Abstract
Objective
Assessment of circulating autoantibodies represents one of the earliest diagnostic procedures in patients with suspected connective tissue disease (CTD), providing important information for disease diagnosis, identification and prediction of potential clinical manifestations. The purpose of this study was to evaluate the ability of multiparametric assay to correctly classify patients with multiple CTDs and healthy controls (HC), independent of clinical features, and to evaluate whether serological status could identify clusters of patients with similar clinical features.
Methods
Patients with systemic lupus erythematosus (SLE), systemic sclerosis (SSc), Sjogren’s syndrome (SjS), undifferentiated connective tissue disease (UCTD), idiopathic inflammatory myopathies (IIM) and HC were enrolled. Serum was tested for 29 autoantibodies. An XGBoost model, exclusively based on autoantibody titres was built and classification accuracy was evaluated. A hierarchical clustering model was subsequently developed and clinical/laboratory features compared among clusters.
Results
908 subjects were enrolled. The classification model showed a mean accuracy of 60.84±4.05% and a mean area under the receiver operator characteristic curve of 88.99±2.50%, with significant discrepancies among groups. Cluster analysis identified four clusters (CL). CL1 included patients with typical features of SLE. CL2 included most patients with SjS, along with some SLE and UCTD patients with SjS-like features. CL4 included anti-Jo1 patients only. CL3 was the largest and most heterogeneous, including all the remaining subjects, overall characterised by low titre or lower-prevalence autoantibodies.
Conclusion
Extended multiparametric autoantibody assay allowed an accurate classification of CTD patients, independently of clinical features. Clustering according to autoantibody titres is able to identify clusters of CTD subjects with similar clinical features, independently of their final diagnosis.
Keywords: autoimmune diseases; lupus erythematosus, systemic; scleroderma, systemic; Sjogren's syndrome
WHAT IS ALREADY KNOWN ON THIS TOPIC
Fully automated multiparametric assays allow the simultaneous detection of multiple autoantigens with high accuracy, potentially providing significant data for patient diagnosis and management.
WHAT THIS STUDY ADDS
The simultaneous quantitative detection of a large number of autoantibodies by a novel digital technique provided informative data for connective tissue disease (CTD) classification and identification of clinically meaningful patients clusters.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
The results support the importance of multiparametric autoantibody detection as a clinical tool to be employed early in the diagnostic work-up. Serological status identifies clusters of CTD patients with similar features, independently of final diagnosis, supporting an approach based on clinical features rather than formal disease classification.
Introduction
Despite the constant increase in technologically advanced research, the diagnosis of systemic connective tissue diseases (CTDs) still represents an undeniable challenge, mainly due to their protean and overlapping clinical manifestations, especially at disease onset. In clinical practice, the diagnosis of patients with CTDs relies on the identification of compatible clinical features in the context of autoimmunity (ie, at least positive antinuclear antibodies (ANA)).1 2 In contrast, the identification of a specific CTD is guided by classification criteria that have been developed for research purposes and, although not directly applicable as diagnostic criteria, they include the most significant features of specific CTDs. This approach does not consider some peculiar aspects of CTDs, such as the wide overlap of multiple clinical features (eg, arthritis, skin rashes, Raynaud’s phenomenon, etc). Additionally, the presence of non-specific features, in the absence of other peculiar clinical or serological elements, leads to the definition of undifferentiated connective tissue disease (UCTD), which is a very heterogeneous condition, including patients with virtually any feature attributable to CTDs.3–5
Nonetheless, although these classification aspects are essential in clinical trials to achieve a minimum level of homogeneity, real-life approach mostly relies on the clinical features of patients, independent of the nosological classification of the CTD. For example, the treatment and follow-up of patients with cytopenia is overall equivalent in systemic lupus erythematosus (SLE), Sjogren’s syndrome (SjS) or UCTD. The same applies to other common clinical aspects of multiple CTDs.
Assessment of circulating autoantibodies represents one of the earliest diagnostic procedures in patients with suspected CTD. It helps in the definition of the diagnosis, as some autoantibodies are rather specific for distinct CTDs, but can also provide very important information in terms of identification of subclinical features or prediction of future disease manifestations, especially for a subset of autoantibodies for which a strong association with clinical features is known (eg, anti-MDA5 and rapidly progressive interstitial lung disease). The identification of specific autoantibodies can guide further diagnostic workup and tailored follow-up.5
The main limitations of testing a wide spectrum of autoantibodies are technical complexity, which often requires specialised personnel and costs. Thus, considerable effort is being dedicated to the development and automation of multiparametric assays, capable of testing multiple autoantigens in a reliable, fast, automatic and cost-efficient manner. In particular, a fully automated digital system using particle-based multi-analyte technology (PMAT), a multiplexed assay in which each different autoantigen is linked to a unique particle, has recently been developed. This novel method allows the simultaneous disclosure of multiple autoantigens and has been demonstrated to accurately detect multiple autoantibodies in specific autoimmune diseases, such as primary biliary cholangitis, idiopathic inflammatory myopathy and ‘seronegative’ antiphospholipid syndrome.6–13
However, all cited studies analysed the validity and accuracy of the PMAT assay in small groups of patients with a single CTD or UCTD, thus precluding its feasibility in patients with different CTDs. To overcome this issue, the PMAT digital system for a panel of 29 autoantibodies has been recently tested for the first time in a cohort of approximately 800 patients with different CTDs, as well as in patients with other disorders and a group of healthy controls (HC).14 The PMAT system demonstrated a very high specificity, ranging between 93.7% and 100%, in the detection of 29 autoantibodies associated with CTDs and, interestingly, a higher diagnostic efficiency than that of individual antibodies or antibodies included in the classification criteria for a specific disease. Moreover, the probability of disease increased with multiple positive autoantibodies, thus opening new potential applications of the PMAT system as a screening tool in patients suspected of having a CTD.14 The objective of our study was to evaluate the ability of a multiparametric assay to correctly classify patients with SLE, SjS, systemic sclerosis (SSc), UCTD, idiopathic inflammatory myopathies (IIM) and HC, independent of clinical features, and to evaluate whether serological status is able to identify clusters of patients with similar clinical features.
