member 3; NDUFC1 = NADH:ubiquinone oxidoreductase subunit C1; NDUFB4 = NADH:ubiquinone oxidoreductase subunit B4; NDUFB10 = NADH:ubiquinone oxidoreductase subunit B10; NDUFA11 = NADH:ubiquinone oxidoreductase subunit A11; NDUFA1 = NADH:ubiquinone oxidoreductase subunit A1; HMGCL = 3-hydroxy-3-methylglutaryl-CoA lyase; HMGCS2 = 3-hydroxy-3-methylglutaryl-CoA synthase isoform 2; BDH1 = β-hydroxybutyrate dehydrogenase-1; PPARA = peroxisome proliferator-activated receptor alpha; ATP5ME = ATP synthase membrane subunit e; ATP5PO = ATP synthase peripheral stalk subunit OSCP; ATP5PF = ATP synthase peripheral stalk subunit F6.
Author contributions
Yimin Zhuang and Jianmin Chai contributed to animal trials, sample collection, data analysis and primary drafting. Yuze Fu and Mahmoud M. Abdelsattar helped to sample collection. Naifeng Zhang contributed to the experimental design, draft proofing, and project management. All authors have read and approved the final manuscript.
Declaration of competing interest
The authors declared that they have no conflicts of interest to this work. We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Data availability statement
The accession for the sequencing data in this study is NCBI Sequence Read Archive: BioProject PRJNA708150.
Acknowledgments
This study was funded by the National Natural Science Foundation of China (31872385) and the National Key R&D Program of China (2018YFD0501902). The study was conducted in accordance with the guidance of Animal Ethics Committee of the Chinese Academy of Agricultural Sciences (AEC-CAAS-20200605; Approval date: 6 November, 2020). The authors also thank the cooperation of goat farm for animal handling. We also appreciate assistances for sampling collection from our lab members including Yuze Fu and Shiqin Wang. We also thank Hunter Usdrowski, University of Arkansas for critical review and proof-reading of this manuscript.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2023.02.012.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- Abdelsattar M.M., Vargas-Bello-Pérez E., Zhuang Y., Fu Y., Zhang N. Impact of dietary supplementation of β-hydroxybutyric acid on performance, nutrient digestibility, organ development and serum stress indicators in early-weaned goat kids. Anim Nutr. 2022;9:16–22. doi: 10.1016/j.aninu.2021.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu Dawud R., Schreiber K., Schomburg D., Adjaye J. Human embryonic stem cells and embryonal carcinoma cells have overlapping and distinct metabolic signatures. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M.S. Drives and limits to feed intake in ruminants. Anim Prod Sci. 2014;54:1513–1524. [Google Scholar]
- Ang Q.Y., Alexander M., Newman J.C., Tian Y., Cai J., Upadhyay V., Turnbaugh J.A., Verdin E., Hall K.D., Leibel R.L., Ravussin E., Rosenbaum M., Patterson A.D., Turnbaugh P.J. Ketogenic diets alter the gut microbiome resulting in decreased intestinal th17 cells. Cell. 2020;181:1263–1275 e16. doi: 10.1016/j.cell.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito A., Hajji N., O'neill K., Keun H.C., Syed N. Beta-hydroxybutyrate oxidation promotes the accumulation of immunometabolites in activated microglia cells. Metabolites. 2020;10(9):346. doi: 10.3390/metabo10090346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnstock G., Ralevic V. Purinergic signaling and blood vessels in health and disease. Pharmacol Rev. 2014;66:102–192. doi: 10.1124/pr.113.008029. [DOI] [PubMed] [Google Scholar]
- Burnstock G., Verkhratsky A. Long-term (trophic) purinergic signalling: purinoceptors control cell proliferation, differentiation and death. Cell Death Dis. 2010;1(1):e9. doi: 10.1038/cddis.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavini S., Iraira S., Siurana A., Foskolos A., Ferret A., Calsamiglia S. Effect of sodium butyrate administered in the concentrate on rumen development and productive performance of lambs in intensive production system during the suckling and the fattening periods. Small Rumin Res. 2015;126:90. [Google Scholar]
