Abstract
The MIDI automated Microbial Identification System (MIS) uses gas chromatography (GC) analysis of whole-cell fatty acid methyl esters (FAMEs) between 9 and 20 carbons in length to characterize a wide range of bacterial genera and species, including mycobacteria. Mycolic acid cleavage products (MACPs) with chain lengths of C22 to C26 are not released by MIDI sample preparation of mycobacteria. Therefore, the MIS library search report often matches several mycobacterial species without any significant difference in the similarity indices. The problem is solved by adding trimethylsulfonium hydroxide (TMSH) instead of sodium sulfate in the last step of sample preparation, thus allowing the identification of MACPs in addition to FAMEs. Only one GC run parameter has to be changed: the temperature program must be extended from 260 to 310°C. The MIS library search report for the identification of bacteria is not disturbed by TMSH. The combination of conventional library search report with the information of typical MACP patterns yields significantly better discrimination of mycobacterial species than the MIDI method allows.
The Microbial Identification System (MIS; Microbial ID, Newark, Del.) is a well-established fully automated gas chromatography (GC) analytical system which identifies bacteria and fungi based on their unique fatty acid profiles (6, 9, 14). The identification of mycobacteria by using the MIS standard library (M17H10) requires well-defined conditions of growth and sample preparation. Whole-cell fatty acid methyl esters (FAMEs) between 9 and 20 carbons in length are also used for the identification of mycobacteria, but the result of library search is often not discriminative enough for clinical application, i.e., in many cases the system does not allow discrimination between different species. Mycobacterial mycolic acid cleavage products (MACPs) with chain lengths of C22 to C26 are not released by the MIS sample preparation (8). The MACP pattern, however, is often highly significant for the identification of mycobacteria (4, 5, 7).
Trimethylsulfonium hydroxide (TMSH) converts fatty acids bound in biomolecules such as phospholipids and/or glycerides into the corresponding FAMEs. The transesterification can be performed at room temperature in a fast, single-step reaction. Secondary alcohols and MACPs are also released from mycobacteria under these conditions. The samples arising from TMSH treatment can be injected directly into the GC (12).
This paper describes the identification of mycobacteria by using the MIS library search with a TMSH-modified MIS sample preparation technique. The results of the conventional standard library search are compared with those of the TMSH-assisted library search.
MATERIALS AND METHODS
Chromatographic conditions.
Specimens were processed on a Hewlett-Packard (Avondale, Pa.) GC system that included a model 5890 Series II GC equipped with a split injector and a flame ionization detector, a model 6890 automatic sampler, a Vectra XU 5/90C computer, a model 3365 Series II ChemStation (Rev A.03.34), and a fused-silica column (25 m by 0.2 mm by 0.33 μm; 5% phenylmethyl silicone, Ultra 2, HP 19091B-102). For MACP analysis, the column temperature program was increased from 260 to 310°C (starting at 170°C and increasing by 5°C min−1, with a final time of 3 min at 310°C) and two integration events were modified (Baseline All Valleys ON, 20.000 min; Integrator OFF, 30.000 min). All other parameters of chromatography were the following, as recommended in the operational manual of the MIS (8): carrier gas, hydrogen; sample volume, 2 μl; split ratio, 1:100; injector temperature, 250°C; detector temperature, 300°C.
For the identification of mycobacteria, version 3.8 of the MIS M17H10 library (standard MYCO method) was used. Peaks were automatically integrated, fatty acids were identified by equivalent chain length (ECL) (9), and percentages of the total peak area and similarity indices were calculated. External calibration was done by using MIDI calibration mixture 1 (FAMEs of straight-chain saturated fatty acids from 9 to 20 carbons in length and five hydroxy acids).
As a second standard, MIDI calibration mixture 1 was supplemented by the addition of straight-chain FAMEs (C21:0 to C26:0, each 80 mg/liter; FAME Kit 23; Larodan, Malmö, Sweden).
The identification of MACP, FAME, and alcohol (Table 1) was made by matching peak retention times with those of fatty acid standards of 20 carbons or more and of biological specimens derived from the mycobacteria Mycobacterium malmoense (ATCC 29571), M. szulgai (NCTC 10831), M. tuberculosis (ATCC 27294), and M. xenopi (clinical isolate).
TABLE 1.