Methods
Subjects enrolled
Enrolled subjects were selected from a previously described multicentre cohort.14 HC and patients diagnosed with SLE, SSc, SjS, UCTD or IIM were selected. All the included patients fulfilled the latest versions of the respective classification criteria.15–19
Demographic (age and sex), clinical and laboratory parameters were collected retrospectively. Briefly, the presence of clinical or laboratory features was considered at any time from diagnosis to enrolment. With regard to laboratory features, the values were considered abnormal according to local laboratory reference values. ANA were considered positive at titres ≥1:80. Where available, standardised definitions of clinical features according to disease activity scores (ie, ESSDAI and SELENA-SLEDAI)20 21 were applied; otherwise, a clinical diagnosis was considered.
Autoantibody detection
The detection protocol and methods have been described in detail elsewhere.14 Briefly, a novel multiparametric detection system based on PMAT (Aptiva) was employed. One serum sample per patient was tested at the Pordenone (Italy) Laboratory of the FIRMA group using the following three multiparametric antigenic panels: CTD IgG Essential (dsDNA, DFS70, U1RNP, Sm, Ro60, Ro52, La, Scl70, Jo1, CENP-B, Ribo-P), CTD IgG Comprehensive (Research Use Only (RUO)) (RNA pol III, Fibrillarin, Th/To Rpp25, Th/To Rpp38, Ku, BICD2, PM/Scl) and Autoimmune Myopathy IgG (RUO) (Mi-2, HMGCR, NXP2, MDA5, PL-7, PL-12, EJ, SRP, TIF1γ, SAE and OJ) (Inova Diagnostics, San Diego, CA).
This system provides quantitative measurements for each autoantibody. Part of the following analysis was performed by applying cut-off values at five arbitrary units (AU)/mL for the CTD IgG Essential panel and 1 AU/mL for both the CTD IgG Comprehensive and Autoimmune Myopathy IgG panels, as recommended by the manufacturer.
Data analysis
Classification models and data clustering
To evaluate the ability of serological status to correctly classify the enrolled subjects among the six groups, a classification model that exclusively considered the results of the CTD IgG Essential, CTD IgG Comprehensive and Autoimmune Myopathy IgG panels was built. Specifically, a multi-class Extreme Gradient Boosting (XGBoost) model with stratified 10-fold cross-validation was employed, accounting for unbalanced data. The model was optimised by hyperparameter tuning with a Grid Search and fivefold cross-validation.
The mean overall balanced accuracy and mean one-versus-rest area under the receiver operator characteristic (ROC) curve (AUC) were calculated as the mean of the balanced accuracy and of the ROC-AUC of the ten cross-validation rounds. The classification accuracy for each group of subjects was calculated from the confusion matrix by using the following formula:
The importance of each variable was evaluated using feature importance analysis.
To further explore the data, an unsupervised machine learning agglomerative hierarchical clustering model was developed, including exclusively the titres of the detected autoantibodies as variables. The number of clusters was determined using the dendrogram method.
Conventional statistics
Comparisons among clusters was performed using the Kruskal-Wallis test for continuous variables and the χ2 test for binary variables. When the omnibus test was significant, post hoc pairwise comparisons were carried out with the Mann-Whitney U test, χ2 or Fisher’s exact test, as appropriate. To avoid type I error, the p value for multiple tests was corrected using the Holm-Bonferroni method. Statistical significance was set at p<0.05.
Analysis was performed with Python V.3.9 and the following packages. Numpy V.1.23.4, Scipy V.1.9.3, Scikit-learn V.1.1.3, Matplotlib V.3.5.3, Seaborn V.0.12.1, Pandas V.1.5.1 and XGBoost 1.5.1
Classification accuracy is shown as percentage of accurately classified subjects±SD. The other data are shown as absolute number (%) and median (IQR), as appropriate.
We used the Strengthening the Reporting of Observational Studies in Epidemiology cross-sectional checklist when writing our report.22
Results
A total of 908 subjects were enrolled, including 124 HC, 166 SLE patients, 133 SSc patients, 276 SjS patients, 103 UCTD patients and 106 IIM patients. Among these, 92.2% of SLE, 98.5% of SSc, 82.2% of SjS, 70.9% of UCTD, 84.0% of IIM and 21.0% of HC had at least one autoantibody above the cut-off suggested by the manufacturer.
All disease-related clinical and laboratory variables were available for all patients enrolled, except for those described in online supplemental table 1.
rmdopen-2023-003365supp001.pdf (134.3KB, pdf)
Classification accuracy of combined multiplex autoantibody assays
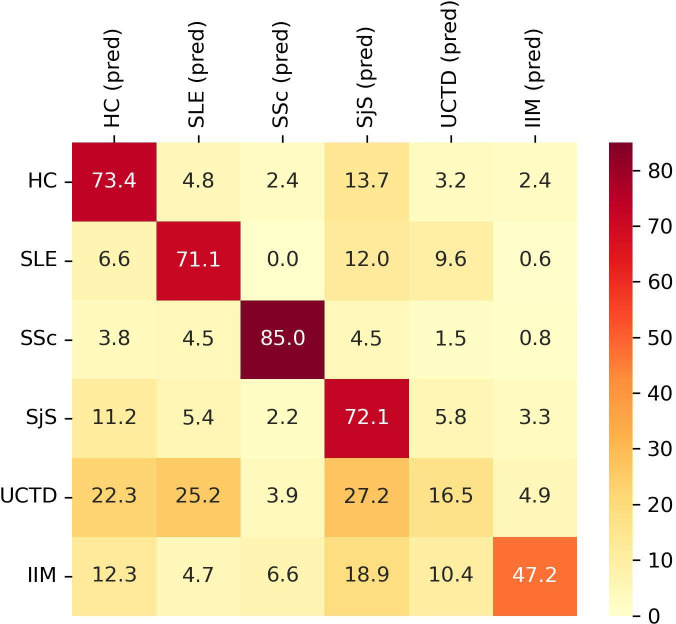
The optimised XGBoost model showed an overall mean classification accuracy of 60.84±4.05% and a mean AUC of 88.99±2.50%. The classification accuracy differed among the six groups. Specifically, 73.4% of HC, 71.1% of SLE, 85.0% of SjS, 72.1% of SSc, 16.5% of UCTD and 47.2% of IIM were correctly classified (figure 1).
Figure 1.