- Cheng C.W., Biton M., Haber A.L., Gunduz N., Eng G., Gaynor L.T., Tripathi S., Calibasi-Kocal G., Rickelt S., Butty V.L., Moreno-Serrano M., Iqbal A.M., Bauer-Rowe K.E., Imada S., Ulutas M.S., Mylonas C., Whary M.T., Levine S.S., Basbinar Y., Hynes R.O., Mino-Kenudson M., Deshpande V., Boyer L.A., Fox J.G., Terranova C., Rai K., Piwnica-Worms H., Mihaylova M.M., Regev A., Yilmaz O.H. Ketone body signaling mediates intestinal stem cell homeostasis and adaptation to diet. Cell. 2019;178:1115–1131 e15. doi: 10.1016/j.cell.2019.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen S.M., Leslie K.E., Steele M.A., Eckert E., Brown H.E., Devries T.J. Validation of a calf-side beta-hydroxybutyrate test and its utility for estimation of starter intake in dairy calves around weaning. J Dairy Sci. 2016;99:7624–7633. doi: 10.3168/jds.2016-11097. [DOI] [PubMed] [Google Scholar]
- Deng Q., Liu G., Liu L., Zhang Y., Yin L., Shi X., Wang J., Yuan X., Sun G., Li Y., Yang W., Guo L., Zhang R., Wang Z., Li X., Li X. Bhba influences bovine hepatic lipid metabolism via ampk signaling pathway. J Cell Biochem. 2015;116:1070–1079. doi: 10.1002/jcb.25062. [DOI] [PubMed] [Google Scholar]
- Diao Q., Zhang R., Fu T. Review of strategies to promote rumen development in calves. Animals. 2019;9(8):490. doi: 10.3390/ani9080490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elustondo P., Martin L.A., Karten B. Mitochondrial cholesterol import. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:90–101. doi: 10.1016/j.bbalip.2016.08.012. [DOI] [PubMed] [Google Scholar]
- Fu S.P., Liu B.R., Wang J.F., Xue W.J., Liu H.M., Zeng Y.L., Huang B.X., Li S.N., Lv Q.K., Wang W., Liu J.X. Beta-hydroxybutyric acid inhibits growth hormone-releasing hormone synthesis and secretion through the gpr109a/extracellular signal-regulated 1/2 signalling pathway in the hypothalamus. J Neuroendocrinol. 2015;27:212–222. doi: 10.1111/jne.12256. [DOI] [PubMed] [Google Scholar]
- Gorka P., Kowalski Z.M., Zabielski R., Guilloteau P. Invited review: use of butyrate to promote gastrointestinal tract development in calves. J Dairy Sci. 2018;101:4785–4800. doi: 10.3168/jds.2017-14086. [DOI] [PubMed] [Google Scholar]
- Han Y.M., Ramprasath T., Zou M.H. Beta-hydroxybutyrate and its metabolic effects on age-associated pathology. Exp Mol Med. 2020;52:548–555. doi: 10.1038/s12276-020-0415-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell J.J., Manning B.D. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends Endocrinol Metabol. 2011;22:94–102. doi: 10.1016/j.tem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D., Kimura I., Wakabayashi M., Tsumoto H., Ozawa K., Hara T., Takei Y., Hirasawa A., Ishihama Y., Tsujimoto G. Short-chain fatty acid receptor gpr41-mediated activation of sympathetic neurons involves synapsin 2b phosphorylation. FEBS Lett. 2012;586:1547–1554. doi: 10.1016/j.febslet.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Kackley M.L., Short J.A., Hyde P.N., Lafountain R.A., Buga A., Miller V.J., Dickerson R.M., Sapper T.N., Barnhart E.C., Krishnan D., Mcelroy C.A., Maresh C.M., Kraemer W.J., Volek J.S. A pre-workout supplement of ketone salts, caffeine, and amino acids improves high-intensity exercise performance in keto-naive and keto-adapted individuals. J Am Coll Nutr. 2020;39:290–300. doi: 10.1080/07315724.2020.1752846. [DOI] [PubMed] [Google Scholar]
- Kashiwaya Y., Sato K., Tsuchiya N., Thomas S., Fell D.A., Veech R.L., Passonneau J.V. Control of glucose utilization in working perfused rat heart. J Biol Chem. 1994;269:25502–25514. [PubMed] [Google Scholar]
- Kersten S., Stienstra R. The role and regulation of the peroxisome proliferator activated receptor alpha in human liver. Biochimie. 2017;136:75–84. doi: 10.1016/j.biochi.2016.12.019. [DOI] [PubMed] [Google Scholar]
- Kuzinski J., Zitnan R., Albrecht E., Viergutz T., Schweigel-Röntgen M. Modulation of vh+-atpase is part of the functional adaptation of sheep rumen epithelium to high-energy diet. Am J Physiol Regul Integr Comp Physiol. 2012;303:R909. doi: 10.1152/ajpregu.00597.2011. [DOI] [PubMed] [Google Scholar]
- Lesmeister K.E., Tozer P.R., Heinrichs A.J. Development and analysis of a rumen tissue sampling procedure. J Dairy Sci. 2004;87:1336–1344. doi: 10.3168/jds.S0022-0302(04)73283-X. [DOI] [PubMed] [Google Scholar]
