Abstract
We compared the sensitivity and accuracy of the NucliSens assay and those of both the standard and modified (addition of a new primer set, primer mix 1, supplied by Roche) Amplicor HIV Monitor assays to quantify human immunodeficiency virus type 1 (HIV-1) RNA in persons infected with HIV-1 subtype A in Abidjan, Côte d’Ivoire. Seventy-one plasma samples from HIV-1-seropositive persons at different stages of HIV infection and 15 samples from HIV antibody-negative persons were analyzed. The HIV-1 genetic subtype was determined either by DNA sequencing or by a restriction fragment length polymorphism assay. Of the 71 samples, 70 (98%) were subtype A and 1 was subtype G. Of the 70 subtype A samples, the proportion of RNA-positive plasma samples and mean HIV-1 RNA levels were significantly higher by the modified HIV Monitor assay (n = 67 [96%]; mean RNA levels, 5.2 log10 HIV-1 RNA copies/ml) than the NucliSens assay (n = 56 [80%]; 4.3 log10 HIV-1 RNA copies/ml) or the standard HIV Monitor assay (n = 44 [63%]; mean RNA levels, 3.8 log10 HIV-1 RNA copies/ml) (all P values were <0.05). The HIV-1 RNA levels by the modified HIV Monitor assay correlated significantly with those by the NucliSens assay (r = 0.76; P < 0.001) and the standard HIV Monitor assay (r = 0.57; P < 0.001), as did the RNA levels by the NucliSens and the standard HIV Monitor assays (r = 0.60; P < 0.001). Lower CD4 cell counts were significantly correlated with higher HIV-1 RNA levels by all three assays (r = −0.47 for the NucliSens assay, −0.45 for the standard HIV Monitor assay, and −0.62 for the modified HIV Monitor assay). These results indicate that the modified HIV Monitor assay has the highest sensitivity and efficiency at quantifying the levels of RNA in persons infected with HIV-1 subtype A and thus constitutes a valuable tool for the monitoring of RNA levels in areas of Africa were HIV-1 subtype A is predominant.
The development of commercial nucleic acid-based assays for the quantification of human immunodeficiency virus type 1 (HIV-1) RNA represents a major advance in the clinical management and follow-up of persons with HIV infection. These assays, which are based on reverse transcriptase PCR, nucleic acid sequence-based amplification (NASBA), or the branched DNA technique, were developed and validated with persons infected with HIV-1 subtype B, the predominant HIV-1 subtype in North America and Europe. In several comparative evaluations, the three commercially available nucleic acid-based techniques have proven to be efficient for the quantification of the HIV-1 RNA load in persons infected with HIV-1 subtype B viruses and are routinely used in clinical practice (2, 3, 4, 6, 10–13). However, the high degree of sequence diversity that characterizes the HIV-1 subtypes may impair the sensitivity of currently available nucleic acid-based assays. To date, limited data exist regarding the sensitivities and accuracies of these assays for the detection of other known subtypes of HIV group M (subtypes A to J) or HIV-1 group O. One study indicates that the detection of HIV-1 subtype A, the predominant subtype in Africa (8), is particularly difficult, particularly by the Roche Amplicor HIV-1 Monitor and NASBA assays (1). The poor sensitivities of these assays for the detection of HIV-1 subtype A limits their clinical application in studies of natural history, pathogenesis, and antiretroviral drug and therapeutic vaccine trials in areas where this subtype predominates. Recently, the Amplicor HIV-1 Monitor test has been modified in an effort to improve its sensitivity for the quantification of non-B subtypes of HIV-1, and a newer version of the NASBA assay, the NucliSens assay, has been produced. In this study, we assessed the sensitivities and accuracies of the standard and modified versions of the Amplicor HIV-1 Monitor and the NucliSens assays for the quantification of the HIV-1 RNA viral load in persons infected with HIV-1 subtype A in Abidjan, Côte d’Ivoire.
MATERIALS AND METHODS
Patient population.
