Abstract
A PCR procedure based on the intergenic region (IR) separating two genes encoding a recently identified mycobacterial two-component system, named SenX3-RegX3, was developed and was shown to be suitable for identifying Mycobacterium bovis BCG. The senX3-regX3 IR contains a novel type of repetitive sequence, called mycobacterial interspersed repetitive units (MIRUs). All tested BCG strains exclusively contained 77-bp MIRUs within the senX3-regX3 IR, whereas all non-BCG M. tuberculosis complex strains contained a 53-bp MIRU, in addition to the 77-bp MIRUs. All 148 strains analyzed so far could be divided into eight different groups according to the copy numbers of the 77-bp MIRU and to the presence or absence of the 53-bp MIRU. BCG strains contained either one, two, or three 77-bp MIRUs. The other strains contained one to five 77-bp MIRUs invariably followed by a 53-bp MIRU. The consistent absence of the 53-bp MIRU in BCG strains and its presence in virulent strains allowed us to develop an enzyme-linked immunosorbent assay using specific capture oligonucleotide probes to distinguish between BCG and other M. tuberculosis complex strains.
Mycobacterium tuberculosis is the most devastating human pathogen, and yet, its virulence mechanisms are poorly understood. It is estimated that one-third of the world’s population is infected with M. tuberculosis and that 3 million people die of tuberculosis each year (22). In addition to M. tuberculosis sensu stricto, other members of what is generally referred to as the M. tuberculosis complex can also cause this disease. The other species of this complex are Mycobacterium africanum, Mycobacterium microti, and Mycobacterium bovis. The latter species includes the Bacille de Calmette et Guérin (BCG) derived from M. bovis. Its virulence has been attenuated through 230 successive passages on media impregnated with beef bile between 1908 and 1921 (3). BCG is today the only available vaccine against tuberculosis.
Since its introduction for clinical use as a tuberculosis vaccine (18) and for immunotherapy against bladder cancer (2), the original BCG vaccine strain has been subcultured many times and has been distributed to a large number of production centers throughout the world. As a consequence, several substrains of the original vaccine strain are now in circulation, and there is evidence that they vary in certain characteristics, such as their antigenic structures (9), their mycolic acid patterns (21), their secreted protein profiles (1), and their IS986 copy numbers (6). The three most commonly used BCG vaccine strains are the Pasteur strain, the Japanese strain, and the Glaxo strain. Other daughter strains include the Moreau (Brazilian), the Montreal, the Russian, the Prague, and the Danish strains.
Although largely considered nonpathogenic, in certain circumstances BCG can nevertheless cause disease in humans (16, 17). This risk may be substantial for immunocompromised hosts (4). However, the prevalence of BCG infection is not known, because most laboratories cannot quickly differentiate between BCG and other members of the M. tuberculosis complex. Several DNA fingerprinting methods which differentiate M. tuberculosis strains are available, but none of them segregate BCG from other M. tuberculosis strains.
We have recently identified an operon coding for new mycobacterial two-component system called SenX3-RegX3 (24). The senX3-regX3 intergenic region (IR) was found to contain mycobacterial interspersed repetitive units (MIRUs) of 77 bp and, in certain strains, MIRUs of 53 bp (19, 24). This senX3-regX3 IR can be used for the detection of M. tuberculosis complex strains by PCR since it was found in all members of the M. tuberculosis complex but in none of the other mycobacterial species tested (19). In the study described in this report, we showed that the senX3-regX3 IR can also be used to differentiate between BCG and virulent strains of the M. tuberculosis complex, because all the virulent strains contained the 53-bp MIRU, whereas none of the BCG strains did.
MATERIALS AND METHODS
Bacterial strains.
Eight M. bovis strains were used. Four M. bovis strains (strains 60 and 63 isolated from goats and strains 76 and 78 isolated from cows) were kindly provided by C. Martin (University of Zaragoza, Zaragoza, Spain). M. bovis AN5 was obtained from G. Marshall (Institut Pasteur, Paris, France). Two M. bovis mtp40-containing strains (strains 89936 and 88367) were provided by B. B. Plikaitis from the Centers for Disease Control and Prevention (Atlanta, Ga.). One M. bovis strain (strain 1) was obtained from the Centre Hospitalier Régional de Lille (Lille, France).
