Abstract
Background
Small cell lung cancer (SCLC) is a highly malignant cancer characterized by metastasis and an extremely poor prognosis. Although combined chemoimmunotherapy improves the prognosis of extensive-stage (ES)-SCLC, the survival benefits remain limited. Furthermore, no reliable biomarker is available so far to predict the treatment outcomes for chemoimmunotherapy.
Methods
This retrospective study included patients with ES-SCLC treated with first-line combined atezolizumab or durvalumab with standard chemotherapy between Janauray 1, 2019 and October 1, 2022 at five medical centers in China as the chemoimmunotherapy group. The patients were divided into one training cohort and two independent external validation cohorts. Additionally, we created a control group of ES-SCLC who was treated with first-line standard chemotherapy alone. The Radiomics Score was derived using machine learning algorithms based on the radiomics features extracted in the regions of interest delineated on the chest CT obtained before treatment. Cox proportional hazards regression analysis was performed to identify clinical features associated with therapeutic efficacy. The log-rank test, time-dependent receiver operating characteristic curve, and Concordance Index (C-index) were used to assess the effectiveness of the models.
Results
A total of 341 patients (mean age, 62±8.7 years) were included in our study. After a median follow-up time of 12.1 months, the median progression-free survival (mPFS) was 7.1 (95% CI 6.6 to 7.7) months, whereas the median overall survival (mOS) was not reached. The TNM stage, Eastern Cooperative Oncology Group performance status, and Lung Immune Prognostic Index showed significant correlations with PFS. We proposed a predictive model based on eight radiomics features to determine the risk of chemoimmunotherapy resistance among patients with SCLC (validation set 1: mPFS, 12.0 m vs 5.0 m, C-index=0.634; validation set 2: mPFS, 10.8 m vs 6.1 m, C-index=0.617). By incorporating the clinical features associated with PFS into the radiomics model, the predictive efficacy was substantially improved. Consequently, the low-progression-risk group exhibited a significantly longer mPFS than the high-progression-risk group in both validation set 1 (mPFS, 12.8 m vs 4.5 m, HR=0.40, p=0.028) and validation set 2 (mPFS, 9.2 m vs 4.6 m, HR=0.30, p=0.012). External validation set 1 and set 2 yielded the highest 6-month area under the curve and C-index of 0.852 and 0.820, respectively. Importantly, the integrated prediction model also exhibited considerable differentiation power for survival outcomes. The HR for OS derived from the low-progression-risk and high-progression-risk groups was 0.28 (95% CI 0.17 to 0.48) in all patients and 0.20 (95% CI 0.08 to 0.54) in validation set. By contrast, no significant differences were observed in PFS and OS, between high-progression-risk patients receiving chemoimmunotherapy and the chemotherapy cohort (mPFS, 5.5 m vs 5.9 m, HR=0.90, p=0.547; mOS, 14.5 m vs 13.7 m, HR=0.97, p=0.910).
Conclusions
The integrated clinical and radiomics model can predict the treatment outcomes in patients with ES-SCLC receiving chemoimmunotherapy, rendering a convenient and low-cost prognostic model for decision-making regarding patient management.
Keywords: Biomarkers, Tumor; Drug Therapy, Combination; Immunotherapy
WHAT IS ALREADY KNOWN ON THIS TOPIC
Landmark studies on extensive-stage small cell lung cancer (ES-SCLC) have demonstrated that combined immunotherapy and chemotherapy as a first-line treatment although improves overall survival, the benefit is limited. No reliable biomarker is available so far to predict the treatment outcomes of chemoimmunotherapy in ES-SCLC.
WHAT THIS STUDY ADDS
We constructed a predictive model using integrated clinical and radiomics information from patients with ES-SCLC and established two external validation cohorts to exclude the potential variations between institutions due to different CT equipment and parameter settings, thereby ensuring the stability and generalizability of our results. Our model can precisely pick out the subgroup of patients who would benefit from chemoimmunotherapy. The risk for disease progression was reduced by ~65% and the risk for death was reduced by ~70% in the low-progression-risk group compared with the high-progression-risk group.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
We developed an economic, non-invasive, valid and effective model for predicting the therapeutic responsiveness to chemoimmunotherapy and the prognosis of patients with ES-SCLC, providing a convenient and cost-effective tool for the management of ES-SCLC. With low input requirements and strong operability, our innovative strategy will have significant clinical significance and profound impacts.
Introduction
Small cell lung cancer (SCLC), a highly aggressive malignancy, accounts for approximately 15% of lung cancers, with around 70% of patients diagnosed with extensive-stage SCLC (ES-SCLC).1–3 Platinum-based chemotherapy has been the standard of care for ES-SCLC for decades, with a 2-year survival rate of approximately 7%.4 Landmark studies on ES-SCLC, such as IMpower133 and CASPIAN, have demonstrated that the combination of programmed death ligand 1 (PD-L1) immunotherapy and chemotherapy as a first-line treatment improves overall survival (OS).5 6 Despite these advancements, a combination of atezolizumab or durvalumab prolongs median progression-free survival (mPFS) by <1 month compared with standard chemotherapy, with a median OS (mOS) benefit of only 2.0–2.7 months.5 6 These findings suggest that a proportion of patients with ES-SCLC exhibit a poor treatment response to immune checkpoint inhibitors (ICIs).
However, no established predictive biomarker is available to guide the use of ICIs in patients with SCLC; patients who will benefit from chemoimmunotherapy in ES-SCLC remain uncertain. Most patients with SCLC exhibit negative PD-L1 expression. Moreover, PD-L1 expression levels are not significantly correlated with response to chemoimmunotherapy.7 8 Previous studies have classified SCLC based on neuroendocrine gene or transcription factor expression as a guide to treatment.9–11 However, the use of multiple transcription factors and microenvironmental indicators for clinical guidance is highly complex, which limits the feasibility of the approach in clinical practice.12–14 Therefore, an accurate, rapid, user-friendly, and cost-effective predictive marker is urgently needed to estimate the treatment outcomes of chemoimmunotherapy.
Artificial intelligence (AI) has been used for monitoring disease progression and predicting disease prognosis. Recent studies have demonstrated that machine learning (ML) analysis of CT can predict the RNA-seq transcriptome of lung cancer tissues, which can be used to evaluate CD8+ T cell tumor infiltration and thus predict immunotherapy response.15 On the other hand, deep learning (DL) algorithms are widely used in cancer diagnosis, precision staging, and efficacy prediction. DL-guided imaging can identify epidermal growth factor receptor mutations, pathological diagnosis, and molecular types of lung cancer.16 17 Random forest classification models can identify patients who are likely to experience immune hyperprogression by analyzing imaging features.18 These findings demonstrate that AI can be used to probe microcosmic imaging features with biological and pathological processes.
Here, this multicenter retrospective cohort study recruited patients with ES-SCLC who received chemoimmunotherapy as first-line treatment. We proposed an ML approach using combined radiomics markers and clinical features to establish an efficacy predictive classifier for the potential benefits of chemoimmunotherapy in patients with ES-SCLC, which is expected to be user-friendly, non-invasive, highly reproducible, and precise.
