Abstract
Differential PCR was performed to determine the copy number of rRNA genes in Pneumocystis carinii f. sp. hominis. Two different reference genes, thymidylate synthase (TS) and beta-tubulin (BTU) genes, were used. Primers for the internal transcribed spacer (ITS) region of nuclear rRNA genes and either the TS or BTU gene were mixed together to perform PCR on seven different bronchoalveolar lavage specimens from patients with P. carinii pneumonia. The radioactivity derived from the incorporated radioactive nucleotides of each PCR product band was then used to calculate the copy number of the ITS relative to that of the TS or BTU gene. The copy number ratio between the ITS and the TS gene was determined to be 0.8, and that between the ITS and the BTU gene was also 0.8. These results suggest that the ITS has the same copy number as the TS or BTU gene. Since the copy number of the TS or BTU gene is presumed to be 1, the results also suggest that P. carinii f. sp. hominis has only one copy of the ITS and thus one copy of the nuclear rRNA genes. Therefore, two types of ITS sequences derived from a specimen would indicate that the patient is infected by two types of P. carinii f. sp. hominis.
Pneumocystis carinii f. sp. hominis causes pneumonia in immunocompromised humans. It is analogous to the form of P. carinii that was first found in rats. P. carinii is a eukaryotic organism with an uncertain taxonomical classification. It was first identified as a form of trypanosome by Chagas (5) but was later reclassified as a different organism by Delanoe and Delanoe (6, 7). P. carinii was traditionally thought to be a protozoan because it has structures common to protozoa and is susceptible to antiprotozoan agents such as pentamidine and trimethoprim-sulfamethoxazole. Currently, P. carinii is considered to be a fungus, based on its ultrastructure, certain elements of its cellular biology, and nucleotide sequences of certain genetic loci. The ultrastructure of the cyst wall of P. carinii resembles that of fungi (30). Furthermore, P. carinii and fungal cell walls share a common epitope which has been identified by a monoclonal antibody (29). P. carinii also lacks some of the characteristic protozoan organelles, such as rhoptries, subpellicular tubules, and conoids (39). The nucleotide sequence of the 18S rRNA gene of P. carinii is more like that of fungi than that of protozoa (11, 35). The gene for elongation factor 3 (EF-3), which is found exclusively in fungi, has been found in P. carinii (40). In addition, thymidylate synthase (TS) and dihydrofolate reductase are two distinct enzymes in P. carinii (12, 14), whereas in protozoa, those activities are contained within a single bifunctional protein (36).
The entire nuclear rRNA gene cluster of P. carinii from rats has been cloned and sequenced (11, 13, 25), and its transcripts have been characterized (24). Like other organisms, P. carinii has three species of nuclear rRNA transcripts: 18S, 5.8S, and 26S. The regions located between 18S and 5.8S and between 5.8S and 26S are referred to as internal transcribed spacer (ITS) regions. The region located between the 18S and 5.8S rRNA genes is called ITS1, and the other is called ITS2. ITS sequences of P. carinii f. sp. hominis have been found to vary among different isolates. This nucleotide sequence variation has been used for typing, and approximately 60 different ITS sequences have been found (23). Some specimens were found to contain P. carinii f. sp. hominis with more than one type of ITS sequence (19–21, 23, 26, 37). It is not clear whether these different ITS sequence types represent different strains of P. carinii f. sp. hominis or whether they are derived from different copies of the rRNA genes of the same organism. In order to answer this question, we have determined the copy number of ITS in P. carinii f. sp. hominis.
MATERIALS AND METHODS
Specimens.
Bronchoalveolar lavage (BAL) fluids from seven different patients with P. carinii pneumonia were obtained from the Clinical Microbiology Laboratory, Indiana University Hospital, after routine diagnostic procedures had been completed. These specimens were collected during the year 1997 and were processed for PCR as described previously (22).
PCR.