A total of 86 plasma samples were tested in this study, including 71 samples obtained from HIV-1-seropositive individuals (47 tuberculosis patients, 17 commercial sex workers, and 7 pregnant women) and 15 samples obtained from HIV antibody-negative commercial sex workers. Whole blood was drawn from these individuals into VACUTAINER CPT tubes (Becton Dickinson, Rutherford, N.J.) containing sodium citrate gel and density gradient media. Plasma was separated from cells within 6 h of the time of collection by centrifugation at 200 × g prior to aliquoting and storage at −70°C. HIV antibody status was determined by enzyme-linked immunosorbent assay testing and was confirmed by either Western blotting or line immunoassays. CD4 lymphocyte cell counts were enumerated by standard flow cytometry with a FACScan flow cytometer (Becton Dickinson, San Jose, Calif.) and commercially available monoclonal antibodies (Becton Dickinson, San Jose, Calif.). HIV-1 subtyping was determined either by DNA sequencing of the C2V3 region of the env gene as described previously or by a restriction fragment length polymorphism assay that is based on the protease gene (5).
HIV-1 RNA assays.
HIV-1 RNA levels in plasma were quantified by the NucliSens assay (Organon Teknika, Boxtel, The Netherlands) and by both the standard (hereinafter referred to as the standard HIV Monitor test) and modified (hereinafter referred to as the modified HIV Monitor test) Amplicor HIV-1 Monitor tests (Roche Diagnostic Systems, Branchburg, N.J.). The modification of the HIV Monitor test consists of 20 μl of a new primer set (SK145 and SK151 [primer mix 1], provided by Roche Diagnostics) that is added to the master mixture prior to amplification. For all assays, HIV-1 RNA extraction, amplification, and detection were performed according to the manufacturer’s instructions. The internal controls for the NucliSens assay consisted of three synthetic RNAs (Qa, Qb, and Qc) of known high, medium, and low concentrations, respectively. These RNAs serve as internal calibrators, each differing from the HIV-1 wild-type RNA by only a small sequence. The HIV Monitor assay uses three internal controls provided by the manufacturer, including a negative and low- and high-positive controls, respectively.
Analysis of data.
All RNA-negative samples were assigned a viral load value equal to the detection limits of the assay: 400 HIV-1 RNA copies/ml (log10 2.6 RNA copies/ml) for the NucliSens assay and 200 copies/ml (log10 2.3 RNA copies/ml) for the HIV Monitor test. All HIV-1 RNA copy numbers were log transformed before statistical analysis. However, the absolute numbers of HIV-1 RNA copies per milliliter were used to calculate coefficients of variations. HIV serologic status was used as a “gold standard” for defining the sensitivities and specificities of the assays. Comparison of the means were carried out by the paired Student t test, and correlation was assessed with the Spearman rank correlation coefficient. Statistical analyses were carried out with EPIINFO, version 6 (Centers for Disease Control and Prevention, Atlanta, Ga.).
RESULTS
Sensitivities and specificities of the quantitative assays for RNA.
Of the 71 HIV-1-positive samples, 70 (98%) were subtype A and 1 was subtype G. Among the 70 subtype A samples, a higher proportion of samples were RNA positive by the modified HIV Monitor test (n = 67 [96%]) than by the NucliSens assay (n = 56 [80%]) and the standard HIV Monitor assay (44 [63%]) (all P values were <0.05). The three samples that were not detected by the modified HIV Monitor assay were also missed by both the standard HIV Monitor and the NucliSens assays. Of the samples that were positive by the corresponding assays, the mean viral load was significantly higher by the modified HIV Monitor assay than by the NucliSens assay (n = 56; 5.1 versus 4.3 log10 HIV-1 RNA copies/ml; P < 0.001) and the standard HIV Monitor assay (n = 44; 5.2 versus 3.8 log10 HIV-1 RNA copies/ml; P < 0.001). Of the 42 samples that were positive by the NucliSens and standard Amplicor Monitor assays, the mean viral load was significantly higher by the NucliSens assay than by the standard Monitor HIV assay (4.5 versus 3.8 log10 HIV-1 RNA copies/ml; P < 0.001). In the one subtype G-positive plasma sample, viral RNA levels were 3.9, 4.0, and 5.5 log10 copies/ml by the standard and modified HIV Monitor assays and the NucliSens assay, respectively.