ECL values for important MACPs, FAMEs, and an alcohol
| MACP, FAME, or alcohol | RTa | ECLMISb | ECLMISplusb |
|---|---|---|---|
| 2-Methyl 20:0 | 17.306 | 20.310 | 20.316 |
| 2-OH-22:0 alcohol | 18.206 | 20.835 | 20.852 |
| 21:0 | 18.454 | 20.989 | 21.000 |
| 22:1 cis 13 | 19.717 | 21.726 | 21.779 |
| 22:0 | 20.075 | 21.944 | 22.000 |
| 2,4,6-Trimethyl 22:0 | 21.174 | 22.587 | 22.704 |
| 23:0 | 21.636 | 22.864 | 23.000 |
| 24:1 cis 15 | 22.809 | 23.555 | 23.790 |
| 24:0 | 23.119 | 23.738 | 24.000 |
| 2,4,6-Trimethyl 24:0 | 24.123 | 24.327 | 24.705 |
| 25:0 | 24.543 | 24.543 | 25.000 |
| 26:0 | 25.913 | 25.384 | 26.000 |
RT, retention time in minutes.
See text for explanation of ECLMIS and ECLMISplus values.
Bacteria and growth conditions.
The examined reference strains were Mycobacterium avium (DSM 43216), M. chelonae (DSM 43804), M. fortuitum (ATCC 6841, DSM 43271, DSM 43075), M. gastri (ATCC 15754), M. gordonae (ATCC 14470), M. intracellulare (DSM 43223), M. kansasii (ATCC 12478), M. malmoense (ATCC 29571), M. marinum (ATCC 927), M. scrofulaceum (ATCC 19981), M. simiae (ATCC 25275), M. smegmatis (DSM 43756, DSM 43061, DSM 43078), M. szulgai (NCTC 10831), M. terrae (DSM 43227), and M. tuberculosis (ATCC 27294), as well as clinical isolates (Table 2). The clinical isolates were identified by conventional biochemical tests, including tests of growth conditions, pigment, catalase, aryl sulfatase, phosphatase, niacin, and amides. Cultures were grown on Middlebrook agar (Difco 0627-01-2) plus Middlebrook OADC enrichment (Difco 0722-64-0) in 5 to 10% CO2 at 35°C as described by MIDI (8).
TABLE 2.
Clinical isolates of mycobacteria analyzed by MIS by using both conventional MIS sample preparation and MIS sample preparation combined with the TMSH technique
| Mycobacterium species | No. of clinical isolates
|
||
|---|---|---|---|
| Total | Recognized by MIS alonea | Recognized by MIS plus TMSHb | |
| M. avium-M. intracellulare-M. scrofulaceum complex | 35 | 2 | 2 |
| M. avium-M. intracellulare group | 31 | 25 | 25 |
| M. avium | 7c | 0 | 0 |
| M. scrofulaceum | 4 | 2 | 2 |
| M. celatum | 1 | 0 | 1 |
| M. chelonae | 6 | 2 | 2 |
| M. fortuitum | 8 | 2 | 2 |
| M. gastri | 2 | 2 | 2 |
| M. gordonae | 10 | 8 | 10 |
| M. kansasii | 9 | 7 | 9 |
| M. malmoense | 4 | 2 | 4 |
| M. marinum | 1 | 1 | 1 |
| M. nonchromogenicum | 3 | 3 | 3 |
| M. smegmatis | 1 | 0 | 0 |
| M. szulgai | 6 | 4 | 6 |
| M. tuberculosis-M. bovis complex | 75 | 4 | 24 |
| M. tuberculosis | 75 | 51 | 51 |
| M. xenopi | 19 | 13 | 19 |
Based on a similarity index of 0.500 or higher with a separation of 0.100 between the first and second choices.
MIS library search completed by including data from the literature.
Part of the M. avium-M. intracellulare group.
Sample preparation.
Sample preparation was done as described in the MIDI operating manual for the MIS (8): each sample was saponified (with sodium hydroxide in methanol), methylated (with hydrochloric acid in methanol), extracted (with hexane in methyl tert-butyl ether), and cleaned by base wash (with sodium hydroxide). After that step in the MIDI protocol, mycolic acids were removed by means of anhydrous sodium sulfate. This is very important to keep the chromatographic system clean; otherwise, the long-chain mycolic acids might lead to disturbances of later GC runs. For MACP analysis, the last step was replaced by adding 10 μl of 0.2 M methanolic TMSH (Macherey-Nagel, Düren, Germany).
RESULTS
Library search by using TMSH-modified sample preparation.