All PCRs for this study were performed under the same conditions. The reaction volume was 20 μl, and the reaction mixture contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 40 μM each deoxynucleoside triphosphate, 5 pmol of each PCR primer, 10 μCi of α-35S-dCTP, 1.2 units of Taq polymerase, and 50 ng of template DNA isolated from BAL specimens. Temperature cycles for each PCR included 1 cycle of 94°C for 2 min; 5 cycles of 94°C for 1 min and 68°C for 2 min; 28 cycles of 94°C for 40 s, 65°C for 40 s, and 68°C for 40 s; and 1 cycle of 72°C for 2 min.
Quantitation of PCR products.
The amplified products were electrophoresed on an 8% polyacrylamide gel. A photograph of the gel was taken. The gel was dried and then exposed to X-ray film in order to obtain an autoradiogram. The gel was also exposed to a Storage Phospho Screen (Kodak, Rochester, N.Y.), and the radioactivity in each PCR product band captured by the screen was determined with a PhosphorImager (Storm 840; Molecular Dynamics, Sunnyvale, Calif.).
RESULTS
Since it was difficult to obtain a sufficient amount of P. carinii f. sp. hominis DNA to perform Southern blot hybridization, we decided to use differential PCR to determine the copy number of the ITS of P. carinii f. sp. hominis. A portion of the ITS regions is amplified simultaneously with a single-copy reference gene in a multiplex PCR. The copy number of the ITS is then determined based on the quantity of the ITS PCR product compared to that of the reference gene. Differential PCR is usually performed with single-step PCR. However, there was no single-step PCR capable of amplifying ITS regions at the time when this study was initiated. We speculated that the inability to amplify the ITS regions by single-step PCR with primers that have been described (23, 26–28) was due to the excessive length of the intended target. Therefore, we decided to develop another PCR method which amplifies a smaller fragment of the target. Upon closer examination of the ITS sequences of different types of P. carinii f. sp. hominis, two areas located at nucleotide positions 16 to 39 and 118 to 142 in the ITS2 region were found to be conserved among all 14 types of ITS2 (23). A pair of primers, FT4 (5′-CAAGCAGAAAAAAGGGGATTGGGC-3′) and RT4 (5′-CTTTCCCAGCGAATTTTTACGACAC-3′), was designed based on the conserved sequences. The FT4 sequence is located at ITS2 nucleotide positions 16 to 39, and the RT4 sequence is located at positions 118 to 142. This pair of primers would amplify a fragment of 127 bp. After many trials of different PCR conditions, the conditions described in Materials and Methods were found to be optimal for amplification.
To perform differential PCR for determination of the ITS copy number, a single-copy reference gene is required. Since the sequence of the TS gene of P. carinii f. sp. hominis is available (14, 31) and this gene is not known to have more than one copy, it was chosen as a single-copy reference gene. Many different pairs of PCR primers were tried, but only one pair, TS-F (5′-CAGGTCAAGGAGTTGACCAACTAG-3′, nucleotide positions 440 to 463) and TS-R (5′-TTAAAGGGAACACCTAGCCCCATG-3′, nucleotide positions 751 to 774), was found to be able to amplify the TS gene of P. carinii f. sp. hominis under the PCR conditions which were determined to be optimal for the single-step ITS PCR. This pair of primers amplifies a fragment of 335 bp.