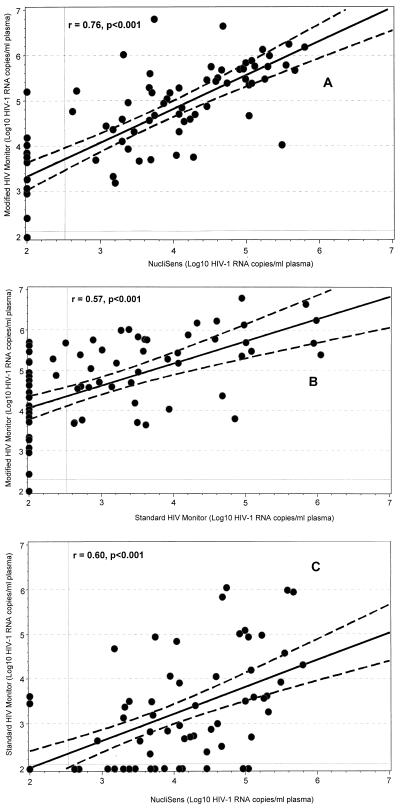
The plasma HIV RNA levels obtained by the modified HIV Monitor assays correlated significantly with those obtained by the NucliSens assays (r = 0.76; P < 0.001) (Fig. 1A) and the standard HIV Monitor assay (r = 0.57; P < 0.001) (Fig. 1B); similarly, a significant correlation was observed between the RNA levels obtained by the NucliSens assay and the standard HIV Monitor assay (r = 0.60; P < 0.001) (Fig. 1C).
FIG. 1.
Correlation between plasma HIV-1 subtype A RNA levels obtained by the modified HIV Monitor, NucliSens, and standard HIV Monitor assays. The diagonal lines indicate the calculated linear regression curves (solid lines) and the 95% confidence limits (dashed lines) for the assays. The vertical and horizontal solid lines represent the detection limits of the assays.
Viral RNA load and CD4 lymphocyte counts.
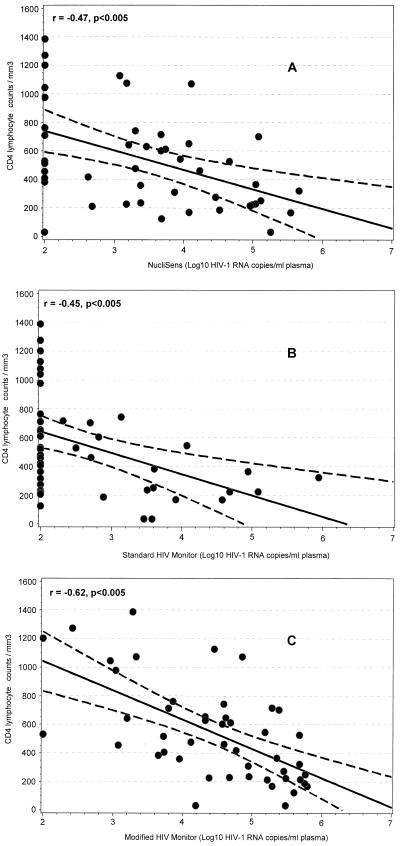
To assess the relationship between the level of immunosuppression and HIV-1 RNA levels as measured by the NucliSens and the HIV Monitor assays, a subset of 47 persons for whom CD4 counts were available were analyzed. The median CD4 cell counts in these persons was 482 cells/mm3 (range, 36 to 1,390 cell/mm3). For all three assays, we observed a similar and significant inverse correlation between decreasing CD4 cell counts and increasing HIV-1 RNA levels (r = −0.47 for NucliSens assay, −0.45 for standard HIV Monitor assay, and −0.62 for the modified HIV Monitor assay (all P values were <0.005) (Fig. 2).
FIG. 2.
Correlation between CD4+ lymphocyte counts and plasma HIV-1 RNA levels. Linear regression curves (solid lines) with 95% confidence limits (dashed lines) are indicated.
Reproducibility and specificity.
Because the standard HIV Monitor assay had the lowest sensitivity, the reproducibilities and specificities of only the NucliSens and modified HIV Monitor assays were assessed. The reproducibility included the variation introduced by all steps of assay performance, including sample preparation, reverse transcription, amplification, and detection of PCR products. Reproducibility was determined in 10 different runs of the HIV Monitor assay by using one specimen whose RNA level (2,159 copies/ml) was known from the first test. In the 10 repeat runs, RNA levels in the specimen ranged from 1,548 to 3,602 copies/ml (mean ± standard deviation, 2,406 ± 674 copies/ml), resulting in a coefficient of variation of 28%. Similarly, for the NucliSens assay, one plasma specimen with initial RNA levels of 8,700 copies/ml was tested in 10 runs; RNA levels for this positive sample ranged from 8,400 to 14,000 copies/ml (mean ± standard deviation, 10,983 ± 2,603 copies/ml), with a coefficient of variation of 24%. The specificities of the modified HIV Monitor and NucliSens assays were assessed by testing 15 HIV-antibody negative samples, all of which gave negative results.