Classification performance of XGBoost model for each group of subjects according to autoantibodies titres. Y axis shows the group subjects belong to. X axis shows the predicted (pred) classification. Data are shown as percentage. HC, healthy controls; IIM, idiopathic inflammatory myopathies; SLE, systemic lupus erythematosus; SjS, Sjogren’s syndrome; SSc, systemic sclerosis; UCTD, undifferentiated connective tissue disease.
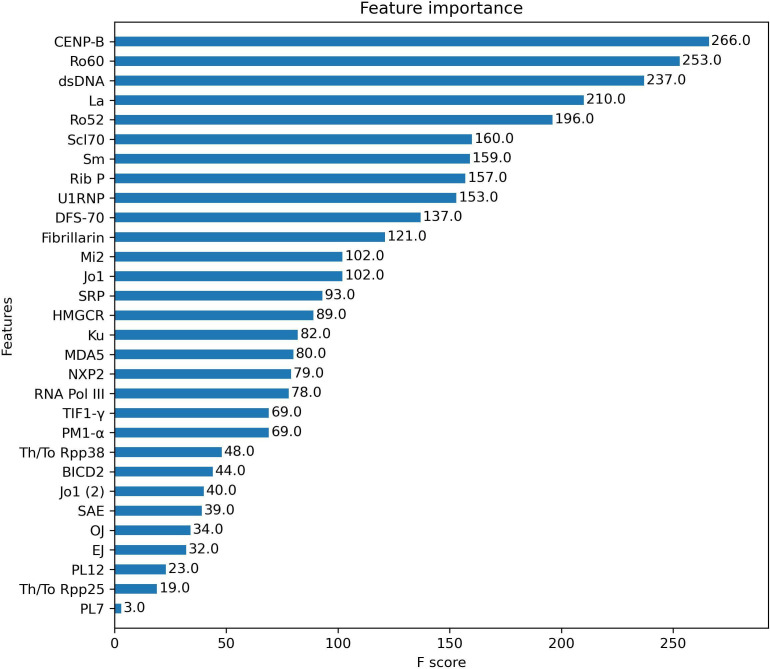
To understand the weight of each autoantibody in the predictive model, the number of times each feature was used to split the data across all trees (F score) was calculated (figure 2). Top five autoantibodies were anti-CENP-B, anti-Ro60, anti-dsDNA, anti-La and anti-Ro52.
Figure 2.
Feature importance analysis. F score of each autoantibody analysed in the XGBoost classification model. F Score represents the number of times each feature was used to split the data across all trees of the model. The higher the F score, the higher the weight of that feature in the model.
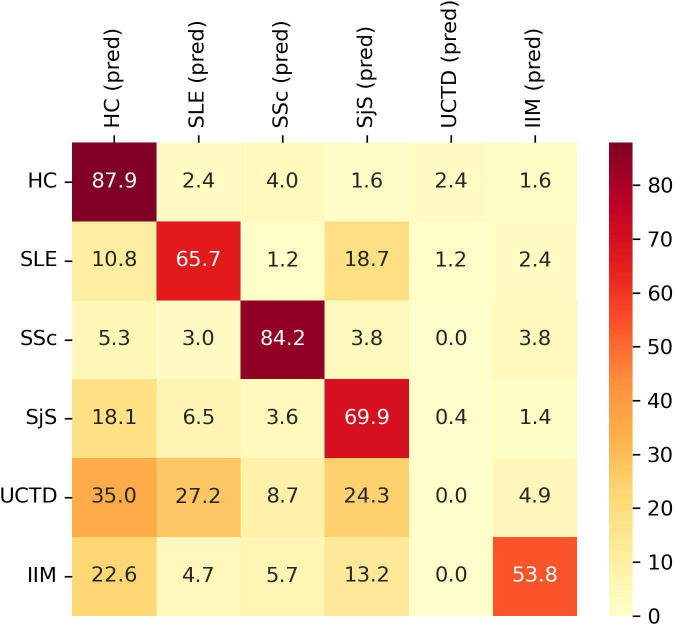
Subsequently, we tested the classification accuracy of an equivalent model built using binary values. Positivity and negativity for each autoantibody were established according to the cut-off values suggested by the assay manufacturer. The overall accuracy was 60.31±6.51%. The classification accuracy for each group is shown in figure 3.
Figure 3.
Classification performance of XGBoost model for each group of subjects according to autoantibodies status (positive vs negative). Y axis shows the group subjects belong to. X axis shows the predicted (pred) classification. Data are shown as percentage. HC, healthy controls; IIM, idiopathic inflammatory myopathies; SLE, systemic lupus erythematosus; SjS, Sjogren’s syndrome; SSc, systemic sclerosis; UCTD, undifferentiated connective tissue disease.