One hundred ninety-nine strains of mycobacteria, including 180 clinical isolates (Table 2) and 19 reference strains, were prepared in accordance with the MIS sample preparation protocol and MIS sample preparation combined with the TMSH technique. The named species in the MIS library report were usually identical for both methods of sample preparation. In particular, the similarity indices showed no relevant difference. This means that MIS sample preparation combined with the TMSH technique allows the use of the MIS library search without modification.
Identification of MACPs.
Identification of MACPs was done by comparing the chromatographic data resulting from the present study with fatty acid profiles reported in the literature (5, 7, 12). All FAME patterns, including those of MACPs and fatty alcohols, show excellent agreement with data in the literature. Table 1 shows the ECL values for important MACPs, long-chain FAMEs, and an alcohol. The ECL values were calculated by MIS after different samples were run. The ECLMIS value is the interpolated ECL value determined by using the conventional MIDI calibration mixture 1 (C9 to C20). The ECLMISplus value is the ECL value determined by using the supplemented MIDI calibration mixture 1 (C9 to C26). The calculated ECL values are the mean values of several (at least six) GC runs done on three independent chromatographic MISs. There was no significant difference between the ECL values of these systems, i.e., there was no mismatch between different runs and all peaks were identified by the systems.
Identification of strains.
Most of the 19 reference strains were identified by MIS alone. M. avium (DSM 43216) and M. intracellulare (DSM 43223) were recognized as M. avium-intracellulare group; M. fortuitum (DSM 43271) was not distinguished from M. chelonae. M. chelonae (DSM 43804) was not distinguished from M. asiaticum. After MIS-plus-TMSH sample preparation, M. asiaticum could be excluded since the MACP C26:0 was not found.
Table 2 shows the results of the analysis of 180 clinical isolates when both MIS sample preparation alone and MIS sample preparation combined with the TMSH technique were used. All strains with unique patterns of FAMEs, MACPs, and alcohols (M. celatum, M. gastri, M. gordonae, M. kansasii, M. malmoense, M. marinum, M. szulgai, M. tuberculosis-M. bovis complex, and M. xenopi) were exactly identified when MIS sample preparation was combined with the TMSH technique. In addition to the data from the MIS library search, data published in literature (5, 7, 12) were considered. The knowledge of MACPs confirmed or extended the results of the MIS library search.
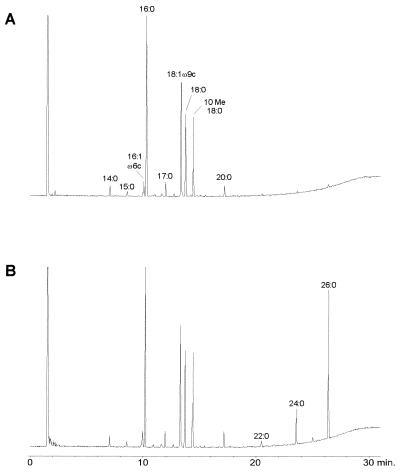
A comparison of FAME profiles of M. tuberculosis resulting from sample preparation by MIS alone (Fig. 1A) with that by MIS combined with the TMSH technique (Fig. 1B) is shown. All typical patterns (5, 7, 12) of FAMEs and MACPs (C26:0 > C24:0) resulting from the use of the TMSH modification of the sample preparation technique are shown. Higher temperatures (260 to 320°C) in the injection port did not generate higher amounts of MACPs. Visual comparison shows that the two profiles in Fig. 1 are similar in the range of the fatty acid chain length, between C10 and C20, which is stressed by the calculated similarity indices (Table 3). Thus, the MIS library search can be used without problems when either of the two sample preparation techniques is used.
FIG. 1.
FAME pattern of M. tuberculosis resulting from the use of conventional MIS sample preparation (A) and MIS sample preparation combined with the TMSH technique (B).
TABLE 3.
MIS library search of one M. tuberculosis strain by using MIS sample preparation and MIS sample preparation combined with the TMSH technique
| Sample prepn method and comparator | Similarity index |
|---|---|
| MIS | |
| Mycobacterium species | 0.850a |
| M. tuberculosis | 0.850a |
| M. bovis | 0.562a |
| Other rapidly growing mycobacteria | 0.450b |
| MIS plus TMSH technique | |
| Mycobacterium species | 0.844a |
| M. tuberculosis | 0.844a |
| M. bovis | 0.535a |
| Other rapidly growing mycobacteria | 0.437b |
Value when compared with the M. tuberculosis-M. bovis complex.