Materials and methods
Study design and participants
In this retrospective study, eligible patients were adults confirmed by definite histopathology of SCLC and classified as ES according to the Veterans Administration Lung Study Group staging system. And the patients should receive first-line standard chemoimmunotherapy (atezolizumab/durvalumab plus chemotherapy). The requirement for informed consent was waived due to the retrospective study design. Finally, we included 379 patients between January 2019 and September 2022 as the chemoimmunotherapy group. The training set (n=253) included patients from The Second Affiliated Hospital of Zhejiang University (n=72), Hunan Cancer Hospital (n=156), and West China Hospital (n=25), whereas the independent external validation sets 1 and 2 included patients from Peking University Cancer Hospital (n=51) and Shanghai Pulmonary Hospital (n=75). In addition, we also collected data from 57 patients with ES-SCLC who received first-line chemotherapy alone as the control group. Baseline chest CT, demographic information (age, sex, ethnicity, and marital status), smoking history, treatment regimen and side effects, follow-up duration, PFS, and OS time were recorded. Disease stage at diagnosis was classified according to the American Joint Committee on Cancer, eighth edition, tumor, node, metastasis staging system; the best objective response was evaluated according to Response Evaluation Criteria in Solid Tumors, V.1.1. Patients who discontinued therapy and had missing follow-up data were excluded. Variables with values of p<0.05 in the univariate Cox proportional hazards regression model were entered into the multivariate Cox regression model; those with values of p<0.05 in the multivariate Cox regression model were included in the final prediction model.
Prediction model construction
The target lung cancer lesions (ie, regions of interest) on the pretreatment lung CT were labeled manually for the remaining patients using 3D-Slicer software. Next, a peripheral ring was constructed by automatically expanding and contracting the tumor border by 2 mm on each side, resulting in a ring that was approximately 4 mm thick inside and outside the border. The lung CT images were labeled manually by two experienced radiologists (QW and MC); disputes were resolved by discussion with a third one (KH). Images were resampled and cropped if needed. The 3D CT images had dimensions ranging from 512×512 × 52 to 512×512 × 512 units. The PyRadiomics package extracted the segmented region of interest images for histological features, including first-order statistical, spatial geometry, texture, and wavelet features. LASSO-Cox proportional risk regression was performed using PFS as the outcome for each eigenvalue of patients in the training set. Features with non-zero coefficients were extracted using 10-fold cross-validation. Radiomics Score (radio-score) was calculated based on a linear combination of the weighted feature coefficients, using the cut-off as the median value. To further optimize the independent variables, correlation and collinearity analyses were performed between the radio-score and clinical features.
Furthermore, we integrated radio-score with significant clinical variables for PFS to apply multivariate Cox regression to construct the combined model. Accordingly, Risk Score was calculated and the median values were chosen to discriminate low-progression-risk/high-progression-risk groups of disease progression.
Statistical analysis
Data were analyzed and plotted using R studio (V.4.2.3). Radiomics features were extracted using Python (V.3.8). Time-dependent receiver operating characteristic (ROC) curve with 6 months and 12 months as cut-offs and Concordance Index (C-index) were used to evaluate the model performance. Decision curve analysis (DCA) was used to assess the clinical benefit of the model. Calibration was evaluated to determine the validity of the model. Differences between low-progression-risk and high-progression-risk groups were assessed using log-rank tests and Kaplan-Meier (K-M) analysis. Correlation was assessed using Spearman’s correlation analysis. Variance inflation factor (VIF) was used to analyze the collinearity between independent variables. Values of p<0.05 were considered statistically significant.
Role of the funding source
The funder of the study played no role in study design, data collection, data analysis, data interpretation, or writing of the article.
Results
Study population
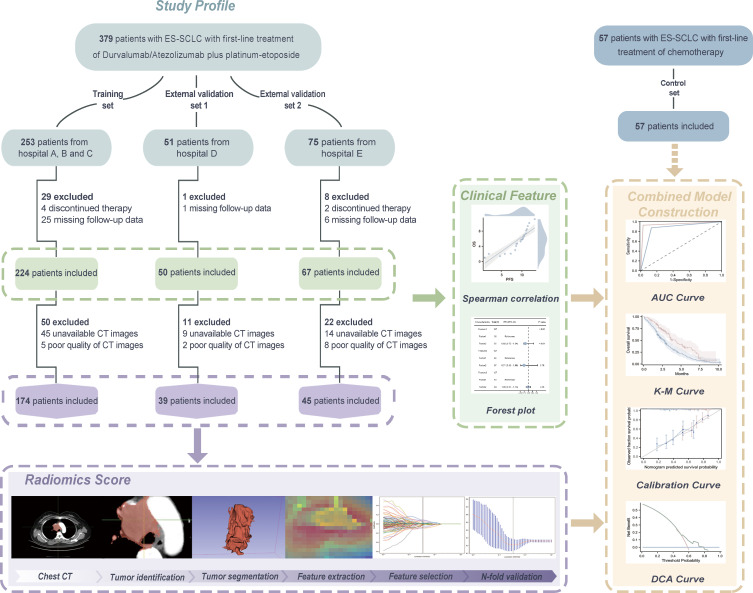
Figure 1 illustrates the study flow. In total, 379 patients in the chemoimmunotherapy group were screened, including 253, in the training set, and 51 and 75 in the external validation sets 1 and 2, respectively. After excluding 38 patients with missing follow-up information and 6 who discontinued treatment, 341 were included in the study. Table 1 presents the baseline characteristics of the participants. Patients were predominantly male (307/341, 90%) and had a mean age of 62±8.7 years, history of smoking (264/341, 77%), stage IV cancer (291/341, 85%), and Eastern Cooperative Oncology Group (ECOG) Score <2 (316/341, 93%). At baseline, 66 (19%) patients had brain metastases and 69 (20%) had liver metastases. Furthermore, 29% (99/341)and 71% (242/341)of patients were treated with durvalumab and atezolizumab, respectively, and the prognosis did not differ between the groups, except for a significantly higher prevalence of immune-related adverse events (irAEs) observed in patients who received durvalumab.
Figure 1.
Workflow of the proposed integrated predictive model and study design. The patients with ES-SCLC treated with combined atezolizumab or durvalumab with standard chemotherapy are included and one training cohort and two independent external validation cohorts are created. Cox regression is employed to screen for therapeutic efficacy associated clinical features. Radiomics Score is constructed using machine learning from the radiomics features extracted in the regions of interest delineated on the chest CT before treatment. Clinical features are further combined with the radiomics score to devise the integrated model, whose outputs create the treatment outcome prediction. AUC, area under the curve; DCA, decision curve analysis; ES-SCLC, extensive-stage small cell lung cancer; K-M, Kaplan-Meier.
Table 1.