Multiparametric autoantibody assays allow the identification of distinct disease clusters
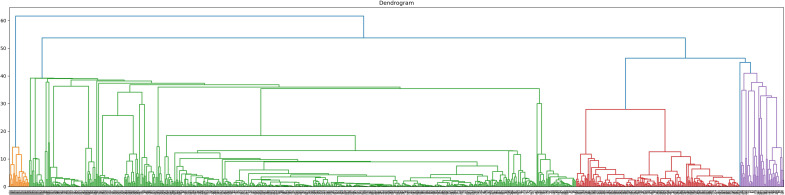
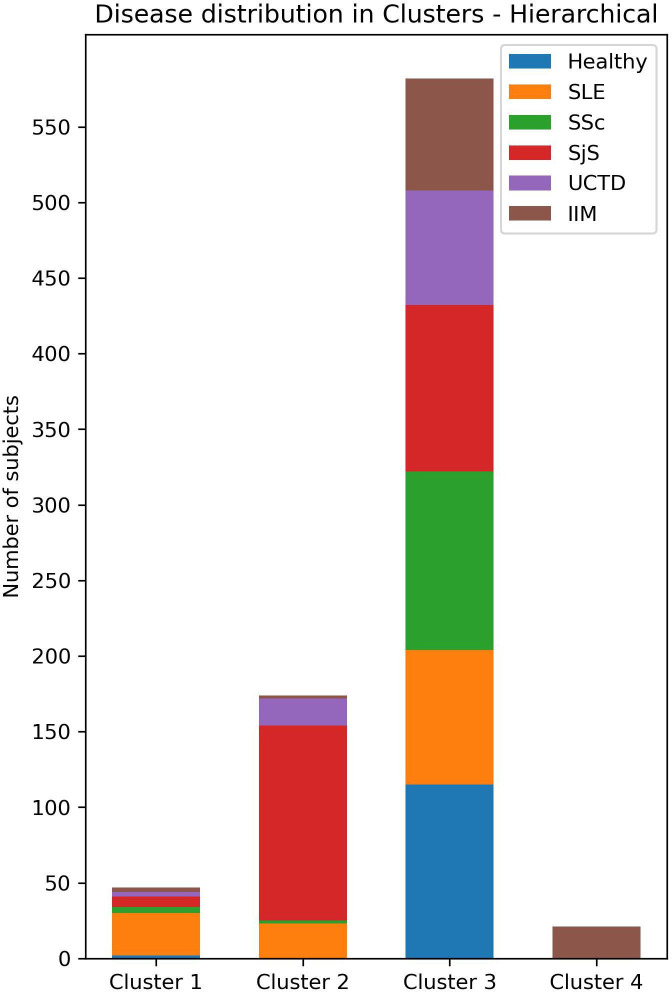
To perform clustering analysis based on the results of the three combined multiparametric assays, subjects with incomplete data were excluded. A total of 824 participants (117 HC, 140 SLE, 124 SSc, 246 SjS, 97 UCTD and 100 IIM) underwent agglomerative hierarchical clustering analysis. The number of clusters was defined according to the dendrogram (figure 4). The cut-off point was selected at four clusters to avoid excessive fragmentation while maintaining a reasonable discriminatory potential. Clusters 1–4 (CL1, CL2, CL3 and CL4) consisted of 47, 174, 582 and 21 subjects, respectively. Of the 47 subjects in CL1, 2 were HC, 28 had SLE, 4 had SSc, 7 had SjS, 3 had UCTD and 3 had IIM. The CL2 group consisted of 23 patients with SLE, 2 with SSc,129 with SjS, 18 with UCTD and 2 with IIM. No HC were clustered in CL2. In CL3, 115 subjects were HC, 89 had SLE, 118 had SSc, 110 had SjS, 76 had UCTD and 74 had IIM. Finally, CL4 exclusively included 21 patients with IIM (figure 5).
Figure 4.
Dendrogram of agglomerative hierarchical clustering model. Cut-off was set at four clusters.
Figure 5.
Distribution of subjects among clusters. IIM, idiopathic inflammatory myopathies; SLE, systemic lupus erythematosus; SjS, Sjogren’s syndrome; SSc, systemic sclerosis; UCTD, undifferentiated connective tissue disease.
The serological characteristics and differences among clusters are shown in online supplemental table 2.
Subsequently, we compared the serological, clinical and laboratory differences among clusters for each group of patients, except for SSc patients who were almost all clustered in CL3. Clusters with fewer than 10 patients were excluded from the analysis to avoid loss of statistical power. Only significantly different and clinically meaningful data are reported in the following paragraphs. The complete data are presented in online supplemental tables 3–6.
Systemic lupus erythematosus
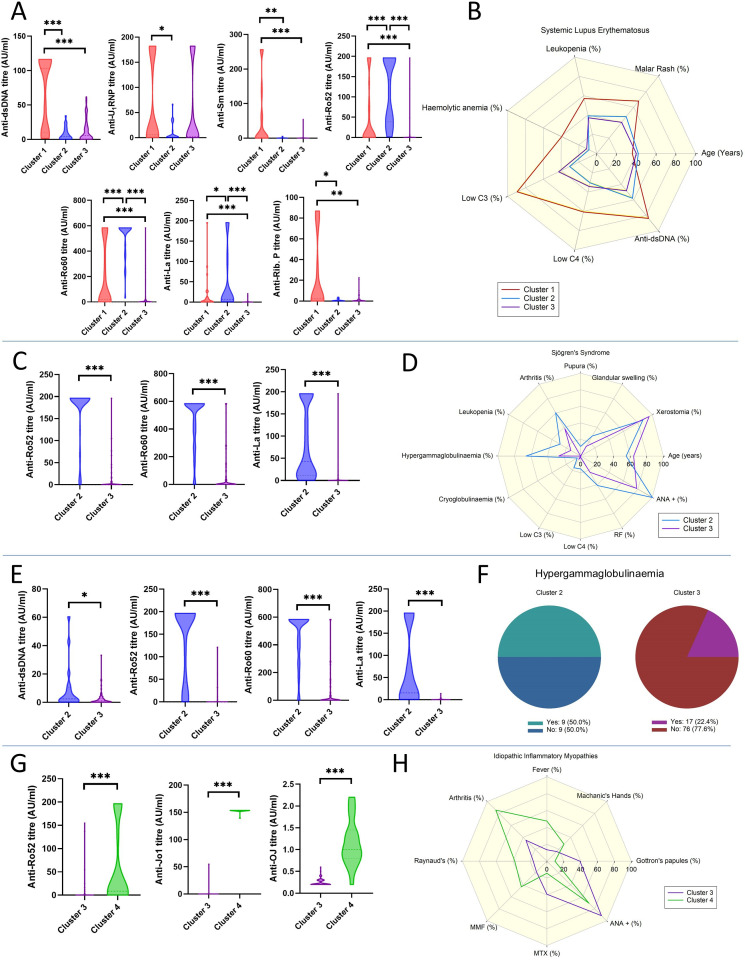
SLE patients in CL1 displayed significantly higher titres of anti-dsDNA (103.1 AU, 9.7–116.3), anti-Sm (1.2 AU, 0.4–67.1) and anti-Ribosomal P (2.1 AU, 0.23–61.4) antibodies compared with both CL2 (4.1 AU, 0.6–13.8, p<0.001; 0.3 AU, 0.2–0.7, p=0.018 and 0.4 AU, 0.1–1.2, p=0.028, respectively) and CL3 (5.5 AU, 1.1–19.6, p<0.001; 0.2 AU, 0.1–0.7, p<0.001 and 0.2 AU, 0.2–1.4, p=0.003, respectively). Patients in CL2 showed significantly higher titres of anti-Ro52 (141.9 AU, 38.5–196.3), anti-SSARo60 (583.7 AU, 453.5–583.7) and anti-La (6.8 AU, 2.7–90.4) compared with CL1 (0.9 AU, 0.43–79.0, p<0.001; 19.7 AU, 1.5–561.4, p<0.001 and 2.3 AU, 0.5–11.6, p=0.038, respectively) and CL3 (0.3 AU, 0.2–0.8, p<0.001; 1.4 AU, 0.4–12.6, p<0.001 and 0.3 AU, 0.2–0.7, p<0.001). Anti-RNP antibodies were also higher in CL1 (5.6 AU, 1.4–182.0) patients compared with CL2 (1.0, 0.4–13.1, p=0.018) (figure 6A). In terms of clinical and laboratory features, CL1 patients were significantly younger (36, 26–43 vs 40, 35–51, p=0.042) and had a higher prevalence of malar rash (67.9% vs 40.4%, p=0.033), leucopenia (57.1% vs 37.1%, p=0.03), haemolytic anaemia (39.3% vs 11.2%, p=0.003), low C3 (89.3% vs 42.7%, p<0.001) and C4 (60.7% vs 34.3%, p=0.003) levels and history of positive dsDNA (84.0% vs 48.2%, p=0.006) compared with CL3. CL1 patients also had a significantly higher prevalence of haemolytic anaemia (39.3% vs 8.7%, p=0.026) and low C3 levels (89.3% vs 30.4%, p<0.001) than CL2 patients (figure 6B).