Value when compared with the Centers for Disease Control group.
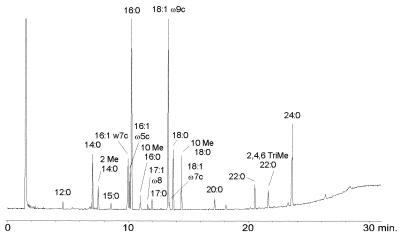
Figure 2 shows the FAME and MACP profile of M. szulgai. The typical MACP profile (finding 2,4,6-trimethyl 22:0 and 24:0 > 22:0) is essential to distinguish M. szulgai from M. kansasii and M. marinum, which are sometimes not sufficiently discriminated after MIS sample preparation alone. The detection of MACPs after TMSH-modified sample preparation allows the discrimination of the three species when the chromatograms are compared with those in the literature (7). A computer-assisted identification by MIS would be possible after updating the MIS library data with patterns from the TMSH technique of sample preparation.
FIG. 2.
FAME and MACP profile of M. szulgai resulting from the use of MIS sample preparation combined with the TMSH technique.
Reproducibility of FAME, alcohol, and MACP analysis.
When the same strain is cultured under standardized conditions and analyzed repeatedly, there is a good reproducibility of the FAME, alcohol, and MACP profiles both qualitatively and quantitatively; similarity indices and blotting dendrograms based on Euclidian distance and principal-component analysis (8) of chromatographic patterns were used for comparison. Samples of the same strain were linked at a Euclidian distance of 2.5 or less (8).
DISCUSSION
The identification of mycobacteria by microbiological culture techniques is difficult to perform and requires up to several weeks. The identification by gene probes is highly specific, but in contrast to chromatographic methods, this method of identification is very expensive and the characterization of one single unknown species requires a series of many gene probes.
Cellular fatty acid compositions are widely used as a basis for the characterization of bacteria. The identification is done by one single chromatographic run, which usually requires less than half an hour. The MIS uses quantitative analyses of fatty acid profiles for reliable identification of many bacteria to the subspecies level. MIS sample preparation is simple and allows the identification of FAMEs and secondary alcohols but not the identification of the MACPs, a characteristic component of the mycobacterial cell wall. Thus, a reliable identification of mycobacteria must take into consideration the quantitative analyses of these molecules (7, 12).
TMSH (3, 10–13) reproducibly converts fatty acids bound in biomolecules such as phospholipids and/or glycerides into the corresponding FAMEs. The transesterification can be performed at room temperature in a fast, single-step reaction. MACPs and secondary alcohols are also released from mycobacteria under these conditions. The profiles of the chromatograms match well those obtained from other sample preparation techniques that need approximately 16 h to prepare MACPs (7).
The simple modification of sample preparation of applying 1 drop of TMSH reagent produces a pattern of FAMEs, alcohols, and MACPs in one single GC run. The results of the conventional MIS library search are not disturbed by the use of TMSH. The identification of long-chain fatty alcohols and MACPs is performed by MIS after ECL values are written into the peak library. The MACP patterns for mycobacteria presented in this study are consistent with those previously reported by others (7). These patterns enable one to differentiate between strains with similar indices of identification or identification of species outside the normal MIS library. Examples of this resolving power are found in differentiating M. avium, M. celatum, and M. xenopi by the MIS system. These species (data not shown) are characterized by similar FAME profiles in the region between C10 and C20 fatty acids (2). The dilemma is that there is no entry for M. celatum in the MIS library and the MIS library search often does not allow reliable discrimination between M. xenopi and M. avium. By using MACP and long-chain fatty alcohol patterns, differentiation of these three species is simple. M. xenopi is characterized by high amounts of C22:0 alcohol and C26:0, M. celatum contains high amounts of C24:0 and C26:0 fatty acids and lacks C22:0 alcohol. M. avium contains high amounts of C24:0; C22:0 alcohol and C26:0 are not found. M. celatum cannot be identified by conventional microbiological testing. Phenotypically, M. celatum is similar to M. avium, but genetic probes for M. avium do not react with it, suggesting that a specific probe is required for its identification (1). In contrast, GC of FAMEs with the enhanced ability to analyze mycolic acid cleavage fragments is less time-consuming and less expensive than DNA methods and can readily differentiate these three species.
