Abstract
Serogroup C strains of Neisseria meningitidis were isolated from a Germany patient with severe meningococcal disease after a trip to the Czech Republic. These strains (case isolates) were characterized by classical and molecular techniques, as were other strains (carrier isolates) isolated from healthy contacts. Five of 10 carrier isolates had switched off the expression of capsular polysaccharide, as demonstrated by a serogroup-specific PCR. The two case isolates were indistinguishable by multilocus sequence typing and belonged to the ET-37 complex. The carrier isolates belonged to four different sequence types, all unrelated to that of the case strains. Pulsed-field gel electrophoresis showed that the case isolates differed from reference ET-37 complex strains from the Czech Republic and Canada as well as from all the carrier isolates. The isolate from the patient’s nasopharynx was indistinguishable from the blood isolate except for a 40,000-bp chromosomal deletion that had occurred during systemic spread.
Bacterial meningitis due to Neisseria meningitidis (the meningococcus) continues to be of global importance for public health authorities. While pandemics affecting China and Africa are usually caused by meningococci of the A capsular serogroup, sporadic meningitis, outbreaks, and hyperendemic disease in Central Europe and the United States are usually caused by serogroups B and C (2). For sporadic meningococcal meningitis, public health efforts often include bacteriological analysis of throat swabs obtained from close contacts of the patient and the treatment of healthy nasopharyngeal carriers with prophylactic antibiotics and/or vaccines (5).
Meningococci isolated from the healthy contacts of a diseased patient (carrier isolates) are not necessarily related to the strain causing disease (case isolates), even if all these isolates express the same capsular serogroup. Various subtyping methods have been used to test relationships among the strains from a cohort, including serotyping and serosubtyping with monoclonal antibodies (MAbs) (11), pulsed-field gel electrophoresis (PFGE) (7), multilocus enzyme electrophoresis (MLEE) (2), ribotyping (22), the randomly amplified polymorphic DNA method (25), and PCR-restriction fragment length polymorphism analysis (16). Recently, a novel portable approach, multilocus sequence typing (MLST), which is based on the DNA sequences of six housekeeping gene fragments, has been developed (18). MLST allows assignment of meningococci to clonal groups within a globally accessible, continuously expanding central database.
In outbreak situations, so many bacteria may be isolated that only some of the carrier strains, usually selected on the basis of their capsular serogroup by latex agglutination, are evaluated in detail. However, the expression of capsular polysaccharide by serogroup B meningococci can undergo phase variation, resulting in the isolation of strains from carriers which are not obviously related to the index strain because they are capsule negative and nonserogroupable (6, 14, 15). The siaD gene encodes a polysialyltransferase which is needed for the synthesis of capsular polysialic acid chains. siaD can be amplified from serogroup B and C meningococci by PCRs (6), enabling the determination of the potential serogroups of bacteria which have become phenotypically nonserogroupable. Here we describe a two-step PCR, an improved version of the siaD PCR (6), which distinguishes between serogroup B, C, W135, and Y meningococci.
Antigenic variation through horizontal genetic exchange can also lead to capsule switching among highly related bacteria (23). Such capsule-switching variants would be classified as unrelated to the parent strain by classical serogrouping. Similarly, antigenic variation can also lead to the switching of other antigens (13), including those used for serological subtyping, such as PorA (21). Thus, serological methods cannot reliably recognize the relatedness of meningococci and epidemiological analyses should rely primarily on molecular techniques, particularly those based on multiple loci scattered around the chromosome (3).
We report here on the molecular investigation of meningococci isolated from individuals who had been in contact with a patient with severe serogroup C meningococcal disease.
MATERIALS AND METHODS
Meningococcal isolates, culture conditions, and identification.