Figure 6.
Selected autoantibody titres differences among clusters in patients with systemic lupus erythematosus (SLE) (A), Sjogren’s syndrome (SjS) (C), undifferentiated connective tissue disease (UCTD) (E) and idiopathic inflammatory myopathies (IIM) (G). Differences in clinical and laboratory features among clusters in patients with SLE (B), SjS (D), UCTD (F) and IIM (H). *p<0.05, **p<0.01, ***p<0.001.
Sjogren’s syndrome
SjS patients in CL2, compared with those in CL3, showed higher titres of anti-Ro52 (196.3, 196.3–196.3 vs 0.4, 0.1–6.3; p<0.001), anti-Ro60 (583.7, 371.9–583.7 vs 0.7, 0.2–31.9; p<0.001) and anti-La (42.9, 11.5,195.8 vs 0.3, 0.1–0.4; p<0.001) (figure 6C), along with a higher prevalence of history of glandular swelling (27.9% vs 13.6%, p=0.007), purpura (11.6% vs 0%, p<0.001), arthritis (60.5% vs 38.2%, p=0.001), leucopenia (28.7% vs 13.6%, p=0.005), hypergammaglobulinaemia (65.9% vs 26.4%, p<0.001), cryoglobulinaemia (4.7% vs 0.0%, p=0.032), rheumatoid factor (41.1% vs 22.7%, p=0.003) and ANA (100% vs 78.2%, p<0.001) positivity. In contrast, CL3 patients were older (64, 54–71 vs 55, 46–69; p<0.001) and had a higher prevalence of history of dry mouth symptoms (95.5% vs 86.8%, p=0.021). (figure 6D).
Undifferentiated connective tissue disease
UCTD patients in CL2 had higher titres of anti-dsDNA (2.4 AU, 0.5–10.4 vs 1.0 AU, 0.3–2.8; p=0.041), anti-Ro52 (196.3 AU, 53.5–196.3 vs 0.2 AU, 0.1–0.5; p<0.001), anti-Ro60 (583.7 AU, 449.2–583.7 vs 0.2 AU, 0.1–0.5; p<0.001) and anti-La (15.5 AU, 1.8–195.8 vs 0.3, 0.1–0.6; p<0.001), compared with patients in CL3 (figure 6E). The only significant difference between clusters in terms of clinical features was the higher prevalence of hypergammaglobulinaemia in CL2 than CL3 (50.0% vs 22.4%, p=0.037) (figure 6F).
Idiopathic inflammatory myopathies
IIM patients in CL4, compared with those in CL3, showed higher titres of anti-Ro52 (8.6 AU, 0.3–161.9 vs 0.2 AU, 0.1–0.6; p<0.001) and anti-OJ (1.0 AU, 0.8–1.5 vs 0.2 AU, 0.2–0.3; p<0.001). Similarly, higher levels of anti-Jo1 antibodies were found in CL4 than in CL3, with an almost perfect concordance between the two anti-Jo1 assays present in two distinct antigen panels (figure 6G).
With regard to other features, patients in CL4 had a higher prevalence of fever (47.6% vs 13.5%, p=0.002), arthritis (85.7% vs 35.1%, p<0.001) and Raynaud’s phenomenon (38.1% vs 16.2%, p=0.038). On the contrary, patients in CL3 showed a higher prevalence of Gottron’s papules (39.2% vs 9.5%, p=0.01) and positive ANA (91.4% vs 70.6%, p=0.035). More CL3 patients were treated with methotrexate compared with subjects in CL4 (39.2% vs 14.3%, p=0.033), and fewer with MMF (17.6% vs 42.9%, p=0.021) (figure 6H).
Discussion
CTDs represent a wide spectrum of systemic disorders, often associated with relevant morbidity and mortality due to their multi-organ involvement. However, their protean clinical presentation, especially at disease onset, the variable diagnostic accuracy of associated specific serological profiles and, in more rare cases, the absence of specific serological markers, often delay diagnosis and, consequently, the introduction of an appropriate treatment.23
The current results expand the findings of our previous study and allow to confirm the good accuracy of Aptiva PMAT system in classifying patients with a specific CTD according to classification criteria.14 A machine learning approach was applied to avoid any confirmation bias due to the selection of autoantigens that are already known to have a high predictive value for specific CTDs. The accuracy of the model is >70% for HC, SLE and SjS and 85% for SSc. As expected, the accuracy is lower (47.2%) for a heterogeneous disease such as IIM, which includes dermatomyositis, polymyositis, necrotising myopathy and anti-synthetase syndrome patients, and even lower for UCTD. Because the assay employed provides a quantitative result and considering that differences in circulating autoantibody levels may play a role in clinical practice for both diagnosis and follow-up, the analysis was performed using antibody titres, rather than binary results (positive vs negative). However, when the analysis was repeated with the latter approach, the results were overall similar, with a slightly higher classification accuracy of HC.