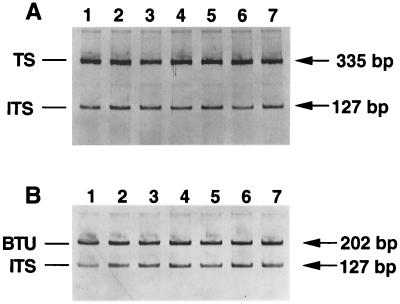
The differential PCR with the ITS as the target and the TS gene as the reference was then applied to seven BAL specimens that were confirmed microscopically to contain P. carinii f. sp. hominis. All seven specimens produced both TS (335 bp) and ITS (127 bp) bands in this differential PCR. Each specimen was assayed three times. A representative photograph of PCR product bands is shown in Fig. 1. The radioactivity derived from incorporated α-35S-dCTP in each band was determined and recorded (Table 1). The copy number of the ITS relative to that of the TS gene was then calculated based on the radioactivity counts of these bands. Since the number of cytosine residues in the amplified TS fragment is 2.8 times that in the ITS (115 versus 41), the ITS copy number was determined by calculating (mean ITS counts × 2.8)/mean TS counts; thus, (50,823 × 2.8)/178,648 (Table 1). In this calculation, the sums of the means of three repeats for all seven TS and ITS bands were used and a value of 0.8 was obtained. This result suggests that the copy number of the ITS is 0.8 times that of the TS gene. The copy number of the TS gene is presumed to be 1, since it is not known to have more than one copy. Therefore, the copy number of the ITS would be 0.8. Because the minimum copy number of a gene is 1, this result implies that P. carinii f. sp. hominis has one copy of the ITS and thus one copy of nuclear rRNA genes.
FIG. 1.
Differential PCR for determination of the ITS copy number, using the TS gene (A) or the BTU gene (B) as the reference gene. Seven different BAL specimens were examined. The sizes of PCR products are given on the right.
TABLE 1.
Radioactivity counts of PCR products from seven different BAL specimens
| Specimen no. | Radioactivity count (mean ± SD)a of product from:
|
|||
|---|---|---|---|---|
| TS-ITS differential PCR
|
BTU-ITS differential PCR
|
|||
| TS | ITS | BTU | ITS | |
| 1 | 26,236 ± 575 | 6,346 ± 163 | 31,320 ± 10,061 | 10,036 ± 2,412 |
| 2 | 24,328 ± 167 | 8,629 ± 339 | 33,771 ± 929 | 13,782 ± 958 |
| 3 | 21,060 ± 3,092 | 9,047 ± 1,250 | 31,032 ± 194 | 16,963 ± 1,004 |
| 4 | 27,174 ± 904 | 6,274 ± 162 | 33,589 ± 2,112 | 17,240 ± 1,822 |
| 5 | 26,121 ± 1,651 | 7,778 ± 318 | 33,922 ± 722 | 14,056 ± 290 |
| 6 | 28,569 ± 652 | 4,820 ± 188 | 32,898 ± 5,696 | 19,682 ± 1,237 |
| 7 | 25,160 ± 674 | 7,929 ± 204 | 35,736 ± 4,650 | 16,357 ± 1,476 |
| Sumb | 178,648 ± 2,393 | 50,823 ± 1,502 | 232,268 ± 1,621 | 108,116 ± 3,116 |
Means are averages of counts from three repeats.
Sum of radioactivity of PCR products of all seven BAL specimens.
To confirm this result, another differential PCR, using a different reference gene, the beta-tubulin (BTU) gene, was performed (9, 10). The same approaches were used for this PCR as for that with the TS gene. Many different pairs of primers were tried, and primers Tu-F (5′-TGGTTCCCTCACCAAAAGTTTCCG-3′, nucleotide positions 1585 to 1608) and Tu-R (5′-GGATCCGGCAATTTCAATGTACGC-3′, nucleotide positions 1763 to 1786) were found to be the most effective in amplifying the intended target under the conditions that were found to be optimal for ITS. Differential PCR with both ITS and BTU gene primers was then performed on all seven BAL specimens. Each specimen was again assayed three times. The BTU gene PCR amplifies a fragment of 202 bp. As in the TS-ITS differential PCR, the radioactivity in each PCR product band was determined and used to calculate the ITS copy number (Table 1). In this system, the number of cytosine residues in the amplified BTU gene fragment is 1.76 times that in the ITS (72 versus 41). A value of 0.8 was obtained when the calculation (mean ITS counts × 1.76)/mean TS counts was performed; thus, (10,816 × 1.76)/232,268 (Table 1).