The method described herein is helpful for users of the MIS since no significant change in sample preparation is necessary. A conventional library search can be done since the FAME pattern of MIS sample preparation is not changed by adding TMSH. The TMSH modification of sample preparation is not time-consuming since the reagents act immediately at room temperature and probes can be introduced directly into the GC. In contrast to conventional MIS sample preparation of mycobacteria, that of the chromatographic system described in this report is not disturbed by any residues of mycolic acids. So, FAME patterns of other bacteria can be investigated directly after running TMSH-treated probes.
Library generation software of MIS enables the creation of a new library for identification of mycobacteria based on TMSH-assisted sample preparation by using qualitative and quantitative profiles of FAMEs, alcohols, and MACPs. As long as such a library tool does not exist, the combination of a conventional library search report and an observation of a typical MACP pattern is an excellent tool for the identification of mycobacteria.
ACKNOWLEDGMENT
We thank W. Bartosik for excellent technical assistance.
REFERENCES
- 1.Böttger E C. Despite longer experiences with fatty acid profiles, DNA-based analysis offers several advantages. ASM News. 1996;62:247–250. [Google Scholar]
- 2.Butler W R, O’Connor S P, Yakrus M A, Smithwick R W, Plikaytis B B, Moss C W, Floyd M M, Woodley C L, Kilburn J O, Vadney F S, Gross W M. Mycobacterium celatum sp. nov. Int J Syst Bacteriol. 1993;43:539–548. doi: 10.1099/00207713-43-3-539. [DOI] [PubMed] [Google Scholar]
- 3.Butte W. Rapid method for the determination of fatty acid profiles from fats and oils using trimethylsulfonium hydroxide for transesterification. J Chromatogr. 1983;261:142–145. [Google Scholar]
- 4.Chou S, Kasatiya S. Misidentification by GLC. ASM News. 1997;63:121–122. [Google Scholar]
- 5.Kaneda K, Imaizumi S, Yano I. Distribution of C22-, C24- and C26- α-unit-containing mycolic acid homologues in mycobacteria. Microbiol Immunol. 1995;39:563–570. doi: 10.1111/j.1348-0421.1995.tb02243.x. [DOI] [PubMed] [Google Scholar]
- 6.Lambert M A, Moss C W, Silcox V A, Good R C. Analysis of mycolic acid cleavage products and cellular fatty acids of Mycobacterium species by capillary gas chromatography. J Clin Microbiol. 1986;23:731–736. doi: 10.1128/jcm.23.4.731-736.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luquin M, Ausina V, Calahorra F L, Belda F, Barceló M G, Celma C, Prats G. Evaluation of practical chromatographic procedures for identification of clinical isolates of mycobacteria. J Clin Microbiol. 1991;29:120–130. doi: 10.1128/jcm.29.1.120-130.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MIDI, Inc. Microbial Identification System, operating manual. Newark, Del: MIDI, Inc.; 1995. [Google Scholar]
- 9.Miller L, Berger T. Hewlett-Packard application note 228-41. Avondale, Pa: Hewlett-Packard; 1985. Bacteria identification by gas chromatography of whole cell fatty acids; pp. 1–8. [Google Scholar]
- 10.Müller K-D, Husmann H, Nalik H P, Schomburg G. Trans-esterification of fatty acids from microorganisms and human blood serum by trimethylsulfonium hydroxide (TMSH) for GC analysis. Chromatographia. 1990;30:245–248. [Google Scholar]
- 11.Müller K-D, Husmann H, Nalik H P. A new and rapid method for the assay of bacterial fatty acids using high resolution capillary gas chromatography and trimethylsulfonium hydroxide. Zentbl Bakteriol. 1990;274:174–182. doi: 10.1016/s0934-8840(11)80100-3. [DOI] [PubMed] [Google Scholar]
- 12.Müller K-D, Nalik H P, Schmid E N, Husmann H, Schomburg G. Fast identification of mycobacterium species by GC analysis with trimethylsulfonium hydroxide (TMSH) for transesterification. J High Resolut Chromatogr. 1993;16:161–165. [Google Scholar]
- 13.Nalik H P, Müller K-D, Ansorg R. Rapid identification of Legionella species from a single colony by gas-liquid chromatography with trimethylsulfonium hydroxide for transesterification. J Med Microbiol. 1992;36:371–376. doi: 10.1099/00222615-36-6-371. [DOI] [PubMed] [Google Scholar]
- 14.Sasser M. MIDI technical note 101. Newark, Del: MIDI, Inc.; 1990. Identification of bacteria by gas chromatography of cellular fatty acids; pp. 1–7. [Google Scholar]