Baseline characteristics of the patients
| Characteristics | Chemoimmunotherapy group, N (%) (n=341) |
Radiomics analysis in the chemoimmunotherapy group, N (%) | Chemotherapy group, N (%) (n=57) |
||
| Training set (n=174) |
Validation set 1 (n=39) | Validation set 2 (n=45) |
|||
| Age, mean (SD), years | 62.0 (8.7) | 62.2 (9.0) | 62.3 (8.8) | 62.6 (8.5) | 62.0 (8.4) |
| Age group, years | |||||
| <65 | 194 (57) | 93 (53) | 19 (49) | 27 (60) | 34 (60) |
| ≥65 | 147 (43) | 81 (47) | 20 (51) | 18 (40) | 23 (40) |
| Sex | |||||
| Male | 307 (90) | 158 (91) | 34 (87) | 40 (89) | 53 (93) |
| Female | 34 (10) | 16 (9) | 5 (13) | 5 (11) | 4 (7) |
| Smoking status | |||||
| Smoker | 264 (77) | 149 (86) | 28 (72) | 23 (51) | 51 (89) |
| Non-smoker | 77 (23) | 25 (14) | 11 (28) | 22 49) | 6 (11) |
| ECOG | |||||
| 0–1 | 316 (93) | 158 (91) | 37 (95) | 42 (93) | 51 (89) |
| 2 | 25 (7) | 16 (9) | 2 (5) | 3 (7) | 6 (11) |
| Disease stage at diagnosis* | |||||
| III | 50 (15) | 29 (17) | 0 (0) | 10 (22) | 14 (25) |
| IV | 291 (85) | 145 (83) | 39 (100) | 35 (78) | 43 (75) |
| Brain metastases | |||||
| Yes | 66 (19) | 33 (19) | 8 (21) | 7 (16) | 10 (18) |
| No | 275 (81) | 141 (81) | 31 (79) | 38(84) | 47 (82) |
| Liver metastases | |||||
| Yes | 69 (20) | 37 (21) | 9 (23) | 8 (18) | 13 (23) |
| No | 272 (80) | 137 (79) | 30 (77) | 37 (82) | 44 (77) |
| LIPI | |||||
| Good | 117 (34) | 59 (34) | 8 (21) | 21 (47) | 16 (28) |
| Intermediate | 143 (42) | 79 (45) | 11 (28) | 15 (33) | 27 (47) |
| Poor | 81 (24) | 36 (21) | 20 (51) | 9 (20) | 14 (25) |
| Drug | |||||
| Durvalumab | 99 (29) | 81 (47) | 3 (8) | 0 (0) | / |
| Atezolizumab | 242 (71) | 93 (53) | 36 (92) | 45 (100) | / |
| Best objective response† | |||||
| CR+PR | 264 (77) | 122 (70) | 33 (84) | 40 (89) | 42 (74) |
| Stable disease | 68 (20) | 46 (26) | 3 (8) | 5 (11) | 9 (16) |
| PD | 9 (3) | 6 (4) | 3 (8) | 0 (0) | 6 (10) |
| irAE | |||||
| Yes | 68 (20) | 39 (22) | 6 (15) | 6 (13) | / |
| No | 273 (80) | 135 (78) | 33 (85) | 39 (87) | / |
| Radiotherapy | |||||
| Chest | 99 (29) | 55 (32) | 10 (26) | 9 (20) | 10 (16) |
| Brain | 63 (18) | 34 (20) | 10 (26) | 5 (11) | 13 (23) |
*Disease stage at diagnosis classified according to American Joint Committee on Cancer, eighth edition, tumor, node, metastasis (TNM) staging system.
†Best objective response was evaluated according to Response Evaluation Criteria in Solid Tumors, V.1.1.
CR, complete response; ECOG, Eastern Cooperative Oncology Group; irAE, immune-related adverse event; LIPI, Lung Immune Prognostic Index; PD, progressive disease; PR, partial response.
Clinical features and treatment outcomes
Over a median follow-up duration of 12.1 months, the mPFS was 7.1 (95% CI 6.6 to 7.7) months, 87 (26%) patients died and the median OS (mOS) was not reached. The 6-month, 9-month, and 12 month PFS rates were 60.7%, 38.2%, and 28.3%, respectively. The 12-month and 18-month OS rates were 75.0% and 58.5%, respectively (table 2). Notably, most patients (264/341, 77%) achieved disease control after receiving chemoimmunotherapy, 1 patient achieved a complete response (CR) and 68 (20%) remained stable, whereas 9 patients (3%) experienced progressive disease (PD) after treatment. The objective response rate (ORR) and disease control rate were 77% and 97%, respectively. Among the 225 patients who have experienced disease progression after treatment, 60% (135/225) suffered disease progression at the primary site, 32% (73/225) developed distant metastasis, and 2% (4/225) demonstrated both types of progression. In total, 68 patients (20%) experienced irAEs, including 20 (6%) with grade ≥3, such as immune-related pneumonia and hepatic injury. During the treatment course, 99 of 341 (29%) patients received chest radiotherapy and 63 (18%) patients received brain radiotherapy (table 1). The K-M curve clearly showed that radiotherapy had minimal effect on PFS, but significantly prolonged mOS (NR vs 17.2 m, 95% CI 14.2-NR, HR=0.48, p=0.001, online supplemental figure S1).
Table 2.
Summary of treatment response
| Chemoimmunotherapy group (n=341) | Radiomics analysis in chemoimmunotherapy group | Chemotherapy group (n=57) | |||
| Training set (n=174) |
Validation set 1 (n=39) | Validation set 2 (n=45) | |||
| ORR | 77.4% | 70.1% | 84.6% | 88.9% | 73.7% |
| DCR | 97.4% | 96.6% | 92.3% | 100.0% | 89.5% |
| Death | 25.5% | 26.4% | 43.6% | 4.4% | 42.1% |
| Follow-up time, median (95% CI), months |
12.1 (11.0 to 13.2) | 12.4 (11.1 to 13.7) | 15.3 (13.6 to 16.9) | 7.1 (6.0 to 8.2) | 11.8 (8.9 to 14.7) |
| PFS, median (95% CI), months | 7.1 (6.6 to 7.7) | 6.9 (6.2 to 7.7) | 6.9 (4.7 to 9.2) | 8.1 (6.3 to 9.9) | 5.9 (5.1 to 6.7) |
| OS, median (95% CI), months | NR | NR | 19.1 (9.9 to 28.3) | NR | 13.7 (8.9 to 18.5) |
| 6-month PFS (%) | 60.7% | 60.3% | 53.8% | 66.1% | 45.5% |
| 9-month PFS (%) | 38.2% | 36.0% | 41.0% | 44.4% | 13.0% |
| 12-month PFS (%) | 28.3% | 25.6% | 38.3% | 22.4% | 4.3% |
| 12-month OS (%) | 75.0% | 73.2% | 62.6% | 86.4% | 59.1% |
| 18-month OS (%) | 58.5% | 60.7% | 51.6% | 86.4% | 30.8% |
DCR, disease control rate; ORR, objective response rate; OS, overall survival; PFS, progression-free survival.