DISCUSSION
In this study, experiments were performed to determine the copy number of the ITS of P. carinii f. sp. hominis. Since ITS regions are parts of the nuclear rRNA genes in P. carinii f. sp. hominis, the copy number of the ITS is also the copy number of rRNA genes. With the TS-ITS differential PCR, the copy number of the ITS was determined to be 0.8 times that of the TS gene. The copy number of the TS gene is presumed to be 1, since it is not known to have more than one copy. Therefore, the copy number of the ITS would be 0.8. Because the minimum copy number of a gene is 1, this result implies that P. carinii f. sp. hominis has one copy of the ITS and thus one copy of nuclear rRNA genes. Using the BTU gene as a reference, we obtained the same result. The copy number ratio between the ITS and the BTU gene was also determined to be 0.8. Since the same results were obtained from two different methods, it is quite certain that the data are reliable.
Copy number determination is normally achieved by Southern blot hybridization. Unfortunately, we were unable to obtain any clinical specimens that contained sufficient numbers of P. carinii f. sp. hominis organisms for Southern blot analysis. Therefore, we used differential PCR, which has been used to determine the copy number of many different genes, including C-myc (1, 32), N-myc (3, 17), erbB (4, 8, 16, 33), HER-2/neu (38), EGFR and MDM-2 (18), and a parathyroid hormone-related peptide gene (34). Since many factors can affect the efficiency of PCR, each specimen was examined under the same conditions three different times, and the mean of radioactivity counts of PCR products derived from incorporated radioactive nucleotides in the three reactions was used to calculate the copy number. We did observe variations in PCR efficiency between runs. For example, the standard deviations of radioactivity counts for the TS PCR of the TS-ITS differential PCR range from 167 to 3,092 (Table 1). Similar degrees of variation were also seen in other PCRs. To minimize the effect of these variations in copy number determination, we used the sum of the mean counts of all seven specimens for the calculation and obtained the same result from the two different differential PCRs.
The determination of the copy number of P. carinii f. sp. hominis rRNA genes has an important implication in typing. As described previously, nucleotide sequence variations in the ITS region can be used to type P. carinii f. sp. hominis isolates. In these typing studies, a number of specimens were found to contain more than one ITS type. It was uncertain whether those specimens actually contained more than one type of P. carinii f. sp. hominis or whether multiple types of ITS represent multiple copies of rRNA genes in the same P. carinii f. sp. hominis isolate. Since the copy number of P. carinii f. sp. hominis rRNA genes has been determined to be 1, multiple types of ITS sequences would represent multiple types of P. carinii f. sp. hominis in a specimen.
The copy number of rRNA genes in rat P. carinii has been determined to be less than 2 (15). In this study, we showed that P. carinii f. sp. hominis has only one copy of nuclear rRNA genes. Both studies reveal an unusual property of P. carinii, since most organisms have more than one copy of nuclear rRNA genes (2). Although we have determined that P. carinii f. sp. hominis has only one copy of nuclear rRNA genes, this result awaits confirmation by sequencing of the entire P. carinii f. sp. hominis genome.
ACKNOWLEDGMENT
This study was supported by NIH grant RO1 AI 34304.