Blood cultures were performed with the BactT/Alert System from Organon Teknika (Durham, N.C.). Cerebrospinal fluid was cultured on Columbia blood agar (Difco, Augsburg, Germany) and chocolate agar (Difco), and enrichment cultures were performed with brain heart infusion medium (Difco) supplemented with factors V and X (Difco). Nasopharyngeal swabs from 60 individuals with contact with the index patient were cultured on chocolate agar plates. After overnight growth at 37°C in a 5% CO2 atmosphere, colonies containing oxidase-positive, gram-negative diplococci were tested with the RAPID NH System (LD Labor Diagnostika, Leiden, Germany) for the biotyping of Neisseria and Haemophilus. Serogrouping was performed by latex agglutination with the Directigen Meningitis Combo Test (Becton Dickinson, Meylan, France) and by an enzyme-linked immunosorbent assay (ELISA) with the following MAbs: MAb 1087 (specific for the serogroup A capsule), MAb 735 (serogroup B), MAbs 924 and 1125 (serogroup C), MAb 1508 (serogroup W135), and MAb 1938 (serogroup Y). For the ELISA, meningococci were grown overnight on chocolate agar, and 20 μl of a bacterial suspension (optical density at 600 nm = 0.15) was added to each well of a microtiter plate (Greiner, Solingen, Germany) which had been coated with poly-d-lysine (Sigma). After drying, the bacteria were fixed with phosphate-buffered saline (PBS)–0.05% glutaraldehyde for 10 min at room temperature. Thereafter, nonspecific binding sites were blocked by incubation with PBS–1% bovine serum albumin for 1 h at 37°C. Binding of the anticapsule antibodies and of a secondary peroxidase-conjugated anti-mouse immunoglobulin antibody (Dianova, Hamburg, Germany) was performed for 1 h in PBS–1% bovine serum albumin. Intermediate wash steps were performed with PBS. ET-37 reference strains were a kind gift of Dominique A. Caugant (World Health Organization Collaborating Centre for Reference and Research on Meningococci, National Institute of Public Health, Oslo, Norway). Serogroup B isolate B1940 has been described elsewhere (9), and the reference serogroup W135 and Y strains were ATCC 750020 and ATCC 55989, respectively.
Serotyping.
The serotypes and serosubtypes of the meningococcal isolates were determined by a previously described ELISA (1) with a set of serotype-specific (2a, 2b, 4, 14, 15, and 21) and serosubtype-specific (P1.1 through P1.15) antibodies, which were provided by J. Suker and I. Feavers (National Institute for Biological Standards and Control, Hertfordshire, United Kingdom).
cps PCR.
The siaA, siaB, and siaC genes of region A of the cps locus (9, 23) yielded a 2-kb product after amplification by colony PCR with primers SH39 and UE16 (Table 1). The PCR conditions were initial denaturation (94°C, 5 min), followed by 36 cycles of annealing (45°C, 1 min), extension (72°C, 90 s), and denaturation (94°C, 1 min) and then one cycle of annealing (45°C, 1 min) and a final extension (72°C, 10 min). The PCR conditions for the amplification of the siaD genes (9, 10, 23) were the same, except that the following annealing temperatures (product sizes) were used: 52°C for primers UE12-UE13 (1.8 kb), 40°C for primers HC2-HC4 (2.1 kb), and 50°C for primers HC39-HC50 (extension time, 2 min; 2.4 kb) and primers HC44-HC50 (extension time, 2 min; 3.2 kb). The recently described siaD (B/C) PCR with primers B/CsiaD1 and B/CsiaD2, which exclusively amplify serogroup B and C genes (6), was also performed as a control.
TABLE 1.