jitc-2023-007492supp002.pdf (89.5KB, pdf)
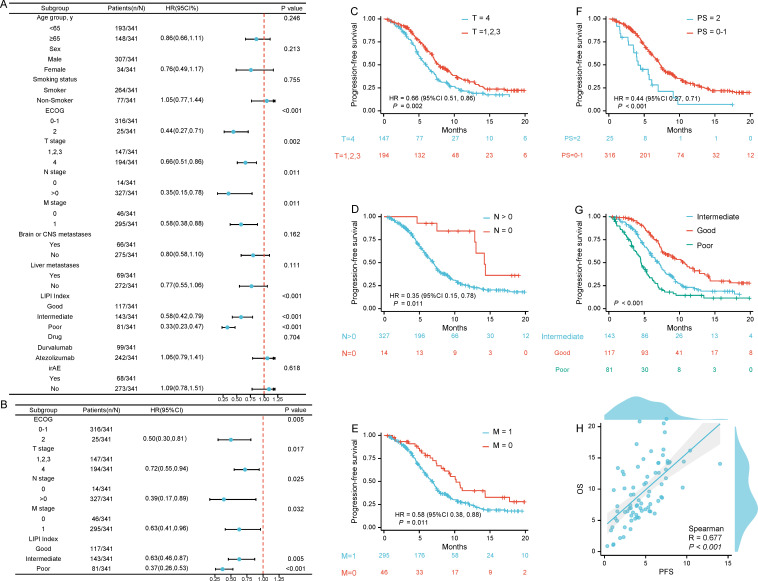
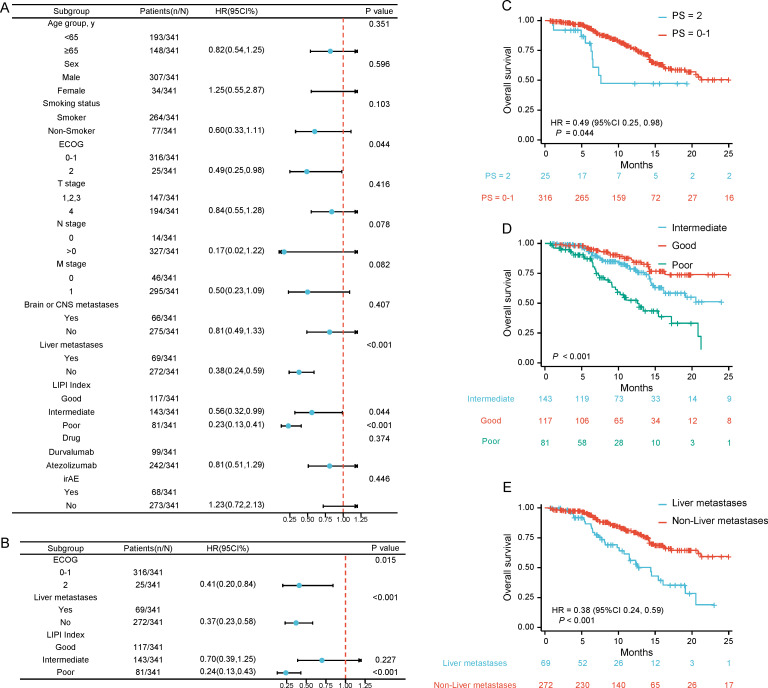
Univariate Cox regression analyses were conducted to investigate the association between clinical features and PFS. ECOG performance status, Lung Immune Prognostic Index (LIPI), T stages, N stages, and M stages significantly impacted the outcome (figure 2). All of the aforementioned were statistically significant in multivariate Cox regression analyses (p<0.05). The K-M curves in figure 2 illustrate the impacts of clinical features on PFS. Of note, Cox regression analyses indicated that ECOG performance status, liver metastasis, and LIPI were significantly associated with survival outcomes (p<0.05; figure 3). The K-M curves in figure 3 illustrate the impacts of clinical features on OS. Furthermore, Spearman’s correlation analyses indicated a significant correlation between PFS and OS (R=0.677, p<0.01) (figure 2H).
Figure 2.
Clinical features associated with PFS. Forest plot for PFS of (A) Univariate Cox proportional hazard model of clinical variables and (B) Multivariate Cox proportional hazard model of clinical variables; (C-G) The K-M curve of PFS-associated clinical variables: (C) T stage; (D) N stage; (E) M stage; (F) ECOG and (G) LIPI; (H) Spearman’s correlation of PFS and OS. CNS, central nervous system; PFS, progression-free survival; K-M, Kaplan-Meier; OS, overall survival; ECOG, Eastern Cooperative Oncology Group; PS, performance status; LIPI, Lung Immune Prognostic Index; irAE, immune-related adverse event.
Figure 3.
Clinical features associated with overall survival. (A–C) Forest plot for OS of (A) Univariate Cox proportional hazard model of clinical variables and (B) Multivariate Cox proportional hazard model of clinical variables; (C–E) The K-M curve of OS associated clinical variables: (C) ECOG, (D) LIPI and (E) Liver metastases. CNS, central nervous system; K-M, Kaplan-Meier; OS, overall survival; ECOG, Eastern Cooperative Oncology Group; PS, performance status; LIPI, Lung Immune Prognostic Index; irAE, immune-related adverse event.
Construction of the radiomics prediction model
After excluding patients with missing imaging data or inadequate image quality, we analyzed imaging omics of 258 patients from three cohorts. The cohorts included 174 patients in the training set and 39 and 45 patients in the external validation sets 1 and 2, respectively. Tables 1 and 2 present the detailed clinical features of each group.