The M. bovis BCG strains included eight vaccine strains and 10 clinical isolates from BCG-osis patients. The Glaxo, Russian, Moreau (Brazilian), Danish, Montreal, and Prague M. bovis BCG vaccine strains were provided by M. Lagranderie (Institut Pasteur, Paris, France), and the 1173P2 (Pasteur) and Japanese strains were obtained from the World Health Organization collection in Stockholm, Sweden. The 10 clinical isolates from BCG-osis patients were from the Institut Pasteur de Lille (strains 1 to 5) and the Centre Hospitalier Régional (Lille, France) (strains 6 to 10). The M. tuberculosis strains included the following: three M. tuberculosis strains isolated in Vietnam and lacking IS6110 (strains V.729, V.761, and V.808) were kindly provided by G. Marshall (Institut Pasteur, Paris, France), M. tuberculosis 2296207 was described previously (19, 24), and six clinical isolates (strains 1033, 1035, 1036, 1037, 1038, and 1039) came from the Centre Hospitalier Régional (Lille, France).
Isolation of chromosomal DNA.
The mycobacterial genomic DNA was isolated as described previously (19). Briefly, the mycobacteria were grown in the medium described by Sauton (23), and protoplasts were obtained by incubating the mycobacteria in a lysozyme-containing solution. The protoplasts were then lysed with proteinase K, and the DNA was extracted with phenol-chloroform, precipitated with isopropanol, resuspended, and treated with RNase. The DNA was extracted again with phenol-chloroform and chloroform, precipitated with ethanol, and resuspended in double-distilled water. The final DNA concentration was approximately 0.33 μg/μl.
PCR amplification of the senX3-regX3 IR and analysis of the PCR products.
The senX3-regX3 IRs of the various mycobacterial strains were amplified by PCR as described previously (19), with oligonucleotides C5 (5′-GCGCGAGAGCCCGAACTGC-3′) and C3 (5′-GCGCAGCAGAAACGTCAGC-3′) used as primers. The negative controls contained the PCR mixture without template DNA. The positive controls contained pRegX3Mt1 or pRegX3Bc1 (24) as template DNA. The PCR products were analyzed by electrophoresis on a 2.5% agarose gel and were visualized with ethidium bromide. The PCR products were then cloned and sequenced with the Zero background/kan cloning kit (Invitrogen Corporation, San Diego, Calif.) as described previously (19).
Hybridization analysis of the senX3-regX3 IR PCR products.
The senX3-regX3 IR PCR products were also analyzed by hybridization with the digoxigenin (DIG)-labeled senX3-regX3 IR of M. tuberculosis 2296207 isolated from pRegX3Mt2 as described previously (19). Alternatively, specific DIG-labeled oligonucleotides were used for hybridization. These oligonucleotides were 3′ end labeled by using the Boehringer Mannheim 3′ end labeling kit (catalog no. 1362372) as specified by the supplier (Boehringer Mannheim, Mannheim, Germany) and had the following sequences: 5′-CCACTCCTCCTCATC-3′ for oligonucleotide O53, which is specific for the 53-bp MIRU present in the senX3-regX3 IR, and 5′-GGGTGGTGCCCCCAC-3′ for oligonucleotide O77, which is specific for the 77-bp MIRU present in the senX3-regX3 IR.
ELISA.
For the detection of the 77-bp or the 53-bp MIRU by enzyme-linked immunosorbent assays (ELISAs), two kits provided by Boehringer Mannheim were used: the PCR ELISA DIG labeling kit (catalog no. 1636120) and the PCR ELISA DIG detection kit (catalog no. 1636111). First, the senX3-regX3 IR was amplified by using the PCR ELISA DIG labeling kit to obtain DIG-labeled PCR products. The second kit was used to immobilize biotinylated oligonucleotides (O53, O53R, or O77) onto streptavidin-coated microtiter plates. The sequence of the O53R oligonucleotide, specific for the 53-bp MIRU, was 5′-GCGCCACTCCTCCTCATC-3′ and was thus three bases longer than that of O53. The DIG-labeled senX3-regX3 IR PCR products were added to the captured oligonucleotide probes in the microtiter plates, and the plates were incubated at room temperature or at 40°C. The bound hybrids were detected by incubation with an anti-DIG-peroxidase conjugate and the colorimetric substrate 2,2′-azino-di(3-ethylbenzthiazoline sulfonate) (ABTS) (Boehringer Mannheim) at 37°C for 1 to 4 h. The absorbence at 405 nm was then measured with an automated microplate reader (model EL340; Bio-Tek Instrument, Inc., Winooski, Vt.).
RESULTS
Amplification of the senX3-regX3 IR.