Oligonucleotides used for the sia specific PCRs
| Oligo- nucleotide | Sequence (5′-3′) | Target | Positiona | Reference |
|---|---|---|---|---|
| SH39 | ATATCATTAGCATCTACC | siaA | 864–881b | 9, 10 |
| UE16 | CGGACAACAGTTCTCCT | siaC | 2916–2900b | 9, 10 |
| UE12 | CGCCTTTGCATCTGTCGTAGC | siaC | 2861–2881b | 12 |
| UE13 | GGAGATCAGAAGTCATAGTA | siaD downstream region (serogroup B) | 4641–4622b | 12 |
| HC2 | AAATCTATAAATTGACTC | siaC-siaD intergenic region (serogroup C) | 94–75 ntc upstream of siaDd | 23 |
| HC4 | GGAGATTTGTTTAGCT | siaD downstream region (serogroup C) | 552–537 nt downstream of siaD | Unpublished |
| B/CsiaD1 | AYATWTTGCATGTMSCYTTYCCTGA | siaD (serogroups B and C) | 3702–3726b; 572–596d | 6 |
| B/CsiaD2 | AGACATTGGGTWGWRGGKGARAGTAA | siaD (serogroups B and C) | 4161–4136b; 1031–1006d | 6 |
| HC39 | GTGTATGATATTCCAATC | siaD (serogroup W135) | 1103–1124e | 9 |
| HC44 | GGTTATATATTTCTAGA | siaD (serogroup Y) | 1079–1095f | 9 |
| HC50 | GGACAGTGGTGAGCTTTG | siaD downstream region (serogroups W135 and Y) | 250–233 nt (W135) and 967–950 nt (Y) downstream of the siaD stop codon | Unpublished |
PFGE.
PFGE was performed as described previously (19) after digestion of the DNA blocks with the restriction endonucleases SpeI and NheI.
MLST.
The sequences of fragments from the abcZ, adk, aroE, gdh, pdhC, and pgm genes were determined as described previously (18) and were compared to the sequences of formerly known alleles. Novel allelic variants were assigned the allelic designations adk13 (ST54), aroE20 (ST54), and gdh18 (ST55). New combinations of the six alleles were assigned the designations ST54 through ST56 (see http://mlst.zoo.ox.ac.uk).
RESULTS
Epidemiological description and properties of strains.
A German male (age, 17 years) developed severe meningococcal septicemia and skin hemorrhaging on his return to Germany from Prague, Czech Republic, where he had attended an international youth gathering with a group of German scouts for 3 days. Serogroup C N. meningitidis was isolated from cultures of both blood (strain 2120) and nasopharyngeal swabs (strain 2121) from the patient; no bacteria could be cultivated from the cerebrospinal fluid. Strains 2120 and 2121 were nonserotypeable and were of the P1.5,2 serosubtype. Disease outbreaks in the Czech Republic since 1993 have been caused by serogroup C meningococci of the ET-37 complex, usually serotype 2a and serosubtype P1.5,2 (17). Such Czech strains belong to a particular MLEE electrophoretic type (ET) called ET-15 (17), which has also caused hyperendemic disease in Canada (4). The patient recovered without complications after treatment with penicillin.
Nasopharyngeal swabs were obtained by general practitioners and the local health authorities from 60 individuals who had been in contact with the patient. Of those, 47 were scouts aged 15 to 21 years who had also attended the youth gathering. Thirteen other swabs were obtained from individuals who had accompanied the scouts, from the families of two of the scouts (scouts 1 and 2, Table 2), or from unspecified sources. Ten meningococci were isolated from 9 of the 60 individuals who had been swabbed (Table 2). Five of the isolates were nonserogroupable, three were serogroup C, and two were serogroup B. The serotypes and serosubtypes of the carrier isolates are also given in Table 2. Only strain 2125 exhibited a serosubtype identical to that of the index strain. However, that strain was serotype 4, whereas the index strain was nonserotypeable.
TABLE 2.
Properties of meningococci isolated from healthy contactsa
| Strain source and no. | Source | Serogroup | Serotype: serosubtype | siaA to siaC | siaD | siaD (B/C) | ST |
|---|---|---|---|---|---|---|---|
| Family members | |||||||
| 2122 | Scout 2 | NG | 4:P1.15 | + | C | + | 55 |
| 2123 | Scout 2 | C | 4:P1.15 | ND | ND | ND | 55 |
| 2127 | Scout 1 | C | NT:P1.15 | ND | ND | ND | 55 |
| 2131 | Mother of scouts 1 and 2 | C | NT:P1.15 | ND | ND | ND | 55 |
| 2130 | Father of scout 1 and 2 | NG | 4:P1.3,6 | + | Y | − | 56 |
| Others | |||||||
| 2118 | Scout 3 | B | 15:P1.16 | ND | ND | ND | 32 |
| 2124 | Scout 4 | NG | 14:P1.13 | + | C | + | 54 |
| 2129 | Subject 5 | B | NT:NST | ND | ND | ND | ND |
| 2125 | Scout leader | NG | 4:P1.5,2 | − | − | − | ND |
| 2126 | Scout 6 | NG | NT:NST | − | − | − | ND |
Abbreviations: NG, nongroupable; NT, nontypeable; NST, nonsubtypeable; ND, not done; siaA to siaC, PCR for serogroup B, C, W135, or Y (this study); siaD, specific PCR for each serogroup (this study); siaD (B/C), PCR for serogroup B or C (6).