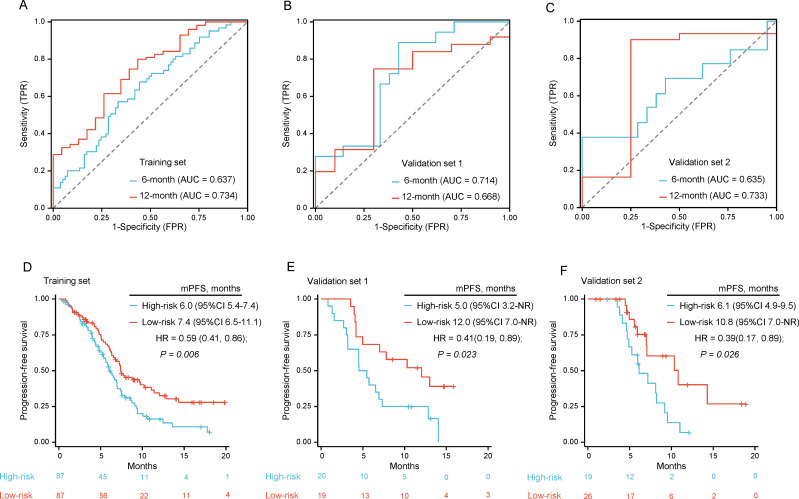
For radiomics analysis, we extracted 1218 radiomics features from the region of interest (ROI) of CT images of the lung. LASSO-Cox regression was performed for the training set using PFS as the outcome variable, which identified eight radiomics features with non-zero coefficients. We multiplied the weight of feature coefficients by the sum of feature values to calculate the radio-score (online supplemental file 1). We divided patients into low-progression-risk and high-progression-risk groups using the median cut-off value of the training set. The time-dependent ROC curve and the C-index were used to evaluate the efficiency of the model, with 6 months and 12 months as the time nodes. The training set demonstrated an area under the curve (AUC) of 0.637 and 0.734 at 6 months and 12 months, respectively, and a C-index of 0.585. The external validation set 1 demonstrated an AUC of 0.714 and 0.668 at 6 months and 12 months, respectively, and a C-index of 0.634. The external validation set 2 demonstrated an AUC of 0.635 and 0.733 at 6 months and 12 months, and a C-index of 0.617 (table 3). The K-M curves for low-progression-risk and high-progression-risk were significantly separated, with an mPFS of 7.4 (95% CI 6.5 to 11.1) months vs 6.0 (95% CI 5.4 to 7.4) months, respectively, in the training set (HR=0.59, 95% CI 0.41 to 0.86, p=0.006). The classification method was applied to external validation sets 1 and 2, and the results showed an mPFS of 12.0 (95% CI 7.0-NR) months vs 5.0 (95% CI 3.2-NR) months (HR=0.41, 95% CI 0.19 to 0.89, p=0.023) and 10.8 (95% CI 7.0-NR) months vs 6.1 (95% CI 4.9 to 9.5) months (HR=0.39, 95% CI 0.17 to 0.89, p=0.026), respectively. The K-M curves illustrating these findings are shown in figure 4.
Table 3.
Performance of different models in predicting PFS
| 6 month AUC (95% CI) |
12 month AUC (95% CI) |
C-index (95% CI) |
|
| Radiomics model | |||
| Training set (n=174) | 0.637 (0.548 to 0.727) | 0.734 (0.623 to 0.845) | 0.585 (0.527 to 0.643) |
| Validation set 1 (n=39) | 0.714 (0.551 to 0.877) | 0.668 (0.466 to 0.870) | 0.634 (0.540 to 0.728) |
| Validation set 2 (n=45) | 0.635 (0.427 to 0.844) | 0.733 (0.397 to 1.000) | 0.617 (0.468 to 0.766) |
| Control set (n=57) | 0.461 (0.297 to 0.624) | NA | 0.534 (0.440 to 0.628) |
| Integrated model | |||
| Training set (n=174) | 0.744 (0.663 to 0.825) | 0.836 (0.753 to 0.919) | 0.699 (0.651 to 0.748) |
| Validation set 1 (n=39) | 0.820 (0.689 to 0.952) | 0.726 (0.525 to 0.928) | 0.688 (0.590 to 0.785) |
| Validation set 2 (n=45) | 0.852 (0.706 to 0.998) | 0.751 (0.372 to 1.000) | 0.820 (0.721 to 0.918) |
| Control set (n=57) | 0.394 (0.233 to 0.555) | NA | 0.556 (0.456 to 0.656) |
AUC, area under the curve; C-index, Concordance Index; NA, Not applicable; PFS, progression-free survival.
Figure 4.
Machine learning based Radiomics model performance. (A–C) The ROC curve of the Radiomics prediction model in (A) training set, (B) validation set 1 and (C) validation set 2; (D–F) K-M curves of PFS in (D) training set, (E) validation set 1 and (F) validation set 2 stratified as low-progression-risk and high-progression-risk using the Radiomics model. AUC, area under the curve; FPR, false positive rate; K-M, Kaplan-Meier; mPFS, median PFS; PFS, progression-free survival; TPR, true positive rate; ROC, receiver operating characteristic.
jitc-2023-007492supp001.pdf (40.7KB, pdf)
Establishment of the integrated prediction model
Although the ML-based Radiomics model can be used as an independent predictor for PFS, its accuracy and efficacy need further improvement. According to results of Spearman’s correlation analysis between radio-score and clinical features, we found there was no correlation between radio-score and clinical features except LIPI (R=0.691, p<0.01) (online supplemental figure S2). Further collinearity analysis showed VIFs of LIPI and radio-score were 1.789 and 3.709, respectively, indicating the absence of collinearity in between (online supplemental table S1). Therefore, we integrated all the PFS-related clinical features related to the Radiomics model to develop a combined prediction model.
jitc-2023-007492supp003.pdf (324.6KB, pdf)
jitc-2023-007492supp005.pdf (115.3KB, pdf)
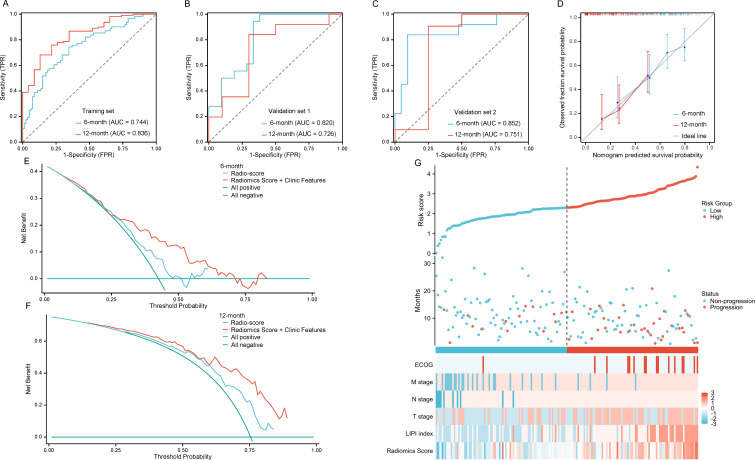
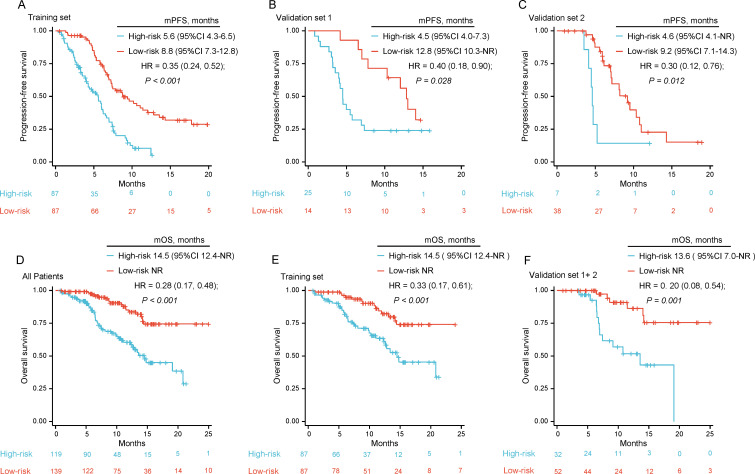
We used Cox regression to model the selected clinical features and radio-scores in the training set. The regression coefficient was used to calculate the comprehensive prediction model score. The median cut-off value was used to divide the low-progression-risk and high-progression-risk groups. Compared with the simple Radiomics model, the AUC at 6 months and 12 months and C-index of the training and validation sets were significantly improved in the combined prediction model (training set: 0.744, 0.836, and 0.699; external validation set 1: 0.820, 0.726, and 0.688; external validation set 2: 0.852, 0.751, and 0.820) (table 3). The calibration curve, DCA plots, and risk factor diagram are shown in figure 5. The DCA plot suggests our prediction model can obtain satisfactory clinical benefits, and the integrated model has a higher net benefit level than the sole Radiomics model. We can also find intuitively that the high- progression-risk group had a higher rate of disease progression from risk factor diagram, compared with the low-progression-risk group. Furthermore, the integrated model significantly increased the degree of separation between the two curves in the low-progression-risk and high-progression-risk groups (figure 6), as well as substantially improved the HR, resulting in a mPFS of 8.8 (95% CI 7.3 to 12.8) months vs 5.6 (95% CI 4.3 to 6.5) months (HR=0.35, 95% CI 0.24 to 0.52, p<0.001) in the training set and 12.8 (95% CI 10.3-NR) months vs 4.5 (95% CI 4.0 to 7.3) months (HR=0.40, 95% CI 0.18 to 0.90, p=0.028) and 9.2 (95% CI 7.1 to 14.3) months vs 4.6 (95% CI 4.1-NR) months (HR=0.30, 95% CI 0.12 to 0.76, p=0.012) in the external validation sets 1 and 2, respectively. These findings indicate that the integrated prediction model dramatically improved the prediction efficiency compared with the sole radiomics model.