When the weight of each autoantibody in the classification model was evaluated, we observed that those with the highest F-score were the most prevalent autoantibodies in CTDs and also those included in classification criteria. This was expected because of the characteristics of the classification model, but also due to the fact that the identification of patients to be included in the study relied on classification criteria including the very same autoantibodies. It is important to underline that, despite other antigens are very specific for certain CTDs (such as tRNA synthetases), their F-score is low in this model due to their very low prevalence in an unselected CTD population.
What is more interesting is the ability of the combined multiparametric assays to identify four distinct clusters of patients. CL1 mostly consists of SLE patients with very few patients with other diseases and HC. CL2 mostly includes SjS patients and a few patients with SLE, and with UCTD. CL4 only includes anti-Jo1 positive patients. Interestingly, CL3 is very heterogeneous. Not only it includes almost all HC and SSc but also a significant proportion of SLE, SjS, UCTD and IIM patients. By looking at the dendrogram, it is interesting to underline that even if a more fragmented clustering were applied, the majority of subjects would still be included in a single large heterogeneous cluster, resembling CL3. This observation suggests that CL3 actually represents a population with distinct serological features.
The inclusion of SSc patients in CL3 requires some additional comments. The vast majority of these subjects display a high titre positivity for anti-CENP (43/124) or anti-Scl70 (46/124). Only two had positive anti-RNA pol III. However, the prevalence of anti-CENP and anti-Scl70 in the overall cohort is quite low (7.3% and 9.8%, respectively). Their weight in the clustering model is therefore inevitably lower than that of more common autoantibodies, such as anti-dsDNA, anti-Ro and anti-La, despite being very specific for SSc diagnosis. In fact, when looking at the XGBoost classification model that was trained with the final diagnosis, anti-CENP and anti-Scl70 are at the first and sixth places in terms of F-score (figure 2) and SSc is the group that was classified with the highest accuracy (figure 1). Additionally, a small but significant proportion of these subjects also had concomitant positivity for anti-Ro52, anti-Ro60, anti-La and anti-U1-RNP autoantibodies, usually at low titre. These are the most likely reasons why the clustering model was not able to effectively distinguish the two main subgroups of SSc patients in this specific cohort, including them in the most heterogeneous cluster. Likely, if SSc patients had represented a larger portion of the cohort, an equivalent model would have been able to cluster most SSc patients independently.
Apart from SSc patients, the subjects in CL3, overall, display very low median titres of all autoantibodies tested. This is due to the fact that they are seronegative (such as most HC), have low-titre autoantibodies, or have low to very-low prevalence autoantibodies, with a limited impact on the clusterisation model. As previously mentioned, this aspect is a consequence of the composition of this specific cohort which, however, is representative of a real-world population.
By observing the effect of the model on the various subpopulations, the clusterisation identifies a younger SLE cluster (CL1), characterised by a typical serological profile (positive anti-dsDNA, anti-Sm and anti-Ribosomal P), with signs of active disease (malar rash, haemolytic anaemia, leucopenia and low complement levels). Differently, patients in CL2 have a higher prevalence of high-titre anti-Ro60/52 and anti-La antibodies, along with low-titre anti-dsDNA antibodies. Finally, SLE patients in CL3 are mostly seronegative, or with low-titre autoantibodies. Consequently, SLE patients in CL2 and CL3 have a significantly lower prevalence of the typical disease features present in CL1 patients.24 25
Similarly, when SjS subpopulation is observed, patients included in CL2 have the typical serological and clinical features of the disease (high-titre anti-Ro60/52, anti-La, a higher prevalence of glandular swelling, arthritis, purpura, leucopenia, hypergammaglobulinaemia, low complement and cryoglobulinaemia). On the contrary, patients clustered in CL3 are mostly seronegative and display the typical ‘mild’ SjS phenotype with fewer systemic manifestations and older age.26–29
Due to its wide heterogeneity, very few differences can be detected in the UCTD sub-population, overall overlapping those of SLE and SjS.
As far as IIM is concerned, because anti-Jo1 anti-synthetase syndrome patients are all clustered in CL4, they show higher prevalence of typical features, such as fever, arthritis, Raynaud’s phenomenon and a lower prevalence of skin features typical of dermatomyositis. Additionally, the more frequent use of methotrexate and mycophenolate mofetil is likely indirect evidence of the extra-muscular involvement in antisynthetase syndrome.30
The results of the clustering analysis seem to suggest that the serological status of CTD patients is able to identify clusters of disease that more closely reflect a real-life clinical approach, rather than a classification criteria-based identification of patients. Independently of the formal definition of patients as having a specific CTD, it seems reasonable to say that CL1 depicts a typical SLE phenotype, CL2 a typical SjS phenotype, while CL3 is more heterogeneous, showing milder disease features and likely including patients that often do not require any immunosuppressive treatment.
This is true not only in terms of a serological pattern, but also in terms of clinical features. In fact, by observing the prevalence of the most commonly shared clinical features among different CTDs, it is clear that the prevalence of hypocomplementemia and leucopenia in CL2 SLE patients is very close to that of CL2 SjS patients. Similarly, the prevalence of hypergammaglobulinaemia in CL2 and CL3 is very similar among SjS and UCTD patients.
We acknowledge that a major limit of the study is its retrospective nature, which may generate bias and incomplete data. However, all variables were well characterised and homogeneous and all patients included fully satisfied the classification criteria for the specific disease. Moreover, although patients were not specifically selected and are overall representative of a real-life CTD population, the external validity of the results may not be the same for all included populations. In particular, while the results of the SLE, SjS and UCTD cohorts seem very consistent and reliable, the clustering model was not able to distinguish subpopulations of SSc patients, such as anti-Scl70 from anticentromere subjects. This is likely due to the number of subjects included and to the prevalence of these autoantibodies in the cohort. A similar problem applies to the heterogeneous IIM, mostly because of the low prevalence of some autoantibodies.
Similarly, the clustering model was not able to identify HC as an independent cluster. However, this was an expected result because a clustering model based exclusively on serological status would not be able to distinguish HC from seronegative CTD patients. The inclusion of HC in the model as an internal control confirmed the goodness of both classification and clustering algorithms to adequately detect this subgroup and to assign them to a single cluster.
We expect that by analysing an even larger cohort of subjects that allowed a more accurate clustering without excessive fragmentation, a more detailed stratification would be possible and additional statistically significant clinical differences would emerge.