Serogroup-specific PCR.
In order to determine the capsular genotypes of the nongroupable isolates, PCRs which can amplify the siaA to siaC and siaD genes, respectively, of region A from serogroup B, C, W135, or Y meningococci were developed. Region A of the cps locus contains the genes required for the synthesis of activated sialic acid (siaA to siaC) and for the polymerization of sialic acid (siaD). The siaA to siaC genes are identical in serogroups B, C, W135, and Y (9, 10, 23). Thus, the siaA to siaC PCR allows the differentiation of serogroups B, C, W135, and Y from the other serogroups of meningococci. In contrast, the siaD genes of serogroup B and C meningococci are 64% identical and differ considerably from the siaD genes of serogroups W135 and Y, which are 98% identical (9, 10, 23). Primers which bind to unique upstream and downstream regions flanking siaD were developed to distinguish serogroups B and C. The homology between the siaD genes of serogroup W135 and Y strains is so high that a common reverse primer was used together with serogroup-specific forward primers which bind within the siaD gene (Table 1). The siaD PCR yielded a larger DNA fragment with serogroup Y strains than with serogroup W135 strains due to a 630-bp insertion in the region downstream of siaD which is present among all serogroup Y strains analyzed to date (11a). Thus, these serogroups can be distinguished among reference strains by siaD PCRs.
We evaluated whether these PCR tests can reliably distinguish capsular genotypes B, C, W135, and Y with a small series of representative meningococcal isolates from our strain collection (serogroups B [seven strains], C [six strains], W135 [three strains], Y [four strains], and A [six strains]) and with two strains of Neisseria lactamica. The sensitivity of the siaA to siaC PCR was 100% because it yielded a product with all the strains of serogroups B, C, W135, and Y but with none of the strains of serogroup A or the N. lactamica strains. With the same set of strains, the sensitivity of the siaD PCR was 85% (seven of seven serogroup B strains, five of six serogroup C strains, two of three serogroup W135 strains, and three of four serogroup Y strains were positive).
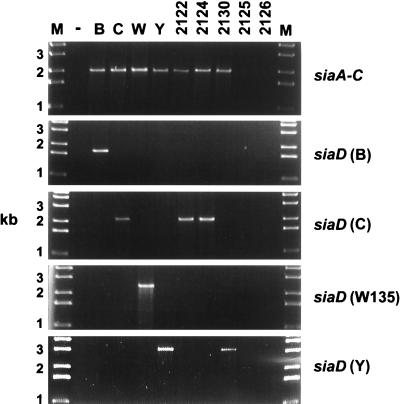
The isolates from healthy contacts analyzed in this study were tested by the siaA to siaC and siaD PCRs and as confirmation, by the siaD (B/C) PCR (6), which amplifies internal fragments of the siaD gene from all serogroup B and C meningococci with a single primer pair. Three of the five nonserogroupable meningococci yielded a positive result in the siaA to siaC PCR (Fig. 1), while the other two, strains 2125 and 2126, were negative by all PCR tests and do not contain genes for capsular polysaccharide B, C, W135, or Y. One of the three strains with a positive siaA to siaC PCR result yielded a specific product by the serogroup Y siaD PCR (genotype Y) (Fig. 1), while the other two strains were positive by the serogroup C siaD PCR (genotype C). One of the latter two strains (strain 2122) had been isolated from a swab which also yielded a typical serogroup C strain (strain 2123).