Figure 5.
Model performance of the integrated model. (A–C) The ROC curve of the combined prediction model in (A) training set, (B) validation set 1 and (C) validation set 2; (D) The calibration curve of the combined prediction model at 6 months and 12 months in predicting PFS. (E–F) DCA curve of the Radiomics model and combined model at 6 months (E) and 12 months (F) in predicting PFS; (G) Risk factor diagram of the combined model in predicting PFS. AUC, area under the curve; TPR, true positive rate; FPR, false positive rate; DCA, decision curve analysis; ECOG, Eastern Cooperative Oncology Group; LIPI, Lung Immune Prognostic Index; PFS, progression-free survival; ROC, receiver operating characteristic.
Figure 6.
Kaplan-Meier analysis in the cohorts. (A–C) K-M curves of PFS in (A) the training set, (B) validation set 1 and (C) validation set 2 stratified as low-progression-risk and high-progression-risk using the integrated model. (D–F) K-M curves of OS in (D) all patients, (E) training set and (F) validation set stratified as low-progression-risk and high-progression-risk using the integrated model. K-M, Kaplan-Meier; mOS, median OS; mPFS, median PFS; OS, overall survival; PFS, progression-free survival.
Of importance, we plotted the OS K-M curve of low-progression-risk and high-progression-risk groups (n=139 and n=119, respectively) predicted by our integrated model (figure 6). There were significant differences in OS between the two groups for the entire population (NR vs 14.5 (95% CI 12.4-NR) months, HR=0.28, 95% CI 0.17 to 0.48), p<0.001. The median OS for the low-progression-risk and high-progression-risk groups in the training set was NR vs 14.5 (95% CI 12.4-NR) months, and HR=0.33, 95% CI 0.17 to 0.61, p<0.001. For the combined external validation sets 1 and 2, the mOS was NR vs 13.6 months (95% CI 7.0-NR), HR=0.20, 95% CI 0.08 to 0.54, p=0.001. In addition, the 12-month OS rates in the low-progression-risk and high-progression-risk groups were 82.3% vs 63.5% in the training set, and 86.4% vs 51.8% in the validation sets 1 plus 2. The 18-month OS rates in the low-progression-risk and high-progression-risk groups were 74.1% vs 45.4% in the training set, and 75.6% vs 43.2% in the validation sets 1 plus 2. Again, these findings demonstrate that our comprehensive model can predict the risk of disease progression and survival outcomes, allowing for the precise identification of patients with ES-SCLC who will benefit most from chemoimmunotherapy.
Performance of the integrated model for the control group
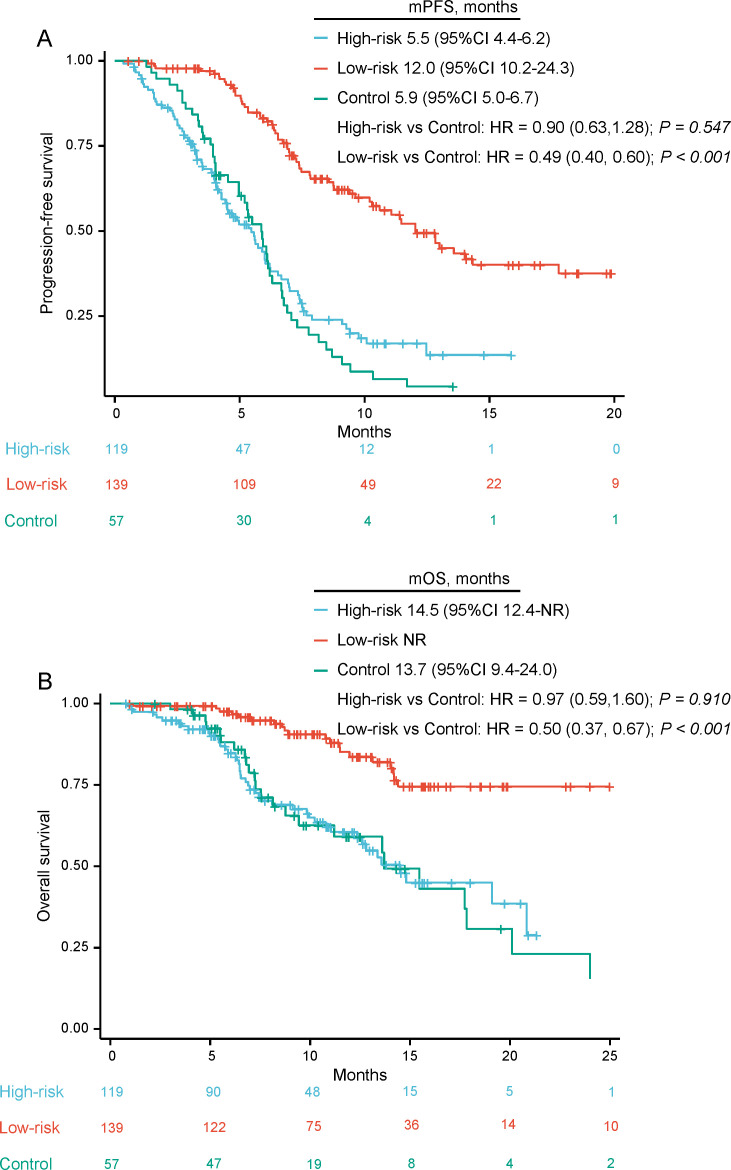
The demographic features of patients in the chemotherapy cohort are shown in table 1. Over a median follow-up time of 11.8 months, the mPFS was 5.9 (95% CI 5.1 to 6.7) months, and mOS was 13.7 (95% CI 8.9 to 18.5) months. No statistically significant differences were observed in PFS or OS between high-progression-risk patients receiving chemoimmunotherapy and the chemotherapy cohort (mPFS, 5.5 (95% CI 4.4 to 6.2) months vs 5.9 (95% CI 5.0 to 6.7) months, HR=0.90, 95% CI 0.63 to 1.28, p=0.547; mOS, 14.5 (95% CI 12.4-NR) months vs 13.7 (95% CI 9.4 to 24.0) months, HR=0.97, 95% CI 0.59 to 1.60, p=0.910) (figure 7). On the other hand, we tried to apply clinical and radiomics information of the chemotherapy cohort into the integrated model. The 6-month AUC and C-index were only 0.394 and 0.556, respectively. The PFS and OS K-M curves of low-progression-risk and high-progression-risk groups indicated that the integrated model could not predict the prognosis in the chemotherapy group (online supplemental figure S3).