- Lin L., Xie F., Sun D., Liu J., Zhu W., Mao S. Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome. 2019;7:83. doi: 10.1186/s40168-019-0701-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Wang Y., Xu L., Liu J., Zhu W., Mao S. Microbiome–host co-oscillation patterns in remodeling of colonic homeostasis during adaptation to a high-grain diet in a sheep model. Animal Microbiome. 2020;2(1):22. doi: 10.1186/s42523-020-00041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund T.M., Ploug K.B., Iversen A., Jensen A.A., Jansen-Olesen I. The metabolic impact of beta-hydroxybutyrate on neurotransmission: reduced glycolysis mediates changes in calcium responses and katp channel receptor sensitivity. J Neurochem. 2015;132:520–531. doi: 10.1111/jnc.12975. [DOI] [PubMed] [Google Scholar]
- Lv X., Chai J., Diao Q., Huang W., Zhuang Y., Zhang N. The signature microbiota drive rumen function shifts in goat kids introduced to solid diet regimes. Microorganisms. 2019;31(11):516. doi: 10.3390/microorganisms7110516. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmuthuge N., Liang G., Guan L.L. Regulation of rumen development in neonatal ruminants through microbial metagenomes and host transcriptomes. Genome Biol. 2019;20:172. doi: 10.1186/s13059-019-1786-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C.M., Walker A.F. Comparison of a short method for kjeldahl digestion using a trace of selenium as catalyst, with other methods. J Sci Food Agric. 1978;29:940–942. [Google Scholar]
- Martin L.A., Kennedy B.E., Karten B. Mitochondrial cholesterol: mechanisms of import and effects on mitochondrial function. J Bioenerg Biomembr. 2016;48:137–151. doi: 10.1007/s10863-014-9592-6. [DOI] [PubMed] [Google Scholar]
- Mikami D., Kobayashi M., Uwada J., Yazawa T., Kamiyama K., Nishimori K., Nishikawa Y., Nishikawa S., Yokoi S., Taniguchi T., Iwano M. Beta-hydroxybutyrate enhances the cytotoxic effect of cisplatin via the inhibition of hdac/survivin axis in human hepatocellular carcinoma cells. J Pharmacol Sci. 2020;142:1–8. doi: 10.1016/j.jphs.2019.10.007. [DOI] [PubMed] [Google Scholar]
- Müller W.A., Aoki T.T., Flatt J.P., Blackburn G.L., Egdahl R.H., Cahill G.F., Jr. Effects of β-hydroxybutyrate, glycerol, and free fatty acid infusions on glucagon and epinephrine secretion in dogs during acute hypoglycemia. Metabolism. 1976;25:1077–1086. doi: 10.1016/0026-0495(76)90015-9. [DOI] [PubMed] [Google Scholar]
- Newman J.C., Verdin E. Beta-hydroxybutyrate: much more than a metabolite. Diabetes Res Clin Pract. 2014;106:173–181. doi: 10.1016/j.diabres.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedro J.M.B., Sica V., Madeo F., Kroemer G. Acyl-coa-binding protein (acbp): the elusive 'hunger factor' linking autophagy to food intake. Cell Stress. 2019;3:312–318. doi: 10.15698/cst2019.10.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of rna-seq experiments with hisat, stringtie and ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M., Muhammad S., Khan M.A., Chen H., Ridder D.A., Muller-Fielitz H., Pokorna B., Vollbrandt T., Stolting I., Nadrowitz R., Okun J.G., Offermanns S., Schwaninger M. The beta-hydroxybutyrate receptor hca2 activates a neuroprotective subset of macrophages. Nat Commun. 2014;5:3944. doi: 10.1038/ncomms4944. [DOI] [PubMed] [Google Scholar]
- Schaff C.T., Rohrbeck D., Steinhoff-Wagner J., Kanitz E., Sauerwein H., Bruckmaier R.M., Hammon H.M. Effects of colostrum versus formula feeding on hepatic glucocorticoid and alpha(1)- and beta(2)-adrenergic receptors in neonatal calves and their effect on glucose and lipid metabolism. J Dairy Sci. 2014;97:6344–6357. doi: 10.3168/jds.2014-8359. [DOI] [PubMed] [Google Scholar]
- Shah T.D., Hickey M.C., Capasso K.E., Palenchar J.B. The characterization of a unique trypanosoma brucei beta-hydroxybutyrate dehydrogenase. Mol Biochem Parasitol. 2011;179:100–106. doi: 10.1016/j.molbiopara.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Shakery A., Pourvali K., Ghorbani A., Fereidani S.S., Zand H. Beta-hydroxybutyrate promotes proliferation, migration and stemness in a subpopulation of 5FU treated SW480 cells: evidence for metabolic plasticity in colon cancer. Asian Pac J Cancer Prev APJCP. 2018;19:3287–3294. doi: 10.31557/APJCP.2018.19.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L.R., Imai S. The dynamic regulation of nad metabolism in mitochondria. Trends Endocrinol Metabol. 2012;23:420–428. doi: 10.1016/j.tem.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker E., Rowland N., Saller C., Friedman M.J.S. Homeostasis during hypoglycemia: central control of adrenal secretion and peripheral control of feeding. Science. 1977;196:79. doi: 10.1126/science.841345. [DOI] [PubMed] [Google Scholar]