We have previously shown that MIRUs are present in the senX3-regX3 IR of all 122 strains of the M. tuberculosis complex tested so far but that their copy numbers vary between different strains (19). The previous study principally focused on clinical M. tuberculosis isolates, and only two BCG strains were analyzed in that study. Interestingly, the senX3-regX3 IRs of these two BCG strains were composed of only 77-bp MIRUs, whereas those of the other strains of the M. tuberculosis complex contained an additional 53-bp MIRU (19, 24). To assess whether this difference could generally distinguish BCG strains from other M. tuberculosis complex strains, the senX3-regX3 IRs of a total of 26 M. bovis strains including 18 BCG strains were analyzed by PCR. In all cases the senX3-regX3 IR PCR gave rise to a DNA fragment, as detected by electrophoresis on a 2.5% agarose gel, which hybridized with the senX3-regX3 IR of M. tuberculosis 2296207, confirming the specificity of the PCR products that were obtained. Results of electrophoresis and hybridization analyses of representative samples are shown in Fig. 1. Although a PCR fragment was amplified for each strain, their sizes varied somewhat. On the basis of this size variation, as well as on the basis of this size variation, as well as on the basis of that observed previously (19), all the strains of the M. tuberculosis complex could be subdivided into eight different groups (groups I to VIII, from the longest to the shortest DNA fragment) (Table 1). In addition to the strains tested in this study, Table 1 presents data for several M. tuberculosis strains that have been studied previously. The majority of the M. tuberculosis strains are included in group V, and strain 2296207 is shown as a representative. Three M. tuberculosis strains lacking IS6110 correspond to groups I and II.
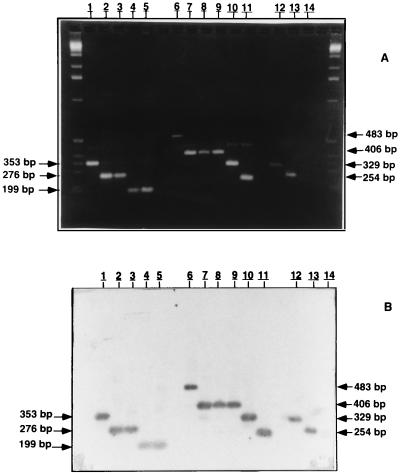
FIG. 1.
Agarose gel electrophoresis and Southern blot analysis of senX3-regX3 IR PCR products. The PCR products were analyzed by electrophoresis and ethidium bromide staining with a 2.5% agarose gel (A) and by Southern blot hybridization with a DIG-labeled 221-bp DNA fragment of M. tuberculosis 2296207 containing the senX3-regX3 IR as a probe (B). The senX3-regX3 IR PCR products were obtained from the Japanese (lanes 1), Russian (lanes 2), Glaxo (lanes 3), Montreal (lanes 4), and Prague (lanes 5) M. bovis BCG strains and from non-BCG M. bovis strains 60 (lanes 6), 63 (lanes 7), 78 (lanes 8), AN5 (lanes 9), 76 (lanes 10), and 1 (lanes 11). Positive (pRegX3Mt1 [lanes 12] and pRegX3Bc1 [lanes 13]) and negative (lanes 14) controls are also shown, as are molecular size markers (1-kb ladder; left and right lanes). The sizes of the PCR products that were obtained are indicated in the left and right margins.
TABLE 1.
Distribution of mycobacterial strains within eight groups by numbers of MIRUs
| Group | Strain(s) | senX3-regX3 IR PCR product length (bp) | No. of 77-bp MIRUs | No. of 53-bp MIRUs |
|---|---|---|---|---|
| I | M. tuberculosis V.808 and V.761 | 560 | 5 | 1 |
| II | M. tuberculosis V.729 and M. bovis 60 | 483 | 4 | 1 |
| III | M. bovis 63, 78, AN5, and 89936 | 406 | 3 | 1 |
| IV | BCG Japanese vaccine strain and BCG 1, 2, 3, 6, 7, and 8 (from patients with BCG-osis) | 353 | 3 | 0 |
| V | M. bovis 76 and 88367 and M. tuberculosis 2296207 | 329 | 2 | 1 |
| VI | BCG 1173P2, Glaxo, Russian, Moreau, and Danish (vaccine strains) and BCG 4, 5, 9, and 10 (from patients with BCG-osis) | 276 | 2 | 0 |
| VII | M. bovis 1 | 252 | 1 | 1 |
| VIII | BCG Prague and Montreal (vaccine strains) | 199 | 1 | 0 |
The 26 M. bovis strains tested in this study were distributed throughout seven of the eight groups (groups II to VIII; Table 1). The 18 BCG strains analyzed were restricted to three different groups (groups IV, VI, and VIII) (Fig. 1; Table 1). The Japanese BCG vaccine strain and six BCG strains isolated from patients with BCG-osis (strains 1, 2, 3, 6, 7, and 8) yielded a 353-bp PCR fragment (group IV). Another group of nine BCG strains (the 1173P2, Glaxo, Russian, Moreau, and Danish strains, as well as the remaining four BCG strains isolated from patients with BCG-osis) yielded a 276-bp PCR fragment (group VI). Finally, two BCG strains (strains Prague and Montreal) yielded a PCR fragment of 199 bp (group VIII).