Figure 7.
Kaplan-Meier curves: (A) PFS, (B) OS, of patients with low- progression- risk and high- progression- risk who received first-line chemoimmunotherapy and patients who received first-line chemotherapy. mOS, median OS; mPFS, median PFS; OS, overall survival; PFS: progression-free survival.
jitc-2023-007492supp004.pdf (204.1KB, pdf)
Discussion
This is the largest multicenter retrospective analysis to date and is the first to employ ML to generate a convincing predictive marker for the treatment outcomes in patients with ES-SCLC. Our findings demonstrate that radiomics can be an independent prognostic predictor for patients treated with chemoimmunotherapy, regardless of other clinical or molecular biomarkers. The addition of clinical features markedly further improved the predictive efficacy of the model. Most importantly, the eventual survival outcome was significantly different between the low-progression-risk and high-progression-risk groups. The risk for disease progression was reduced by ~65% and the risk for death was reduced by ~70% in the low-progression-risk group compared with high-progression-risk group.
Chemoimmunotherapy is the recommended first-line treatment for ES-SCLC. Our retrospective study demonstrated an mPFS of 7.1 months, ORR of 77%, and irAE rate of 20%. The therapeutic efficacy was not correlated with the prevalence of irAEs, drug agents, or the presence of pure baseline brain metastases. These findings are in general consistent with the randomized control studies, such as IMpower133 and CASPIAN, as well as the recent emerging CAPSTONE-1 and ASTRUM-005,5 6 19 20 except for a relatively longer OS, which could partially be explained by the contributions of radiotherapy.21 We did observe that radiotherapy played a crucial role in the improvement of survival outcomes of the ES-SCLC group patients, which warrants validation by the randomized trials in future. LIPI was an independent prognostic predictor, which is in line with previous observations in both limited-stage SCLC and patients with ES-SCLC.22 23 Our proposed ML Radiomics model, to some extent, is capable of identifying the beneficial population with ES-SCLC, achieving 6-month and 12 month AUCs of 0.635–0.733 in the validation sets. Furthermore, by integrating the clinical features with the radio-score, we devised the more valid and accurate ML-based predictive model. The patients were divided into low-progression-risk and high-progression-risk groups by the integrated classifier. The K-M curves for both PFS and OS demonstrated a statistically significant separation beginning at an early stage and persisting until the final observation date. The AUC and C-index for the training set and external validation sets at 6 months and 1 year all approach or even exceed 0.8. The OS HRs for OS (0.2–0.3) were clinically sound.
SCLC, a highly heterogeneous and immunologically ‘cold’ tumor, is characterized by the downregulation of histocompatibility complex class I and reduced numbers of natural killer (NK) cells and CD8+T lymphocytes in the tumor microenvironment.24–27 A series of biomarkers, such as PD-L1 status and tumor mutation burden, have failed to demonstrate an acceptable predictive ability.28 Gay et al10classified SCLC into SCLC-A, N, P, and I subtypes based on the expression of the transcription factors of ASCL1, NEUROD1, and POU2F3. They found SCLC-I tumors derive greater survival benefit from immune checkpoint blockade. However, notably, the OS HRs derived from our model are markedly greater than the ones observed in SCLC-I. Moreover, the assessment of the aforementioned biomarkers often requires invasive procedures and is complex and expensive. Also, the heterogeneity of SCLC poses challenges for biomarker testing using small specimens. Hence, these characteristics make the use of current biomarkers challenging in the clinical setting.
Our ML-based integrated prognostic model, using the information from chest CT and clinical characters, is economic and non-invasive, and could accurately identify patients with ES-SCLC who would well respond to immunotherapy. Notably, to validate the generalizability of the proposed model, we trained the model using the combined data from three hospitals and tested it using two databases from two other hospitals. Our model performed well in these different populations. On the other hand, unlike the commonly used DL model with controversial clinical application due to the relatively poor interpretation, transparency, and repeatability, our ML model based on ROI segmentation had high interpretability, stability, and accuracy. Of note, the PFS K-M curve of the high-progression-risk group closely overlapped with that of the control group, the ones who received first-line chemotherapy. In concert, not only mPFS, but the kinetics of the PFS K-M curve of the high-progression-risk group were quite comparable to the mPFS observed in the control groups (who received chemotherapy) from the IMpower133 and CASPIAN Trials.5 6 These observations suggest that the patients with a high progression risk judged by our integrated model, highly mimic the responsiveness of the ones who received chemotherapy alone. Thus, we believe our model does pick out the subgroup of patients who would benefit from immunotherapy.
There were some limitations in our study. First, the OS data of our chemoimmunotherapy group were immature; second, a C-index of 0.6 suggests that the sole radiomics model did play a role, but was not the dominant determinant for PFS prediction. However, we will continue to follow-up these patients and develop novel algorithms, and we believe an updated and clinically applicable model will be launched in the near future.
Collectively, this retrospective study of patients with ES-SCLC included 341 patients from multicenters and established two external validation cohorts to exclude the potential variations between the institutions due to different CT equipment and parameter settings, thereby ensuring the the stability and generalizability of our results. We developed an economic, non-invasive, valid, and effective model for predicting the therapeutic responsiveness to chemoimmunotherapy and the prognosis of patients with ES-SCLC, providing a convenient and cost-effective tool for the management of ES-SCLC. With low input requirements and strong operability, our innovative strategy will have significant clinical significance and profound impacts.
Footnotes
Twitter: @yxia
JZ, YH, XY, PT, LZ and KH contributed equally.
Contributors: Conceptualization: YX, WL, YG, YZha; Investigation: JieZha, YH, XY, PT, LZ, KH, JinZha, JZho, QW, MC; Data curation: LZ, KH, JieZha, JinZha, QW, MC, YZhu; Writing of the original draft: JieZha, YX; Writing of the review and editing: YH, XY, PT, LZ, KH, WL, YG, YZha; Supervision: WL, YG, YX; Funding acquisition: YX. YX is responsible for the overall content as guarantor.