REFERENCES
- 1.Abou-Elella A, Gramlich T, Fritsch C, Gansler T. c-myc amplification in hepatocellular carcinoma predicts unfavorable prognosis. 1996. Mod Pathol. 1996;9:95–98. [PubMed] [Google Scholar]
- 2.Birnstiel M L, Chipchase M, Speirs J. The ribosomal RNA cistrons. Prog Nucleic Acid Res Mol Biol. 1971;11:351–389. doi: 10.1016/s0079-6603(08)60332-3. [DOI] [PubMed] [Google Scholar]
- 3.Boerner S, Squire J, Thorner P, McKenna G, Zielenska M. Assessment of MYCN amplification in neuroblastoma biopsies by differential polymerase chain reaction. Pediatr Pathol. 1994;14:823–832. doi: 10.3109/15513819409037680. [DOI] [PubMed] [Google Scholar]
- 4.Brandt B, Vogt U, Harms F, Bosse U, Zanker K S, Assmann G. Double-differential PCR for gene dosage estimation of erbB oncogenes in benign and cancer tissues and comparison to cellular DNA content. Gene. 1995;159:29–34. doi: 10.1016/0378-1119(94)00651-8. [DOI] [PubMed] [Google Scholar]
- 5.Chagas C. Nova tripanozomiaza humana. Mem Inst Oswaldo Cruz. 1909;1:159–218. [Google Scholar]
- 6.Delanoe P, Delanoe M. Sur les rapports des kystos de carinii le Trypanosoma lewis. C R Acad Sci. 1912;155:658–660. [Google Scholar]
- 7.Delanoe P, Delanoe M. De la rareté de Pneumocystis carinii chez cobayes de la région de Paris; absence de kysts chez d’autres animaux lapin, grenouille, zanguilles. Bull Soc Pathol Exot. 1914;7:271–274. [Google Scholar]
- 8.Deng G, Yu M, Chen L C, Moore D, Kurisu W, Kallioniemi A, Waldman F M, Collins C, Smith H S. Amplifications of oncogene erbB-2 and chromosome 20q in breast cancer determined by differentially competitive polymerase chain reaction. Breast Cancer Res Treat. 1996;40:271–281. doi: 10.1007/BF01806816. [DOI] [PubMed] [Google Scholar]
- 9.Dyer M, Volpe F, Delves C J, Somia N, Burns S, Scaife J G. Cloning and sequence of a beta-tubulin cDNA from Pneumocystis carinii: possible implications for drug therapy. Mol Microbiol. 1992;6:991–1001. doi: 10.1111/j.1365-2958.1992.tb02165.x. [DOI] [PubMed] [Google Scholar]
- 10.Edlind T D, Bartlett M S, Weinberg G A, Prah G N, Smith J W. The beta-tubulin gene from rat and human isolates of Pneumocystis carinii. Mol Microbiol. 1992;6:3365–3373. doi: 10.1111/j.1365-2958.1992.tb02204.x. [DOI] [PubMed] [Google Scholar]
- 11.Edman J C, Kovacs J A, Masur H, Santi D V, Elwood H J, Sogin M L. Ribosomal RNA sequence shows Pneumocystis carinii to be a member of the fungi. Nature. 1988;334:519–522. doi: 10.1038/334519a0. [DOI] [PubMed] [Google Scholar]
- 12.Edman J C, Edman U, Cao M, Lundgren B, Kovacs J A, Santi D V. Isolation and expression of the Pneumocystis carinii dihydrofolate reductase gene. Proc Natl Acad Sci USA. 1989;86:8625–8629. doi: 10.1073/pnas.86.22.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edman J C, Kovacs J A, Masur H, Santi D V, Elwood H J, Sogin M L. Ribosomal RNA genes of Pneumocystis carinii. J Protozool. 1989;36:18S–20S. doi: 10.1111/j.1550-7408.1989.tb02672.x. [DOI] [PubMed] [Google Scholar]
- 14.Edman U, Edman J C, Lundgren B, Santi D V. Isolation and expression of Pneumocystis carinii thymidylate synthase gene. Proc Natl Acad Sci USA. 1989;86:6503–6507. doi: 10.1073/pnas.86.17.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giuntoli D, Stringer S L, Stringer J R. Extraordinarily low number of ribosomal RNA genes in P. carinii. J Eukaryot Microbiol. 1994;41:88S. [PubMed] [Google Scholar]
- 16.Gramlich T L, Cohen C, Fritsch C, DeRose P B, Gansler T. Evaluation of c-erbB-2 amplification in breast carcinoma by differential polymerase chain reaction. Am J Clin Pathol. 1994;101:493–499. doi: 10.1093/ajcp/101.4.493. [DOI] [PubMed] [Google Scholar]