The other eight M. bovis strains which are not BCG fell into four different classes (groups II, III, V, and VII) (Fig. 1; Table 1). Four of these eight strains (strains 63, 78, AN5, and 89936) were found in group III on the basis of the presence of a 406-bp PCR fragment. One M. bovis strain (strain 60) was in group II and had a 483-bp fragment, two strains (strains 76 and 88367) were in group V and each had a 329-bp fragment, and one strain (strain 1) was in group VII and had a 252-bp fragment.
Sequence analysis of the senX3-regX3 IR.
The senX3-regX3 IR PCR products obtained from all 26 M. bovis strains were cloned and sequenced (Fig. 2; Table 1). The senX3-regX3 IR was always found to contain one or several copies of the 77-bp MIRU, whereas the 53-bp MIRU was present only in groups I, II, III, V, and VII. In all cases, the nucleotide sequences of the 77-bp and the 53-bp MIRUs were identical to those reported previously (19, 24). Interestingly, all the BCG strains invariably fell within those groups (groups IV, VI and VIII) that did not contain the 53-bp MIRU. They only contained either one (group VIII), two (group VI), or three (group IV) 77-bp MIRUs. All the other M. bovis strains, as well as the previously analyzed M. tuberculosis, M. microti, and M. africanum strains, contained a 53-bp MIRU.
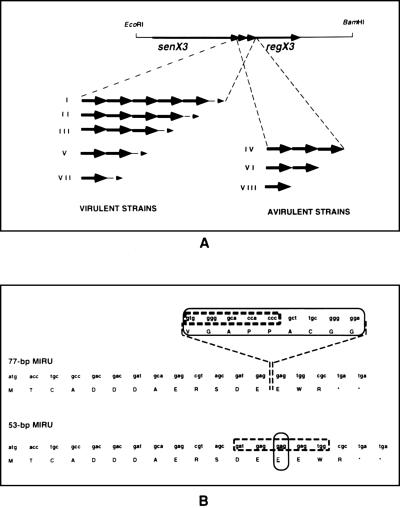
FIG. 2.
Schematic representation of the mycobacterial senX3-regX3 IR. (A) The 3.2-kb EcoRI-BamHI fragment containing the senX3-regX3 operon is shown by the thin line. The senX3 and regX3 genes are indicated by the thicker arrows. The IRs of the different strains can be divided into eight groups. Five groups (groups I, II, III, V, and VII) correspond to virulent, non-BCG strains and contain variable copies of the 77-bp MIRU (thick arrows) and one copy of the 53-bp MIRU (thin arrow). Three groups (groups IV, VI, and VIII) correspond to avirulent, BCG strains and contain exclusively one to three 77-bp MIRUs. (B) The nucleotide and amino acid sequences of the 77- and 53-bp MIRUs are indicated by the single-letter code. The sequence differences between the 77- and 53-bp MIRUs are circled. The O77 oligonucleotide specific for the 77-bp MIRU and O53 specific for the 53-bp MIRU are boxed.
Differentiation between BCG and other strains of the M. tuberculosis complex by hybridization.
The invariable presence of the 53-bp MIRU in all strains except BCG and its invariable absence in BCG strains prompted us to use specific probes to distinguish between the 53-bp and the 77-bp MIRUs in the senX3-regX3 IR (Fig. 2). For this purpose oligonucleotides O53 and O77, specific for the 53-bp and the 77-bp MIRUs, respectively, were designed for hybridization with the senX3-regX3 IR PCR products used as a matrix (Fig. 2B). On dot blotted PCR products, hybridization with O53 at 45 or 40°C, followed by washing at room temperature or at 40°C, respectively, yielded positive signals for PCR fragments obtained from non-BCG strains only (data not shown). Hybridization and washing at 45°C were too stringent, and the signal was lost. The strongest signal was observed after hybridization at 45°C and washing at room temperature. When the same conditions were used after Southern blotting of the senX3-regX3 IR PCR products, O53 was also found to give a positive signal only for PCR fragments obtained from non-BCG strains (Fig. 3A).
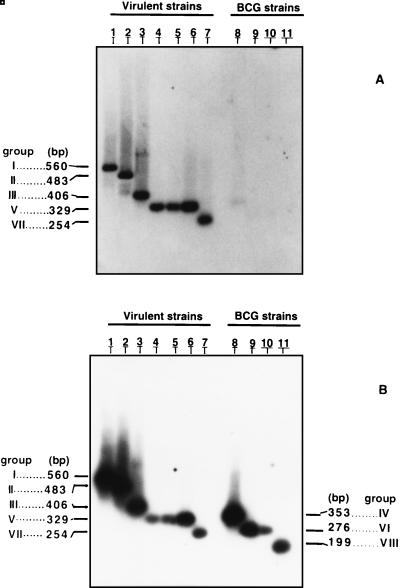
FIG. 3.