FIG. 1.
Determination of the capsular genotype of nonserogroupable carrier strains by PCR amplification of sia genes. The strains tested are listed at the top, and the molecular sizes of the standards are given on the left. Lanes: −, negative control without template DNA; B, serogroup B strain B1940 (9); C, index strain 2120 (serogroup C, this study); W, serogroup W135 strain (ATCC 750020); Y, serogroup Y strain (ATCC 55989), other numbers at the top are nonserogroupable carrier isolates.
MLST.
The MLST procedure, which consists of the sequencing of six independent gene fragments (18), was applied to the two strains isolated from the patient, seven of the carrier isolates, and two representative ET-15 strains of the ET-37 complex (strain 259/43 from the Czech Republic and strain 88/048 from Canada). Both case isolates and both representative ET-15 strains were ST11, the sequence type (ST) most commonly associated with strains of the ET-37 complex.
All carrier isolates except strain 2118 belonged to novel STs, labelled 54 through 56; serogroup B strain 2118 was ST32, which is characteristic of the ET-5 complex (Table 2). Thus, none of the carrier strains was related to the strains from the patient. Four of the carrier strains, three serogroup C strains and one genotype C strain, were ST55 and NT:P1.15 or 4:P1.15. These strains had been isolated from two brothers who had also visited the youth gathering and from their mother. The boys’ father carried the unrelated genotype Y strain of ST56. ST56 is related to ST12 (five of six alleles are identical) and ST13 (four of six alleles are identical), which were isolated from healthy carriers in Norway (18). STs 54 and 55 are not closely related to previously defined STs since they differed from all of them by at least two alleles.
PFGE.
MLST is a highly conservative genotyping method that is insensitive to microevolution and that is therefore highly suitable for determining relationships between bacterial strains and for long-term epidemiology. In contrast, PFGE is very sensitive to microevolution and can reveal minor differences between related strains (19). Bacterial DNA was analyzed by PFGE after digestion with SpeI and NheI to determine if minor differences distinguished any of the related strains (Fig. 2). Several bands differed between the ST11 strains isolated in Canada and the Czech Republic and between either of those bacteria and the ST11 isolates from the throat and blood of the German patient. In contrast, the latter two strains were almost identical, except that after digestion with SpeI, the DNA from the throat isolate yielded a fragment of 170 kb, while the DNA from the blood isolate yielded a fragment of 130 kb. Similarly, after NheI digestion, the throat isolate yielded a 160-kb band while the blood isolate yielded a 120-kb band (Fig. 2). These results suggest that this strain had suffered a 40-kb deletion during invasion from the throat to the bloodstream. No differences were observed between the four ST55 isolates after digestion with NheI, and three of those isolates, including strains 2122 (nongroupable, genotype C) and 2123 (serogroup C) isolated from a carrier (scout 2) were also indistinguishable after digestion with SpeI. The other three carrier isolates (ST32, ST54, and ST56, respectively) yielded unique PFGE patterns, as expected.
FIG. 2.
PFGE analysis of case and carrier strains after digestion with SpeI or NheI. The strains tested and their STs are indicated at the top of each panel, and the positions of molecular weight (MW) markers are given on the left. Representative ET-15 strains from Canada (strain 88/048) and the Czech Republic (strain 259/43) are next to the isolates from the blood (strain 2120) and throat (strain 2121) from the index patient. Other strains were isolated from carriers and are described in Table 2. The positions of the bands which differ between strains 2120 and 2121 are indicated by asterisks.