- 17.Huddart S N, Mann J R, McGukin A G, Corbett R. MYCN amplification by differential PCR. Pediatr Hematol Oncol. 1993;10:31–34. doi: 10.3109/08880019309016525. [DOI] [PubMed] [Google Scholar]
- 18.Hunter S B, Abbott K, Varma V A, Olson J J, Barnett D W, James C D. Reliability of differential PCR for the detection of EGFR and MDM2 gene amplification in DNA extracted from FFPE glioma tissue. J Neuropathol Exp Neurol. 1995;54:57–64. doi: 10.1097/00005072-199501000-00007. [DOI] [PubMed] [Google Scholar]
- 19.Keely S P, Stringer J R. Sequences of Pneumocystis carinii f. sp. hominis strains associated with recurrent pneumonia vary at multiple loci. J Clin Microbiol. 1997;35:2745–2747. doi: 10.1128/jcm.35.11.2745-2747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latouche S, Ortona E, Mazars E, Margutti P, Tamburrini E, Siracusano A, Guyot K, Nigou M, Roux P. Biodiversity of Pneumocystis carinii hominis: typing with different DNA regions. J Clin Microbiol. 1997;35:383–387. doi: 10.1128/jcm.35.2.383-387.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latouche S, Poirot J-L, Bernard C, Roux P. Study of internal transcribed spacer and mitochondrial large-subunit genes of Pneumocystis carinii hominis isolated by repeated bronchoalveolar lavage from human immunodeficiency virus-infected patients during one or several episodes of pneumonia. J Clin Microbiol. 1997;35:1687–1690. doi: 10.1128/jcm.35.7.1687-1690.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C H, Lu J J, Bartlett M S, Durkin M M, Liu T H, Wang J, Jiang B, Smith J W. Nucleotide sequence variation in Pneumocystis carinii strains that infect humans. J Clin Microbiol. 1993;31:754–757. doi: 10.1128/jcm.31.3.754-757.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C H, Helweg-Larsen J, Lundgren B, Lundgren J D, Tang X, Jin S, Li B, Bartlett M S, Lu J J, Olsson M, Luvas S B, Roux P, Cargnel A, Atzori C, Matos O, Smith J W. Update on Pneumocystis carinii f. sp. hominis typing based on nucleotide sequence variations in the internal transcribed spacer regions of rRNA Genes. J Clin Microbiol. 1998;36:734–741. doi: 10.1128/jcm.36.3.734-741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin H, Niu M T, Yoganathan T, Buck G A. Characterization of the rRNA-encoding genes and transcripts, and a group-I self-splicing intron in Pneumocystis carinii. Gene. 1992;119:163–173. doi: 10.1016/0378-1119(92)90268-t. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Rocourt M, Pan S, Liu C, Leibowitz M J. Sequence and variability of the 5.8S and 26S rRNA genes of Pneumocystis carinii. Nucleic Acids Res. 1992;20:3763–3772. doi: 10.1093/nar/20.14.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J J, Bartlett M S, Shaw M M, Queener S F, Smith J W, Ortiz-Rivera M, Leibowitz M J, Lee C H. Typing of Pneumocystis carinii strains that infect humans based on nucleotide sequence variations of internal transcribed spacers of rRNA genes. J Clin Microbiol. 1994;32:2904–2912. doi: 10.1128/jcm.32.12.2904-2912.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu J J, Bartlett M S, Smith J W, Lee C H. Typing of Pneumocystis carinii strains with type-specific oligonucleotide probes derived from nucleotide sequences of internal transcribed spacers of rRNA genes. J Clin Microbiol. 1995;33:2973–2977. doi: 10.1128/jcm.33.11.2973-2977.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J J, Chen C H, Bartlett M S, Smith J W, Lee C H. Comparison of six different PCR methods for the detection of Pneumocystis carinii. J Clin Microbiol. 