Southern blot analysis of the senX3-regX3 IR PCR products. DIG-labeled O53 (A) and O77 (B) were used as probes for Southern blot analysis of the senX3-regX3 IR PCR products obtained from M. tuberculosis V.808 (lane 1), V.729 (lanes 2), and 2296207 (lane 6); from non-BCG M. bovis AN5 (lane 3), 76 (lane 4), 88367 (lane 5), and 1 (lane 7); and from the Japanese (lane 8), 1173P2 (lane 9), Glaxo (lane 10), and Prague (lane 11) BCG strains. The group numbers and sizes of the amplified DNA fragments are given in the left and right margins.
In contrast to O53, oligonucleotide O77 yielded strong signals for the senX3-regX3 IR PCR products obtained from all strains of the M. tuberculosis complex tested (Fig. 3B) after hybridization at 55°C and washing at room temperature. The combination of O53 and O77 therefore allowed us to develop a test which includes a positive control in itself, since hybridization with O77 yielded a positive signal for each amplified senX3-regX3 IR, whereas hybridization with O53 yielded a positive signal only for the senX3-regX3 IR PCR products of the non-BCG strains. This allowed us to easily distinguish between BCG and non-BCG strains of the M. tuberculosis complex.
ELISA-based distinction between BCG and non-BCG strains.
Since the combined use of O77 and O53 allowed us to distinguish between BCG and non-BCG strains, we developed an ELISA using specific oligonucleotides as immobilized capture probes. When biotinylated O77 was immobilized on streptavidin-coated microtiter plates, DIG-labeled senX3-regX3 IR PCR fragments of all tested strains hybridized to the probe at room temperature or at 40°C, yielding optical density at 450 nm (OD450) values of 1.5 or more for 10 μl of the PCR mixture (Fig. 4). In contrast, when the biotinylated oligonucleotide O53 was immobilized, only the DIG-labeled senX3-regX3 IR PCR fragments of non-BCG strains hybridized to the probe (data not shown). However, this DNA hybridized only weakly at room temperature or at 40°C, which was not sufficient for a clear distinction between BCG DNA and non-BCG DNA.
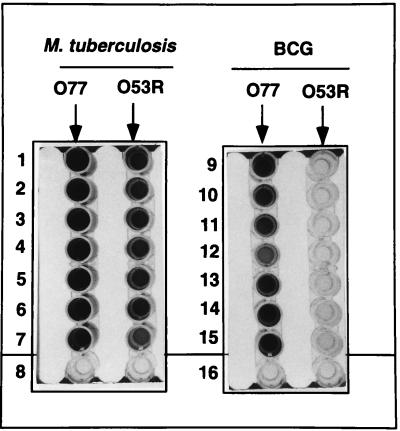
FIG. 4.
ELISA of the senX3-regX3 IR PCR products. Biotinylated oligonucleotides O77 and O53R were immobilized on streptavidin-coated microtiter plates and incubated with 10 μl of the DIG-labeled senX3-regX3 IR PCR products obtained from M. tuberculosis strains 1033 (wells 1), 1035 (wells 2), 1036 (wells 3), 1037 (wells 4), 1038 (wells 5), 1039 (wells 6), and 2296207 (wells 7) and the Japanese (wells 9), 1173P2 (wells 10), Glaxo (wells 11), Russian (wells 12), Danish (wells 13), Prague (wells 14) and Montreal (wells 15) BCG strains. Wells 8 contained no PCR product, and wells 16 contained neither the PCR product nor the oligonucleotides.
To increase the sensitivity of the assay, oligonucleotide O53R, which had a higher melting temperature, was thus designed. When this oligonucleotide was used, hybridization with the PCR fragments from 10 μl of the reaction mixture of the non-BCG strains gave OD450 values of more than 1.5 at 40°C, whereas the values for BCG strains were less than 0.2 (Fig. 4). However, at room temperature, this assay could not sufficiently discriminate BCG strains (OD450, 0.8) from non-BCG strains (OD450, 1.5). Thus, by using O77 and O53R in parallel at a hybridization temperature of 40°C with as little as 10 μl of the PCR mixture, the ELISA includes a positive control to permit the discrimination between BCG and non-BCG strains.
The OD450 values were routinely measured 1 to 4 h after the addition of the ABTS substrate. Overnight incubation with the substrate did not change the sensitivity or the specificity of the test.