DISCUSSION
The field evaluation described here included a large panel of HIV-1 subtype A samples, the predominant HIV-1 subtype in Africa, and indicates that the modified HIV Monitor assay has a significantly higher sensitivity (96%) than the standard HIV monitor and NucliSens assays for the quantification of virus RNA levels in these samples. The sensitivities that we observed for the standard HIV Monitor (63%) and NucliSens (80%) assays were relatively higher than those reported previously for these assays (44 and 56%, respectively) by Alaeus and colleagues (1). This higher sensitivity may be explained by the fact that more than half of our study population were tuberculosis-positive HIV-1-infected persons, and thus, they had high viral RNA loads, whereas in the previously reported study of 27 persons infected with subtype A, most subjects either were asymptomatic or were undergoing antiretroviral therapy (1).
In addition to the sensitivity, the accuracy of RNA assays is critical. Because no gold standard for HIV-1 RNA quantification is currently available, the assessment of accuracy is difficult. However, several observations suggest that both the NucliSens and the modified HIV Monitor assays can accurately quantify RNA levels in HIV-1 subtype A-infected persons. First, both assays showed similar and significant inverse relationships between viral load and CD4 counts (Fig. 2), which is consistent with what has been reported for persons with HIV-1 subtype B infection (2, 3, 6, 7, 9, 11–13). Second, the RNA levels obtained by both the HIV Monitor and the NucliSens assays were significantly correlated. Lastly, although only a limited number of HIV-negative plasma samples were tested, the specificities of the assays were high (100%), and the high reproducibility observed was within the range reported for other viral RNA load assays. For instance, our observed coefficients of variation of 24 and 28% for the NucliSens and the modified HIV Monitor assays, respectively, are consistent with previously reported values, which ranged from 10 to 50% for the standard HIV Monitor assay, 20 to 30% for the branched DNA assay, and 15 to 60% for the NASBA test (2, 3, 6, 11–13).
An important question for investigators carrying out HIV-related laboratory research in Africa is the choice of the most sensitive and accurate assays for quantifying the viral RNA loads in the plasma of persons infected with the non-subtype B HIV-1 subtypes. A number of factors must be considered in addressing this question. Although sensitivity, specificity, and reproducibility are the most critical parameters, minimum sample volume, commercial availability, cost, ease of performance, shelf life, potential for contamination, turnaround time, and the need for refrigeration of reagents during shipping must also be taken into account. None of the available commercial assays satisfy all the considerations outlined above; however, each assay has its strengths and limitations. Both the NucliSens and HIV Monitor assays have the same ease of performance, use small samples volumes (100 to 200 μl), and have similar turnaround times (5 to 6 h) and high costs (US$ 50 to 60 per sample). Some strengths of the modified HIV Monitor assay include its higher sensitivity and low detection limit of 200 HIV-1 RNA copies/ml. Furthermore, the assay incorporates dUTP and uracil-N-glycosylase to prevent carryover contamination (contamination can easily be achieved by the NucliSens assay, given the numerous openings and closings of the tubes involved in the procedure), can be run on standard PCR and enzyme-linked immunosorbent assay equipment, and does not require refrigeration of reagents during shipment. The NucliSens assay has the advantage that it can be used with serum specimens as well as plasma samples collected on heparin (which is inhibitory to the HIV Monitor test), but some of its reagents require refrigeration during shipment, and its performance necessitates specialized instrumentation.
A possible limitation of our study is that the HIV-1 subtype designations that we used do not exclude the possibility for the occurrence of recombinant viruses, since the subtypes were based on either the env C2V3 region or the protease gene (for the restriction fragment length polymorphism assay), which are different from the gag region used in both the HIV Monitor and NucliSens assays. However, the proportion of recombinant HIV-1 is likely to be very low in Côte d’Ivoire; we have shown previously that more than 95% of HIV-1 infections are caused by subtype A viruses (5, 8).
In summary, we have shown that compared with the NucliSens and standard HIV Monitor tests, the modified HIV Monitor test has a significantly higher sensitivity for the quantification of HIV-1 RNA levels in persons infected with HIV-1 subtype A, thereby constituting a valuable tool for the monitoring of HIV-1 RNA levels in areas of Africa were HIV-1 subtype A is predominant. Similar comprehensive evaluations are needed from other parts of Africa where HIV-1 subtypes C and D predominates.