In conclusion, this study confirms that a multiparametric assay performed on a fully automated digital system using PMAT may be a reliable technique, allowing a clinically relevant stratification of CTD patients. The system seems to be able to identify clusters of subjects with similar clinical features, independently of their final diagnosis. These results support the importance of obtaining a wide autoantibody assay in CTD patients. The combination and titres of multiple autoantibodies may in fact characterise a patient with a higher or lower probability of having or developing specific clinical manifestations, independently of its formal classification and diagnosis, thus informing clinical management.
It may be argued that routinely testing very rare autoantibodies in all patients with suspected CTD, independently of their clinical features may not be reasonable due to their very low predictive value in case of low a priori probability. While a two-step approach (more prevalent autoantigens as a first-line test and less frequent autoantigens tested subsequently when clinically relevant) is perfectly reasonable and is currently commonly applied in clinical practice, automated multiparametric assays may be able to change this paradigm.
Footnotes
Twitter: @GiaCafaro
GC and EB contributed equally.
DV and NB contributed equally.
Collaborators: Onelia Bistoni, Carlo Perricone, Fabiana Topini, Manuela Sebastiano: Rheumatology Unit, Department of Medicine and Surgery, University of Perugia, Perugia, Italy; Paola Migliorini: Clinical Immunology, University of Pisa, Pisa, Italy; Silvia Piantoni, Ilaria Cavazzana, Micaela Fredi and Stefania Masneri: Rheumatology and Clinical Immunology Unit, Department of Clinical and Experimental Sciences, ASST Spedali Civili and University of Brescia, Brescia, Italy; Emirena Garrafa: Laboratory of Clinical Chemistry, Department of Molecular and Translational Medicine, ASST Spedali Civili and University of Brescia, Brescia, Italy; Francesca Bellisai, Sara Cheleschi and Maria-Romana Bacarelli: Rheumatology Unit, Department of Medicine, Surgery and Neuroscience, Azienda Ospedaliera Universitaria Senese, Policlinico Le Scotte, Siena, Italy; Marilina Tampoia: Clinical Pathology, Presidio Ospedaliero SS. Annunziata, Taranto, Italy; Margherita Zen: Rheumatology Unit, Department of Medicine, University of Padua, Padua, Italy; Paola Parronchi and Daniele Cammelli: Department of Experimental and Clinical Medicine, University of Florence, Florence, Italy; Maurizio Benucci: Rheumatology Unit, San Giovanni di Dio Hospital, Florence, Italy; Mariangela Manfredi: Laboratory of Immunology and Allergology, San Giovanni di Dio Hospital, Florence, Italy; Roberto Giacomelli and Luisa Arcarese: Clinical Immunology and Rheumatology, University Campus Biomedico, Rome, Italy; Patrizia Rovere Querini and Valentina Canti: IRCCS San Raffaele, Milan, Italy.
Contributors: GC: conceptualisation, methodology, formal analysis, writing – original draft, visualisation; EB: conceptualisation, writing – original draft; CB, FF, VR, AF, MF, AG, BP, MI, AR, SDR: investigation, resources, writing – review and editing; RG: conceptualisation, writing – review and editing, supervision; DV, NB: conceptualisation, investigation, resources, data curation, writing – review and editing, project administration, guarantor.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: NB has received lecture fees from Inova Diagnostics.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Contributor Information
FIRMA Collaborators:
Onelia Bistoni, Carlo Perricone, Fabiana Topini, Manuela Sebastiano, Paola Migliorini, Silvia Piantoni, Ilaria Cavazzana, Micaela Fredi, Stefania Masneri, Emirena Garrafa, Francesca Bellisai, Sara Cheleschi, Maria-Romana Bacarelli, Marilina Tampoia, Margherita Zen, Paola Parronchi, Daniele Cammelli, Maurizio Benucci, Mariangela Manfredi, Roberto Giacomelli, Luisa Arcarese, Patrizia Rovere Querini, and Valentina Canti
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by Comitato Etico Regionale Umbria (3574/19) and was conducted in accordance with the principles of the Declaration of Helsinki. Due to its retrospective observational nature, the need for informed consent was waived by the Ethics Committee.
References
- 1. Bossuyt X, De Langhe E, Borghi MO, et al. Understanding and interpreting antinuclear antibody tests in systemic rheumatic diseases. Nat Rev Rheumatol 2020;16:715–26. 10.1038/s41584-020-00522-w [DOI] [PubMed] [Google Scholar]
- 2. Irure-Ventura J, López-Hoyos M. The past, present, and future in antinuclear antibodies (ANA). Diagnostics 2022;12:647. 10.3390/diagnostics12030647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Didier K, Bolko L, Giusti D, et al. Autoantibodies associated with connective tissue diseases: what meaning for clinicians? Front Immunol 2018;9:541. 10.3389/fimmu.2018.00541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dyball S, Rodziewicz M, Mendoza-Pinto C, et al. Predicting progression from undifferentiated connective tissue disease to definite connective tissue disease: a systematic review and meta-analysis. Autoimmun Rev 2022;21:103184. 10.1016/j.autrev.2022.103184 [DOI] [PubMed] [Google Scholar]
- 5. Mulhearn B, Tansley SL, McHugh NJ. Autoantibodies in connective tissue disease. Best Pract Res Clin Rheumatol 2020;34:101462. 10.1016/j.berh.2019.101462 [DOI] [PubMed] [Google Scholar]
- 6. Cavazzana I, Richards M, Bentow C, et al. Evaluation of a novel particle-based assay for detection of autoantibodies in idiopathic inflammatory myopathies. J Immunol Methods 2019;474:112661. 10.1016/j.jim.2019.112661 [DOI] [PubMed] [Google Scholar]