DISCUSSION
A serogroup C strain was isolated from a patient who had returned from the Czech Republic where serogroup C ET-37 complex meningococci have recently caused epidemics of meningitis and septicemia (17). Due to the increased risk of disease among the contacts of patients with sporadic cases of meningococcal disease, close contacts were offered prophylactic antibiotic therapy and their carrier status was determined in order to estimate whether the index strain had spread. The data presented here demonstrate that serogrouping of meningococci isolated from healthy carriers is an inadequate criterion for determining spread. The molecular mechanisms of reversible capsule phase variation have recently been elucidated for serogroup B meningococci (14, 15), and the data presented here suggest that capsule phase variation (whose mechanism remains to be elucidated) may also occur among serogroup C meningococci. Two highly related meningococci were isolated from one healthy carrier, and the meningococci were identical by MLST and PFGE analysis but differed in their expression of the C polysaccharide. Two other carrier isolates possessed genes for the expression of the C or the Y polysaccharide, although they were nonserogroupable. If the results for serogroup B meningococci can be extrapolated, these nonserogroupable strains are potentially capable of reversion to encapsulation and are potential sources of disease. These observations indicate that serogrouping of carrier isolates should rely on molecular techniques as well as on classical serological methods. The PCR methods described here allow determination of the B, C, W135, and Y capsular genotypes within 2 days after colony isolation and could readily be implemented in most laboratories for this purpose. Informed decisions on whether to offer prophylactic therapy to unrelated contacts could then be reached after both microbiological and capsular genotyping data were available.
Molecular typing methods.
Both short-term and long-term epidemiological studies depend on reliable typing methods which can be used to determine whether bacteria spread between individuals. The results obtained by serological methods are not conclusive because many isolates are not typeable and because the serotype and serosubtype are not uniform even within highly related bacteria such as those of the ET-37 complex (24) due to frequent horizontal genetic exchange (21). Similarly, the ST55 bacteria described here were variably serotype 4 and nontypeable (Table 2). Phylogenetic methods based on multiple loci scattered around the chromosome are necessary for reliable analysis of the relationships between bacteria (3). Of these, the MLST method has been shown to be as reliable as MLEE and even more conservative (18). The MLST analysis showed unambiguously that the strains from the index patient belonged to the ET-37 complex; almost all ET-37 complex strains are ST11, and all ST11 strains belong to the ET-37 complex. MLST also showed that none of the strains isolated from the 60 contacts were of the ET-37 complex. Inefficient spread of ET-37 complex bacteria has also been found in other investigations (1a, 20). Four strains isolated from three carriers within one family all belonged to the novel ST ST55, and the three other carrier strains investigated belonged to three other STs. One of the latter was serogroup B, ET-5 complex, which has been associated with hyperendemic disease in many countries (8).
PFGE is a typing method that is highly sensitive to microevolution and was used to determine whether differences could be found between the strains analyzed by MLST. The sensitivity of this method is demonstrated by the observation that only a few bands were identical between the representative ET-37 meningococci from Canada and the Czech Republic or between those bacteria and the strains isolated from the German patient. The results showed that the four carrier strains of ST55 were almost identical, as were the two isolates from the patient. The results also showed that a deletion of 40 kb of unknown significance seemed to have occurred during invasion from the throat to the bloodstream.
Relationships among ET-37 bacteria.
By the MLEE scheme ET-37 complex strains from patients with hyperendemic disease in Canada differ from other ET-37 complex strains by one enzyme and have therefore been referred to as “ET-15” (4). ET-37 strains from recent outbreaks in the Czech Republic are indistinguishable by MLEE from the Canadian strains (17), suggesting that there might be an epidemiological link between disease in both countries. The index patient described here became sick on the way home from a trip to the Czech Republic and may therefore have been colonized with ET-37 complex bacteria during that trip.
The data presented here do not support the differentiation of ET-15 strains and other strains of the ET-37 complex. All were identical by MLST, but the Canadian and Czech strains differed markedly by PFGE. The differences between these strains greatly exceed the differences found by PFGE during clonal spread of serogroup A meningococci (19), suggesting that these strains do not share a recent common ancestor and that there may be no direct epidemiological link between disease in Canada and disease in the Czech Republic. Equally great differences were found between the German isolate and the representative Czech strain, and the data do not clarify where the patient was colonized. The genetic variability of the ET-37 complex has not yet been analyzed by PFGE, and numerous ET-37 complex strains isolated in the Czech Republic and Germany might need to be investigated if the origin of the German disease strain were to be clarified. However, such analyses might fail if genomic rearrangements such as the 40-kb deletion described here for the otherwise identical throat and blood isolates of the patient were common among ET-37 meningococci.