Funding: This study was funded by the Natural Science Foundation of Zhejiang Province (LTGY23H010004)
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. Individual patient data are available on request addressed to the corresponding author. After approval of the proposal, the data can be shared within a secure online platform. A data-sharing agreement will be needed.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by the ethics committees of the Second Affiliated Hospital of Zhejiang University School of Medicine (2022-0237) and the joint hospitals where applicable. Due to the respective nature of our study, the ethics committee agreed with the waiver of informed consent.
References
- 1.Bernhardt EB, Jalal SI. Small cell lung cancer. Cancer Treat Res 2016;170:301–22. 10.1007/978-3-319-40389-2_14 [DOI] [PubMed] [Google Scholar]
- 2.Nicholson AG, Chansky K, Crowley J, et al. The International Association for the study of lung cancer lung cancer staging project: proposals for the revision of the clinical and pathologic staging of small cell lung cancer in the forthcoming eighth edition of the TNM classification for lung cancer. J Thorac Oncol 2016;11:300–11. 10.1016/j.jtho.2015.10.008 [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Zimmermann S, Parikh K, et al. Current diagnosis and management of small-cell lung cancer. Mayo Clinic Proceedings 2019;94:1599–622. 10.1016/j.mayocp.2019.01.034 [DOI] [PubMed] [Google Scholar]
- 4.Amarasena IU, Chatterjee S, Walters JAE, et al. Platinum versus non-platinum chemotherapy regimens for small cell lung cancer. Cochrane Database Syst Rev 2015;2015:CD006849. 10.1002/14651858.CD006849.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paz-Ares L, Dvorkin M, Chen Y, et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 2019;394:1929–39. 10.1016/S0140-6736(19)32222-6 [DOI] [PubMed] [Google Scholar]
- 6.Horn L, Mansfield AS, Szczęsna A, et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med 2018;379:2220–9. 10.1056/NEJMoa1809064 [DOI] [PubMed] [Google Scholar]
- 7.Longo V, Catino A, Montrone M, et al. What are the biomarkers for Immunotherapy in SCLC. Int J Mol Sci 2021;22:11123. 10.3390/ijms222011123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iams WT, Porter J, Horn L. Immunotherapeutic approaches for small-cell lung cancer. Nat Rev Clin Oncol 2020;17:300–12. 10.1038/s41571-019-0316-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, Girard L, Zhang Y-A, et al. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl Lung Cancer Res 2018;7:32–49. 10.21037/tlcr.2018.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gay CM, Stewart CA, Park EM, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 2021;39:346–360. 10.1016/j.ccell.2020.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu S, Fetsch P, Thomas A, et al. Molecular subtypes of primary SCLC tumors and their associations with neuroendocrine and therapeutic markers. J Thorac Oncol 2022;17:141–53. 10.1016/j.jtho.2021.08.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang WZ, Shulman A, Amann JM, et al. Small cell lung cancer: subtypes and therapeutic implications. Semin Cancer Biol 2022;86:543–54. 10.1016/j.semcancer.2022.04.001 [DOI] [PubMed] [Google Scholar]
- 13.Stewart CA, Gay CM, Xi Y, et al. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat Cancer 2020;1:423–36. 10.1038/s43018-019-0020-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim JS, Ibaseta A, Fischer MM, et al. Intratumoural heterogeneity generated by notch signalling promotes small-cell lung cancer. Nature 2017;545:360–4. 10.1038/nature22323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018;19:1180–91. 10.1016/S1470-2045(18)30413-3 [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Yu H, Gan Y, et al. Mining whole-lung information by artificial intelligence for predicting EGFR genotype and targeted therapy response in lung cancer: a multicohort study. Lancet Digit Health 2022;4:e309–19. 10.1016/S2589-7500(22)00024-3 [DOI] [PubMed] [Google Scholar]
- 17.Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018;24:1559–67. 10.1038/s41591-018-0177-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaidya P, Bera K, Patil PD, et al. Novel, non-invasive imaging approach to identify patients with advanced non-small cell lung cancer at risk of hyperprogressive disease with immune checkpoint blockade. J Immunother Cancer 2020;8:e001343. 10.1136/jitc-2020-001343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Zhou C, Yao W, et al. Adebrelimab or placebo plus carboplatin and etoposide as first-line treatment for extensive-stage small-cell lung cancer (CAPSTONE-1): a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2022;23:739–47. 10.1016/S1470-2045(22)00224-8 [DOI] [PubMed] [Google Scholar]
- 20.Cheng Y, Han L, Wu L, et al. Effect of first-line serplulimab vs placebo added to chemotherapy on survival in patients with extensive-stage small cell lung cancer: the ASTRUM-005 randomized clinical trial. JAMA 2022;328:1223–32. 10.1001/jama.2022.16464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian Y, Ma J, Jing X, et al. Radiation therapy for extensive-stage small-cell lung cancer in the era of immunotherapy. Cancer Lett 2022;541:215719. 10.1016/j.canlet.2022.215719 [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Ai X, Xu H, et al. Treatment patterns and outcomes of immunotherapy in extensive-stage small-cell lung cancer based on real-world practice. Thorac Cancer 2022;13:3295–303. 10.1111/1759-7714.14684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun B, Hou Q, Liang Y, et al. Prognostic ability of lung immune prognostic index in limited-stage small cell lung cancer. BMC Cancer 2022;22:1233. 10.1186/s12885-022-10351-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bern MD, Parikh BA, Yang L, et al. Inducible down-regulation of MHC class I results in natural killer cell tolerance. J Exp Med 2019;216:99–116. 10.1084/jem.20181076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Best SA, Hess JB, Souza-Fonseca-Guimaraes F, et al. Harnessing natural killer immunity in metastatic SCLC. J Thorac Oncol 2020;15:1507–21. 10.1016/j.jtho.2020.05.008 [DOI] [PubMed] [Google Scholar]
- 26.Doyle A, Martin WJ, Funa K, et al. Markedly decreased expression of class I histocompatibility antigens, protein, and mRNA in human small-cell lung cancer. J Exp Med 1985;161:1135–51. 10.1084/jem.161.5.1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen T, Rodriguez BL, Chen L, et al. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov 2019;9:646–61. 10.1158/2159-8290.CD-18-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu SV, Reck M, Mansfield AS, et al. Updated overall survival and PD-L1 subgroup analysis of patients with extensive-stage small-cell lung cancer treated with atezolizumab, carboplatin, and etoposide (Impower133). J Clin Oncol 2021;39:619–30. 10.1200/JCO.20.01055 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007492supp002.pdf (89.5KB, pdf)
jitc-2023-007492supp001.pdf (40.7KB, pdf)
jitc-2023-007492supp003.pdf (324.6KB, pdf)
jitc-2023-007492supp005.pdf (115.3KB, pdf)
jitc-2023-007492supp004.pdf (204.1KB, pdf)
Data Availability Statement
Data are available upon reasonable request. Individual patient data are available on request addressed to the corresponding author. After approval of the proposal, the data can be shared within a secure online platform. A data-sharing agreement will be needed.