- Stubbs B.J., Cox P.J., Evans R.D., Santer P., Miller J.J., Faull O.K., Magor-Elliott S., Hiyama S., Stirling M., Clarke K. On the metabolism of exogenous ketones in humans. Front Physiol. 2017;8:848. doi: 10.3389/fphys.2017.00848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., Van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by rna-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soest P.J., Robertson J.B., Lewis B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci. 1991;74(10):3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- Veech R.L. The therapeutic implications of ketone bodies: the effects of ketone bodies in pathological conditions: ketosis, ketogenic diet, redox states, insulin resistance, and mitochondrial metabolism. Prostaglandins Leukot Essent Fatty Acids. 2004;70:309–319. doi: 10.1016/j.plefa.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Vonmeyenn F., Porstmann T., Gasser E., Selevsek N., Schmidt A., Aebersold R., Stoffel M. Glucagon-induced acetylation of foxa2 regulates hepatic lipid metabolism. Cell Metabol. 2013;17:436–447. doi: 10.1016/j.cmet.2013.01.014. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhou Y., Rychahou P., Fan T.W., Lane A.N., Weiss H.L., Evers B.M. Ketogenesis contributes to intestinal cell differentiation. Cell Death Differ. 2017;24:458–468. doi: 10.1038/cdd.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.H., Liu C.L., Chiu W.C., Twu Y.C., Liao Y.J. Hmgcs2 mediates ketone production and regulates the proliferation and metastasis of hepatocellular carcinoma. Cancers. 2019;26(12):1876. doi: 10.3390/cancers11121876. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.F. Oxidative phosphorylation: regulation and role in cellular and tissue metabolism. J Physiol. 2017;595:7023–7038. doi: 10.1113/JP273839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won Y.J., Lu V.B., Puhl H.L., Ikeda S.R. Β-hydroxybutyrate modulates n-type calcium channels in rat sympathetic neurons by acting as an agonist for the g-protein-coupled receptor ffa3. J Neurosci. 2013;33:19314–19325. doi: 10.1523/JNEUROSCI.3102-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Gu J., Guo R., Huang Y., Yang M. Structure of mammalian respiratory supercomplex i1iii2iv1. Cell. 2016;167:1598–1609 e10. doi: 10.1016/j.cell.2016.11.012. [DOI] [PubMed] [Google Scholar]
- Yamazaki D., Kito H., Yamamoto S., Ohya S., Imaizumi Y. Contribution of k(ir)2 potassium channels to atp-induced cell death in brain capillary endothelial cells and reconstructed hek293 cell model. Am J Physiol Cell Physiol. 2011;300(1):C75–C86. doi: 10.1152/ajpcell.00135.2010. [DOI] [PubMed] [Google Scholar]
- Yao W., Jiao Y., Zhou Y., Luo X. Klf13 suppresses the proliferation and growth of colorectal cancer cells through transcriptionally inhibiting hmgcs1-mediated cholesterol biosynthesis. Cell Biosci. 2020;10:76. doi: 10.1186/s13578-020-00440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo H., Saitoh T., Shiraishi S., Yanagita T., Sugano T., Minami S., Kobayashi H., Wada A. Distinct effects of ketone bodies on down-regulation of cell surface insulin receptor and insulin receptor substrate-1 phosphorylation in adrenal chromaffin cells. J Pharmacol Exp Therapeut. 2003;304:994–1002. doi: 10.1124/jpet.102.044115. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Chai J., Cui K., Bi Y., Diao Q., Huang W., Usdrowski H., Zhang N. Longitudinal investigation of the gut microbiota in goat kids from birth to postweaning. Microorganisms. 2020;8(8):1111. doi: 10.3390/microorganisms8081111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession for the sequencing data in this study is NCBI Sequence Read Archive: BioProject PRJNA708150.