ACKNOWLEDGMENTS
We gratefully acknowledge the expert technical assistance of Gabriele Heinze, the receipt of strains from D. A. Caugant, and the receipt of MAbs from J. Suker.
REFERENCES
- 1.Abdillahi H, Poolman J T. Neisseria meningitidis group B serosubtyping using monoclonal antibodies in whole-cell ELISA. Microb Pathog. 1988;4:27–32. doi: 10.1016/0882-4010(88)90045-9. [DOI] [PubMed] [Google Scholar]
- 1a.Achtman, M. Unpublished data from Mali.
- 2.Achtman M. Global epidemiology of meningococcal disease. In: Cartwright K, editor. Meningococcal disease. Chichester, England: John Wiley & Sons, Ltd.; 1995. pp. 159–175. [Google Scholar]
- 3.Achtman, M. A phylogenetic perspective on molecular epidemiology. In M. Sussman (ed.), Molecular medical microbiology, in press. Academic Press, London, United Kingdom.
- 4.Ashton F E, Ryan J A, Borczyk A, Caugant D A, Mancino L, Huang D. Emergence of a virulent clone of Neisseria meningitidis serotype 2a that is associated with meningococcal group C disease in Canada. J Clin Microbiol. 1991;29:2489–2493. doi: 10.1128/jcm.29.11.2489-2493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begg N. Outbreak management. In: Cartwright K, editor. Meningococcal disease. Chichester, England: John Wiley & Sons; 1995. pp. 285–305. [Google Scholar]
- 6.Borrow R, Claus H, Guiver M, Smart L, Jones D M, Kaczmarski E B, Frosch M, Fox A J. Non-culture diagnosis and serogroup determination of meningococcal B and C infection by a sialyltransferase (siaD) PCR ELISA. Epidemiol Infect. 1997;118:111–117. doi: 10.1017/s0950268896007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bygraves J A, Maiden M C. Analysis of the clonal relationships between strains of Neisseria meningitidis by pulsed field gel electrophoresis. J Gen Microbiol. 1992;138:523–531. doi: 10.1099/00221287-138-3-523. [DOI] [PubMed] [Google Scholar]
- 8.Caugant D A, Froholm L O, Bovre K, Holten E, Frasch C E, Mocca L F, Zollinger W D, Selander R K. Intercontinental spread of a genetically distinctive complex of clones of Neisseria meningitidis causing epidemic disease. Proc Natl Acad Sci USA. 1986;83:4927–4931. doi: 10.1073/pnas.83.13.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claus H, Vogel U, Mühlenhoff M, Gerardy-Schahn R, Frosch M. Molecular divergence of the sia locus in different serogroups of Neisseria meningitidis expressing polysialic acid capsules. Mol Gen Genet. 1997;257:28–34. doi: 10.1007/pl00008618. [DOI] [PubMed] [Google Scholar]
- 10.Edwards U, Müller A, Hammerschmidt S, Gerardy-Schahn R, Frosch M. Molecular analysis of the biosynthesis pathway of the alpha-2,8 polysialic acid capsule by Neisseria meningitidis serogroup B. Mol Microbiol. 1994;14:141–149. doi: 10.1111/j.1365-2958.1994.tb01274.x. [DOI] [PubMed] [Google Scholar]
- 11.Frasch C E, Zollinger W D, Poolman J T. Serotype antigens of Neisseria meningitidis and a proposed scheme for designation of serotypes. Rev Infect Dis. 1985;7:504–510. doi: 10.1093/clinids/7.4.504. [DOI] [PubMed] [Google Scholar]
- 11a.Frosch, M. Unpublished data.