DISCUSSION
BCG is one of the world’s most widely used vaccines. It is also used as one of the most effective medications in the immunotherapeutic treatment of human bladder carcinoma. Although considered relatively safe, BCG can occasionally cause disease such as sepsis, osteitis, and meningitis (15–17). Therefore, timely and unequivocal diagnosis of BCG infection is of importance in order to provide appropriate therapy for BCG-mediated illness. However, it has been notoriously difficult to distinguish BCG from the other members of the M. tuberculosis complex. This distinction may be particularly important for patients suffering from cellular immunodeficiencies (4) or for patients undergoing bladder cancer treatment (2). It has been estimated that approximately 5% of BCG-treated cancer patients experience adverse reactions to the treatment (15).
BCG strains can be distinguished from other M. tuberculosis complex strains on the basis of differences in certain biochemical and growth characteristics and on the basis of the resistance and sensitivity to cycloserine. However, these methods and other, nonstandardized methods (5, 8, 12, 13) are time-consuming, may be laborious, sometimes require specialized and expensive equipment, and do not always provide definitive proof that a clinical isolate is BCG. The insertion sequence IS1081 has been proposed as a useful tool that can be used to recognize BCG by restriction fragment length polymorphism analysis (26). However, some M. tuberculosis strains have recently been shown to contain IS1081 also (11). Mahairas et al. (20) have identified genetic differences between BCG and the other members of the M. tuberculosis complex, and on the basis of these differences Talbot et al. (25) have developed a multiplex PCR method for the identification of BCG. However, the method has not been tested with mycobacteria that are not part of the M. tuberculosis complex. Frothingham (7) has used sequevar differences within the major polymorphic tandem repeats to distinguish BCG from other M. tuberculosis complex strains, but these repeats can also be found in mycobacteria that are not part of the complex (10).
In this study, we have developed a PCR method based on the senX3-regX3 IR, which we have recently demonstrated to be specific for M. tuberculosis complex strains (19). This region is composed of two types of novel repetitive sequences, named MIRUs (24). Our results show that BCG strains exclusively contained 77-bp MIRUs in the senX3-regX3 IR, whereas the other strains of the M. tuberculosis complex contained both 77-bp and 53-bp MIRUs. This difference allowed us not only to distinguish BCG from other strains on the basis of size differences of the PCR-amplified DNA fragments but also to develop a discriminative ELISA using two synthetic oligonucleotide probes.
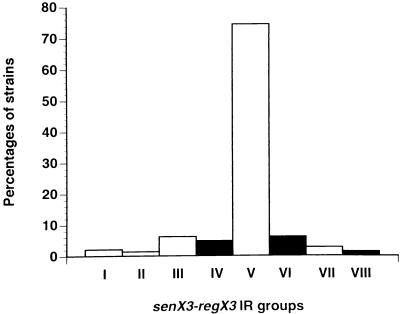
Although all BCG strains contained only the 77-bp MIRU in the senX3-regX3 IR, the copy number of this MIRU varied somewhat between BCG strains. They could therefore be classified into three different groups on the basis of the sizes of the PCR products. These products contained three, two, or one 77-bp MIRU, respectively. None of the other M. tuberculosis complex strains analyzed in this study or in a previous study (19) fell within any of these three groups. On the basis of MIRU copy numbers, all 148 strains of the M. tuberculosis complex analyzed so far could be divided into eight distinct groups, two more than was reported previously (19). Figure 5 depicts the relative frequency of the strains within each group and shows that the majority of the strains fell by far within group V. All the BCG strains fell in group IV, (three 77-bp MIRUs), VI (two 77-bp MIRUs), or VIII (one 77-bp MIRU), whereas all the other strains fell in group I, II, III, V, or VII, each containing one 53-bp MIRU in addition to variable numbers of copies (from one to five) of the 77-bp MIRU.
FIG. 5.
Distribution of the various M. tuberculosis complex strains among eight different groups. The MIRU copy numbers of a total of 148 strains of the M. tuberculosis complex were determined in this study and in a previous study (19). The distributions of the strains are indicated as percentages for each group. Groups IV, VI, and VIII (black bars) contain only BCG strains. Groups I, II, III, V, and VII (white bars) contain only non-BCG strains.
The original BCG strain was initially derived from virulent M. bovis between 1908 and 1921 and was subsequently distributed and cultured under different conditions with various growth media (3, 18). This resulted in the generation of several BCG substrains which differ in antigenic and biochemical properties (20, 21), as well as in their contents of certain insertion sequences (6). Two subgroups have been distinguished (1, 6, 21). Group A comprises the Japanese, Russian, and Moreau strains, derived from the original Pasteur BCG strain prior to 1925, and group B comprises the 1173P2, Glaxo, Danish, Montreal, and Prague strains (Table 2). Interestingly, the BCG strain that possesses the longest senX3-regX3 IR belongs to group A, and the BCG strains possessing the smallest senX3-regX3 IR belong to group B. The BCG strain with an intermediate senX3-regX3 IR length belongs either to group A or to group B.