- 7. Mahler M, Malyavantham K, Seaman A, et al. Profiling of myositis specific antibodies and composite scores as an aid in the differential diagnosis of autoimmune myopathies. Diagnostics 2021;11:2246. 10.3390/diagnostics11122246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Radin M, Cecchi I, Barinotti A, et al. Identifying subsets of patients with undifferentiated connective tissue disease: results from a prospective, real-world experience using particle-based multi-analyte technology. Autoimmun Rev 2023;22:103298. 10.1016/j.autrev.2023.103298 [DOI] [PubMed] [Google Scholar]
- 9. Radin M, Foddai S, Cecchi I, et al. Improving the clinical accuracy in patients with antiphospholipid antibodies using anti-phosphatidylserine/prothrombin and anti-Beta2 glycoprotein I domain and particle-based multi-analyte technology. Thromb Res 2021;202:100–3. 10.1016/j.thromres.2021.03.019 [DOI] [PubMed] [Google Scholar]
- 10. Radin M, Cecchi I, Foddai SG, et al. Validation of the particle-based multi-analyte technology for detection of anti-phosphatidylserine/prothrombin antibodies. Biomedicines 2020;8:622. 10.3390/biomedicines8120622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Richards M, García-De La Torre I, González-Bello YC, et al. Autoantibodies to Mi-2 alpha and Mi-2 beta in patients with idiopathic inflammatory myopathy. Rheumatology 2019;58:1655–61. 10.1093/rheumatology/kez092 [DOI] [PubMed] [Google Scholar]
- 12. Sciascia S, Radin M, Ramirez C, et al. Evaluation of novel assays for the detection of autoantibodies in antiphospholipid syndrome. Autoimmun Rev 2020;19:102641. 10.1016/j.autrev.2020.102641 [DOI] [PubMed] [Google Scholar]
- 13. Villalta D, Seaman A, Tiongson M, et al. Evaluation of a novel extended automated particle-based multi-analyte assay for the detection of autoantibodies in the diagnosis of primary biliary cholangitis. Clin Chem Lab Med 2020;58:1499–507. 10.1515/cclm-2020-0122 [DOI] [PubMed] [Google Scholar]
- 14. Bizzaro N, Villalta D, Bini V, et al. Multiparametric autoantibody analysis: a new paradigm for the diagnosis of connective tissue diseases. Arthritis Res Ther 2022;24:278. 10.1186/s13075-022-02980-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aringer M, Costenbader K, Daikh D, et al. 2019 European League Against Rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Arthritis Rheumatol 2019;71:1400–12. 10.1002/art.40930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lilleker JB, Vencovsky J, Wang G, et al. Response to: 'Antisynthetase syndrome or what else? Different perspectives indicate the need for new classification criteria' by Cavagna et al. Ann Rheum Dis 2018;77:e51. 10.1136/annrheumdis-2017-212382 [DOI] [PubMed] [Google Scholar]
- 17. Shiboski CH, Shiboski SC, Seror R, et al. American College of Rheumatology/European League Against Rheumatism classification criteria for primary Sjögren’s syndrome: a consensus and data-driven methodology involving three international patient cohorts. Arthritis Rheumatol 2017;69:35–45. 10.1002/art.39859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van den Hoogen F, Khanna D, Fransen J, et al. Classification criteria for systemic sclerosis: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis 2013;72:1747–55. 10.1136/annrheumdis-2013-204424 [DOI] [PubMed] [Google Scholar]
- 19. Mosca M, Neri R, Bombardieri S. Undifferentiated connective tissue diseases (UCTD): a review of the literature and a proposal for preliminary classification criteria. Clin Exp Rheumatol 1999;17:615–20. [PubMed] [Google Scholar]
- 20. Petri M, Kim MY, Kalunian KC, et al. Combined oral contraceptives in women with systemic lupus erythematosus. N Engl J Med 2005;353:2550–8. 10.1056/NEJMoa051135 [DOI] [PubMed] [Google Scholar]
- 21. Seror R, Ravaud P, Bowman SJ, et al. EULAR Sjogren’s syndrome disease activity index: development of a consensus systemic disease activity index for primary Sjogren’s syndrome. Ann Rheum Dis 2010;69:1103–9. 10.1136/ard.2009.110619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elm E von, Altman DG, Egger M, et al. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. BMJ 2007;335:806–8. 10.1136/bmj.39335.541782.AD [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lenti MV, Rossi CM, Melazzini F, et al. Seronegative autoimmune diseases: a challenging diagnosis. Autoimmun Rev 2022;21:103143. 10.1016/j.autrev.2022.103143 [DOI] [PubMed] [Google Scholar]
- 24. Kiriakidou M, Ching CL. Systemic lupus erythematosus. Ann Intern Med 2020;172:ITC81–96. 10.7326/AITC202006020 [DOI] [PubMed] [Google Scholar]
- 25. Zucchi D, Elefante E, Schilirò D, et al. One year in review 2022: systemic lupus erythematosus. Clin Exp Rheumatol 2022;40:4–14. 10.55563/clinexprheumatol/nolysy [DOI] [PubMed] [Google Scholar]
- 26. Cafaro G, Gerli R, Bartoloni E. Association between glandular infiltrate and leukopenia in Sjögren syndrome (SS): data from the Italian research group on SS (GRISS). J Rheumatol 2020;47:1840–1. 10.3899/jrheum.200328 [DOI] [PubMed] [Google Scholar]
- 27. Cafaro G, Perricone C, Baldini C, et al. Significance of anti-La/SSB antibodies in primary Sjögren’s syndrome patients with combined positivity for anti-Ro/SSA and salivary gland biopsy. Clin Exp Rheumatol 2020;38 Suppl 126:53–6. [PubMed] [Google Scholar]
- 28. Luo J, Zhang Y, Chen J-Q, et al. Distinct clinical phenotypes of primary Sjögren’s syndrome differ by onset age: a retrospective study of 742 cases and review of the literature. Clin Exp Rheumatol 2022;40:2373–80. 10.55563/clinexprheumatol/b4z1qu [DOI] [PubMed] [Google Scholar]
- 29. Manfrè V, Chatzis LG, Cafaro G, et al. Sjögren’s syndrome: one year in review 2022. Clin Exp Rheumatol 2022;40:2211–24. 10.55563/clinexprheumatol/43z8gu [DOI] [PubMed] [Google Scholar]
- 30. Cavagna L, Nuño L, Scirè CA, et al. Clinical spectrum time course in anti Jo-1 positive Antisynthetase syndrome: results from an international retrospective multicenter study. Medicine (Baltimore) 2015;94:e1144. 10.1097/MD.0000000000001144 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
rmdopen-2023-003365supp001.pdf (134.3KB, pdf)
Data Availability Statement
Data are available upon reasonable request.