- 12.Frosch M, Edwards U, Bousset K, Krauße B, Weisgerber C. Evidence for a common molecular origin of the capsule gene loci in gram-negative bacteria expressing group II capsular polysaccharides. Mol Microbiol. 1991;5:1251–1263. doi: 10.1111/j.1365-2958.1991.tb01899.x. [DOI] [PubMed] [Google Scholar]
- 13.Halter R, Pohlner J, Meyer T F. Mosaic-like organization of IgA protease genes in Neisseria gonorrhoeae generated by horizontal genetic exchange in vivo. EMBO J. 1989;8:2737–2744. doi: 10.1002/j.1460-2075.1989.tb08415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammerschmidt S, Hilse R, van Putten J P, Gerardy-Schahn R, Unkmeir A, Frosch M. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 1996;15:192–198. [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerschmidt S, Müller A, Sillmann H, Mühlenhoff M, Borrow R, Fox A, van Putten J, Zollinger W D, Gerardy-Schahn R, Frosch M. Capsule phase variation in Neisseria meningitidis serogroup B by slipped strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol Microbiol. 1996;20:1211–1220. doi: 10.1111/j.1365-2958.1996.tb02641.x. [DOI] [PubMed] [Google Scholar]
- 16.Knight A I, Cartwright K A, McFadden J. Identification of a UK outbreak strain of Neisseria meningitidis with a DNA probe. Lancet. 1990;335:1182–1184. doi: 10.1016/0140-6736(90)92697-g. [DOI] [PubMed] [Google Scholar]
- 17.Krizova P, Musilek M. Changing epidemiology of meningococcal invasive disease in the Czech Republic caused by new clone Neisseria meningitidis C:2a: P1.2(P1.5), ET-15/37. Cent Eur J Public Health. 1995;3:189–194. [PubMed] [Google Scholar]
- 18.Maiden M C, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morelli G, Malorny B, Muller K, Seiler A, Wang J, del Valle J, Achtman M. Clonal descent and microevolution of Neisseria meningitidis during 30 years of epidemic spread. Mol Microbiol. 1997;25:1047–1064. doi: 10.1046/j.1365-2958.1997.5211882.x. [DOI] [PubMed] [Google Scholar]
- 20.Ronne T, Berthelsen L, Buhl L H, Lind I. Comparative studies on pharyngeal carriage of Neisseria meningitidis during a localized outbreak of serogroup C meningococcal disease. Scand J Infect Dis. 1993;25:331–339. doi: 10.3109/00365549309008507. [DOI] [PubMed] [Google Scholar]
- 21.Suker J, Feavers I M, Achtman M, Morelli G, Wang J F, Maiden M C. The porA gene in serogroup A meningococci: evolutionary stability and mechanism of genetic variation. Mol Microbiol. 1994;12:253–265. doi: 10.1111/j.1365-2958.1994.tb01014.x. [DOI] [PubMed] [Google Scholar]
- 22.Swaminathan B, Matar G M, Reeves M W, Graves L M, Ajello G, Bibb W F, Helsel L O, Morales M, Dronavalli H, el Swify M, DeWitt W, Hunter S B. Molecular subtyping of Neisseria meningitidis serogroup B: comparison of five methods. J Clin Microbiol. 1996;34:1468–1473. doi: 10.1128/jcm.34.6.1468-1473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swartley J S, Marfin A A, Edupuganti S, Liu L J, Cieslak P, Perkins B, Wenger J D, Stephens D S. Capsule switching of Neisseria meningitidis. Proc Natl Acad Sci USA. 1997;94:271–276. doi: 10.1073/pnas.94.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J F, Caugant D A, Morelli G, Koumare B, Achtman M. Antigenic and epidemiologic properties of the ET-37 complex of Neisseria meningitidis. J Infect Dis. 1993;167:1320–1329. doi: 10.1093/infdis/167.6.1320. [DOI] [PubMed] [Google Scholar]
- 25.Woods J P, Kersulyte D, Tolan R W, Jr, Berg C M, Berg D E. Use of arbitrarily primed polymerase chain reaction analysis to type disease and carrier strains of Neisseria meningitidis isolated during a university outbreak. J Infect Dis. 1994;169:1384–1389. doi: 10.1093/infdis/169.6.1384. [DOI] [PubMed] [Google Scholar]