TABLE 2.
Characteristics of the principal BCG strains
| BCG strain | Biochemical characteristicsa | Immuno- genicityb | No. of 77-bp MIRUs in the senX3-regX3 IR |
|---|---|---|---|
| Japanese | A | Low | 3 |
| 1173P2 (Pasteur) | B | High | 2 |
| Glaxo | B | High | 2 |
| Russian | A | High | 2 |
| Moreau (Brazil) | A | NDc | 2 |
| Danish | B | ND | 2 |
| Montreal | B | ND | 1 |
| Prague | B | Low | 1 |
BCG strains in group A have two copies of IS986, secrete the 23-kDa MPB70 protein, and contain methoxycolate, whereas group B strains do not.
Assessment of immunogenicity is based on the data published by Lagranderie et al. (14).
ND, not determined.
Different BCG strains have also recently been evaluated for their capacity to survive in mice and to trigger immune responses. The 1173P2, Glaxo, and Russian strains were found to multiply actively in mice and to persist for several months, whereas the Japanese and Prague strains did not (Table 2). These differences were attributed to differences in the genetic characteristics of the various BCG strains (14). We observed that the more persistent and immunogenic strains (1173P2, Russian, and Glaxo) contained two 77-bp MIRUs in the senX3-regX3 IR, whereas the less persistent strains contained either three (Japanese) or one (Prague) 77-bp MIRU. However, there is no evidence of any direct link between the number of MIRUs and persistence or immunogenicity, and this observation may be fortuitous.
In conclusion, the MIRUs contained within the senX3-regX3 IR can be used as a target to differentiate BCG from other strains of the M. tuberculosis complex either by size analysis of the PCR products or by ELISA with specific capture oligonucleotides.
ACKNOWLEDGMENTS
We thank G. Delcroix, B. B. Plikaytis, G. Marshall, M. Lagranderie, C. Martin, and A. Vachée for the gifts of bacterial strains and E. Fort for photography.
The work was supported by INSERM, Institut Pasteur de Lille, Région Nord-Pas de Calais, and Ministère de la Recherche. J.M. held a fellowship from the Fondation Recherche et Partage and Sidaction, and P.S. is a researcher of CNRS.
REFERENCES
- 1.Abou-Zeid C, Smith I, Grange J, Steele J, Rook G. Subdivision of daughter strains of bacille Calmette-Guérin (BCG) according to secreted protein patterns. J Gen Microbiol. 1986;132:3047–3053. doi: 10.1099/00221287-132-11-3047. [DOI] [PubMed] [Google Scholar]
- 2.Brosman S A. Bacillus Calmette-Guérin immunotherapy. Urol Clin N Am. 1992;19:557–564. [PubMed] [Google Scholar]
- 3.Calmette A, Guérin G, Nègre L, Boquet A. Prémunition des nouveaux-nés contre la tuberculose par le vaccin BCG, 1921–1926. Ann Inst Pasteur (Paris) 1926;40:89–133. [Google Scholar]
- 4.Edwards K M, Kernolde D S. Possible hazards of routine bacillus Calmette-Guérin immunization in human immunodeficiency virus-infected children. Pediatr Infect Dis J. 1996;15:836–838. doi: 10.1097/00006454-199609000-00023. [DOI] [PubMed] [Google Scholar]
- 5.Floyd M M, Silcox V A, Jones W D, Jr, Butler W R, Kilburn J O. Separation of Mycobacterium bovis BCG from Mycobacterium tuberculosis and Mycobacterium bovis by high-performance liquid chromatography of mycolic acids. J Clin Microbiol. 1992;30:1327–1330. doi: 10.1128/jcm.30.5.1327-1330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fomukong N G, Dale J W, Osborn T W, Grange J M. Use of gene probes based on the insertion sequence IS986 to differentiate between BCG vaccine strains. J Appl Bacteriol. 1992;72:126–133. doi: 10.1111/j.1365-2672.1992.tb01813.x. [DOI] [PubMed] [Google Scholar]
- 7.Frothingham R. Differentiation of strains in Mycobacterium tuberculosis complex by DNA sequence polymorphisms, including rapid identification of M. bovis BCG. J Clin Microbiol. 1995;33:840–844. doi: 10.1128/jcm.33.4.840-844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frothingham R, Wilson K H. Molecular phylogeny of the Mycobacterium avium complex demonstrates clinically meaningful divisions. J Infect Dis. 1994;169:305–312. doi: 10.1093/infdis/169.2.305. [DOI] [PubMed] [Google Scholar]