1995;33:2785–2788. doi: 10.1128/jcm.33.10.2785-2788.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundgren B, Kovacs J A, Nelson N N, Stock F, Martinez A, Gill V J. Pneumocystis carinii and specific fungi have a common epitope, identified by a monoclonal antibody. J Clin Microbiol. 1992;30:391–395. doi: 10.1128/jcm.30.2.391-395.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto Y, Matsuda S, Tegoshi T. Yeast glucan in the cyst wall of Pneumocystis carinii. J Protozool. 1989;36:21S–22S. doi: 10.1111/j.1550-7408.1989.tb02674.x. [DOI] [PubMed] [Google Scholar]
- 31.Mazars E, Odberg-Ferragut C, Dei-Cas E, Fourmaux M-E, Aliouat E M, Brun-Pascaud M, Mougeot G, Camus D. Polymorphism of the thymidylate synthase gene of Pneumocystis carinii from different host species. J Eukaryot Microbiol. 1995;42:26–32. doi: 10.1111/j.1550-7408.1995.tb01536.x. [DOI] [PubMed] [Google Scholar]
- 32.Pascale R M, DeMiglio M R, Muroni M R, Simile M M, Daino L, Seddaiu M A, Nufris A, Gaspa L, Deiana L, Feo F. c-myc amplification in pre-malignant and malignant lesions induced in rat liver by the resistant hepatocyte model. Int J Cancer. 1996;68:136–142. doi: 10.1002/(SICI)1097-0215(19960927)68:1<136::AID-IJC24>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 33.Roetger A, Brandt B, Barnekow A. Competitive-differential polymerase chain reaction for gene dosage estimation of erbB-1 (egfr), erbB-2, and erbB-3 oncogenes. DNA Cell Biol. 1997;16:443–448. doi: 10.1089/dna.1997.16.443. [DOI] [PubMed] [Google Scholar]
- 34.Sidler B, Alpert L, Henderson J E, Deckelbaum R, Amizuka N, Silva J E, Goltzman D, Karaplis A C. Amplification of the parathyroid hormone-related peptide gene in a colonic carcinoma. J Clin Endocrinol Metab. 1996;81:2841–2847. doi: 10.1210/jcem.81.8.8768840. [DOI] [PubMed] [Google Scholar]
- 35.Stringer S L, Stringer J R, Blase M A, Walzer P D, Cushion M T. Pneumocystis carinii: sequence from ribosomal RNA implies a close relationship with fungi. Exp Parasitol. 1989;68:450–461. doi: 10.1016/0014-4894(89)90130-6. [DOI] [PubMed] [Google Scholar]
- 36.Toth I, Lazar G, Goodman H. Purification and immunochemical characterization of a dihydrofolate reductase-thymidylate synthase enzyme complex from wild-carrot cells. EMBO J. 1987;6:1853–1858. doi: 10.1002/j.1460-2075.1987.tb02443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsolaki A G, Miller R F, Underwood A P, Banerji S, Wakefield A E. Genetic diversity at the internal transcribed spacer regions of the rRNA operon among isolates of Pneumocystis carinii from AIDS patients with recurrent pneumonia. J Infect Dis. 1996;174:141–156. doi: 10.1093/infdis/174.1.141. [DOI] [PubMed] [Google Scholar]
- 38.Valerón P F, Chirino R, Fernandez L, Torres S, Navarro D, Aguiar J, Cabrera J J, Diaz-Chico B N, Diaz-Chico J C. Validation of a differential PCR and an ELISA procedure in studying HER-2/neu status in breast cancer. Int J Cancer. 1996;65:129–133. doi: 10.1002/(SICI)1097-0215(19960117)65:2<129::AID-IJC1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 39.Vavra J, Kucera K. Pneumocystis carinii Delanöe, its ultrastructure and ultrastructural affinities. Protozoology. 1970;17:463–483. doi: 10.1111/j.1550-7408.1970.tb04715.x. [DOI] [PubMed] [Google Scholar]
- 40.Ypma-Wong M F, Fonzi W A, Sypherd P S. Fungus-specific translation elongation factor 3 gene present in Pneumocystis carinii. Infect Immun. 1992;60:4140–4145. doi: 10.1128/iai.60.10.4140-4145.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]