ACKNOWLEDGMENTS
We thank Kevin De Cock for critical review of the manuscript and for helpful suggestions and Joan-Luis Njampo, James Baggs, and Bharat Parekh for technical assistance.
This work was financially supported by the Division of HIV/AIDS Prevention, National Center for HIV, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, Ga.
REFERENCES
- 1.Alaeus A, Lidman K, Sönnerborg A, Albert J. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS. 1997;11:859–865. doi: 10.1097/00002030-199707000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Cao Y, Ho D, Todd J, Kokka R, Urdea M, Lifson J, Piatak M, Chen S, Hahn B, Saag M, Shaw G. Clinical evaluation of branched DNA signal amplification for quantifying HIV type 1 in human plasma. AIDS Res Hum Retroviruses. 1995;11:353–361. doi: 10.1089/aid.1995.11.353. [DOI] [PubMed] [Google Scholar]
- 3.Dewar R, Highbarger H C, Sarmiento M, Todd J, Vasudevachari M B, Davey R, Kovacs J, Salzman N, Lane H C, Urdea M S. Application of branched DNA signal amplification to monitor human immunodeficiency virus type 1 burden in human plasma. J Infect Dis. 1994;170:1172–1179. doi: 10.1093/infdis/170.5.1172. [DOI] [PubMed] [Google Scholar]
- 4.Dunne A L, Crowe S M. Comparison of branched DNA and reverse transcriptase polymerase chain reaction for quantifying six different HIV-1 subtypes in plasma. AIDS. 1997;11:126–127. [PubMed] [Google Scholar]
- 5.Ellenberger, D., D. Pieniazek, J. Nkengasong, C.-C. Luo, C. Maurice, M. Janini, A. Ramos, D. Hu, I.-M. Coulibaly, E. Ekpini, S. Z. Wiktor, A. E. Greenberg, G. Schochetman, and M. Rayfield. Unpublished data. [DOI] [PubMed]
- 6.Havlir D V, Richman D D. Viral dynamics of HIV: implication for drug development and therapeutic strategies. Ann Intern Med. 1996;124:984–994. doi: 10.7326/0003-4819-124-11-199606010-00006. [DOI] [PubMed] [Google Scholar]
- 7.Henrard D R, Philips J F, Muenz L R, Blattner W A, Wiesner D, Eyster M E, Goedert J J. Natural history of HIV-1 cell free viremia. JAMA. 1995;274:554–558. [PubMed] [Google Scholar]
- 8.Janssens W, Buvé A, Nkengasong J. The puzzle of HIV-1 subtypes in Africa. AIDS. 1997;11:705–712. doi: 10.1097/00002030-199706000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Mellor J W, Rinaldo C R, Gupta P, White R M, Todd J A, Kingsley L A. Progression in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 10.Pachl C, Todd J, Kern D, Sheridan P, Fong S-J, Stempien M, Hoo B, Besemer D, Yeghiazarian T, Irvine B, Kolberg J, Kokka R, Neuwald P, Urdea M. Rapid and precise quantification of HIV-1 RNA in plasma using a branched DNA signal amplification assay. J Acquired Immune Defic Syndr Hum Retrovirol. 1995;8:446–454. doi: 10.1097/00042560-199504120-00003. [DOI] [PubMed] [Google Scholar]
- 11.Revets H, Marissens D, De Wit S, Lacor P, Clumeck N, Lauwers S, Zissis G. Comparative evaluation of NASBA HIV-1 RNA QT, Amplicor-HIV monitor, and Quantiplex HIV RNA assay, three methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:1058–1064. doi: 10.1128/jcm.34.5.1058-1064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Todd J, Yeghiazarian T, Hoo B, Detmer J, Kolberg J, White R, Wilber J, Urdea M. Quantitation of human immunodeficiency virus plasma RNA by branched DNA and reverse transcriptase coupled polymerase chain reaction assay methods: a critical evaluation of accuracy and reproducibility. Serodiagn Immunother Infect Dis. 1994;6:233–239. [Google Scholar]
- 13.Vandamme A M, Schmit J C, Van Dooren S, Van Laethem K, Gobbers E, Kok W, Coubau P, Witvrouw M, Peetermans W, De Clercq E, Desyter J. Quantification of HIV-1 RNA in plasma: comparable results with the NASBA HIV-1 RNA QT and the Amplicor HIV monitor test. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;13:127–139. doi: 10.1097/00042560-199610010-00003. [DOI] [PubMed] [Google Scholar]