- 9.Harboe M, Nagai S. MPB70 a unique antigen of Mycobacterium bovis BCG. Am Rev Respir Dis. 1984;129:444–452. doi: 10.1164/arrd.1984.129.3.444. [DOI] [PubMed] [Google Scholar]
- 10.Hermans P W M, van Soolingen D, van Embden J D A. Characterization of a major polymorphic tandem repeat in Mycobacterium tuberculosis and its potential use in the epidemiology of Mycobacterium kansasii and Mycobacterium gordonae. J Bacteriol. 1992;174:4157–4165. doi: 10.1128/jb.174.12.4157-4165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huh Y J, Ahn D I, Kim S J. Limited variation of DNA fingerprints (IS6110 and IS1081) in Korean strains of Mycobacterium tuberculosis. Tubercle Lung Dis. 1995;76:324–329. doi: 10.1016/s0962-8479(05)80031-0. [DOI] [PubMed] [Google Scholar]
- 12.Jones W D., Jr Differentiation of known strains of BCG from isolates of Mycobacterium bovis and Mycobacterium tuberculosis by using mycobacteriophage 33D. J Clin Microbiol. 1975;1:391–392. doi: 10.1128/jcm.1.4.391-392.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristjansson M, Green P, Manning H L, Slutsky A M, Brecher S M, von Reyn C F, Arbeit R D, Maslow J N. Molecular confirmation of bacillus Calmette-Guérin as the cause of pulmonary infection following urinary tract instillation. Clin Infect Dis. 1993;17:228–230. doi: 10.1093/clinids/17.2.228. [DOI] [PubMed] [Google Scholar]
- 14.Lagranderie M R R, Balazuc A-M, Deriaud E, Leclerc C D, Gheorghiu M. Comparison of immune responses of mice immunized with five different Mycobacterium bovis BCG vaccine strains. Infect Immun. 1996;64:1–9. doi: 10.1128/iai.64.1.1-9.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamm D L. Complications of bacillus Calmette-Guérin immunotherapy. Urol Clin N Am. 1992;19:565–572. [PubMed] [Google Scholar]
- 16.Lotte A, Wasz-Hockert O, Poisson N. Second IUATLD study on complications included by intradermal BCG-vaccination. Bull Int Union Tuberc. 1988;63:47–59. [PubMed] [Google Scholar]
- 17.Lotte A, Wasz-Hockert O, Poisson N, Dumitrescu N, Verron M, Couvet E. BCG complications: estimates for the risks among vaccinated subjects and statistical analysis of their main characteristics. Adv Tuberc Res. 1984;21:107–193. [PubMed] [Google Scholar]
- 18.Lugosi L. Theoretical and methodological aspects of BCG vaccine from the discovery of Calmette and Guérin to molecular biology. A review. Tubercle Lung Dis. 1992;73:252–261. doi: 10.1016/0962-8479(92)90129-8. [DOI] [PubMed] [Google Scholar]
- 19.Magdalena J, Vachée A, Supply P, Locht C. Identification of new DNA region specific for members of Mycobacterium tuberculosis complex. J Clin Microbiol. 1998;36:937–943. doi: 10.1128/jcm.36.4.937-943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahairas G G, Sabo P J, Hickey M J, Singh D C, Stover C K. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minnikin D E, Minnikin S M, Dobson G, Goodfellow M, Portaels F, van den Breen L, Sesardic D. Mycolic acid patterns of four vaccine strains of Mycobacterium bovis BCG. J Gen Microbiol. 1983;129:889–891. doi: 10.1099/00221287-129-3-889. [DOI] [PubMed] [Google Scholar]
- 22.Raviglione M C, Snider D E J, Kochi A. Global epidemiology of tuberculosis-morbidity and mortality of a worldwide epidemic. JAMA. 1995;273:220–226. [PubMed] [Google Scholar]
- 23.Sauton B. Sur la nutrition minérale du bacille tuberculeux. C R Acad Sci. 1912;155:860. [Google Scholar]
- 24.Supply P, Magdalena J, Himpens S, Locht C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol Microbiol. 1997;26:991–1003. doi: 10.1046/j.1365-2958.1997.6361999.x. [DOI] [PubMed] [Google Scholar]
- 25.Talbot E A, Williams D L, Frothingham R. PCR identification of Mycobacterium bovis BCG. J Clin Microbiol. 1997;35:566–569. doi: 10.1128/jcm.35.3.566-569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Soolingen D, Hermans P W M, de Haas P E W, van Embden J D A. Insertion element IS1081-associated restriction fragment length polymorphisms in Mycobacterium tuberculosis complex species: a reliable tool for recognizing Mycobacterium bovis BCG. J Clin Microbiol. 1992;30:1772–1777. doi: 10.1128/jcm.30.7.1772-1777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]