Significance
Viral envelope glycoproteins, such as the SARS-CoV-2 spike protein, are decorated by carbohydrates that hide the protein surface from the host immune system. Here, we identified the Burkholderia oklahomensis agglutinin (BOA) lectin as a protein that can bind to these carbohydrates and inhibit viral entry of SARS-CoV-2. Uniquely, BOA’s inhibitory mechanism involves multivalent interactions, which, when reduced to monovalent contacts, result in abrogation of inhibition. Since BOA is targeting carbohydrates on spike, protein sequence variations in variants of concern do not interfere with BOA’s inhibitory potency. It is anticipated that multivalent glycan targeting can be exploited to create broad-spectrum coronavirus viral entry inhibitors.
Keywords: glycan shield, SARS-CoV-2, lectins
Abstract
SARS-CoV-2 spike harbors glycans which function as ligands for lectins. Therefore, it should be possible to exploit lectins to target SARS-CoV-2 and inhibit cellular entry by binding glycans on the spike protein. Burkholderia oklahomensis agglutinin (BOA) is an antiviral lectin that interacts with viral glycoproteins via N-linked high mannose glycans. Here, we show that BOA binds to the spike protein and is a potent inhibitor of SARS-CoV-2 viral entry at nanomolar concentrations. Using a variety of biophysical approaches, we demonstrate that the interaction is avidity driven and that BOA cross-links the spike protein into soluble aggregates. Furthermore, using virus neutralization assays, we demonstrate that BOA effectively inhibits all tested variants of concern as well as SARS-CoV 2003, establishing that multivalent glycan-targeting molecules have the potential to act as pan-coronavirus inhibitors.
The pandemic of COVID-19 has claimed over 6 million lives to date (1). In the race for pharmacological interventions, the spike protein on SARS-CoV-2, the causative agent of COVID-19, has become the predominant target for vaccine development and early monoclonal antibody therapeutics (2–4). The spike protein forms a trimer, with approximately 25 protruding from the outer surface of the virion in a semistabilized prefusion state (5, 6). Upon engagement with the cellular receptor, angiotensin-converting enzyme-2 (ACE2), spike undergoes a substantial conformational change, triggering fusion between the virus and host cell membranes. This process is initiated by and requires the transition of the receptor binding domain (RBD) from a buried “down” state, to an exposed, active “up” state. Although antibodies are effective weapons in the fight against COVID-19, new variants of SARS-CoV-2 are rapidly evolving in the race between virus and host, rendering many antibodies ineffective (7).
Like other viral envelope proteins, spike is heavily glycosylated, and this glycan shield cloaks the protein, effectively hiding 44% of its solvent-accessible surface area from recognition by the host immune system (8–11). Spike sugars are predominately N-linked and have been mapped to 22 different asparagine (Asn) residues (8, 11, 12). As glycosylation is a nontemplated process, spike glycans can range in glycotype from high mannose to complex glycans, depending on the accessibility of the glycan to the glycan processing enzymes and creating heterogeneity at any given site (8). Although most glycans appear to hide the protein surface, there is growing evidence that some possess additional structural roles (13, 14). In particular, virions produced from CRISPR engineered cells, with altered glycan processing machinery that results in changed glycosylation patterns, exhibit reduced infectivity (15). Furthermore, an all-atom molecular dynamics simulation of glycosylated spike suggested that the N165 and N234 glycans regulate the up/down RBD conformational switch (13), while the N343 glycan acts as a gate to permit the up/down placement of the RBD (14). Subsequent biophysical and structural studies, in which the glycosylation capacity of specific residues was ablated by selective mutation, confirmed a pivotal role for the N165 and N234 glycans in RBD positioning and ACE2 binding (16). Therefore, targeting and exploiting glycans on spike constitutes an attractive strategy for developing SARS-CoV-2 inhibitors.
Glycans are recognized and bound by lectins, a class of proteins that is present in all domains of life, ranging from bacteria and fungi to plants and animals (17). Importantly, most lectins possess exquisite configurational and conformational specificity toward particular glycans. Functionally, lectins play key roles in a wide variety of biological processes, including host–pathogen interactions, cell–cell signaling, and immune modulation (18). Among the antiviral lectins, activities against a wide range of viruses have been reported, including HIV, Ebola virus, SARS-CoV 2003, MERS-CoV, and SARS-CoV-2 (17, 19, 20). As potential therapeutics, however, their uses have not been fully realized, due to interactions with host glycans, as well as immunogenic and mitogenic effects (17, 19–22).
Structurally, lectins adopt a diverse set of architectures (17, 22–25). They can bind glycans mono- or multivalently at single or multiple sites. The anti-HIV activities of lectins are associated with binding to the high-mannose glycans on gp120, the major HIV envelope glycoprotein that is critical for entry of the virus into the host cell. Among the HIV-inactivating lectins, the Oscillatoria agardhii agglutinin (OAA) family exhibits a unique beta barrel topology, in single-domain or multidomain arrangements (22). Each beta barrel contains two glycan binding sites, targeting the core 3α,6α-mannopentaose of N-linked high mannose glycans on the HIV envelope protein and inhibiting HIV infection at nanomolar concentrations (21, 22, 24, 26). For example, the OAA family member Burkholderia oklahomensis agglutinin (BOA) has a dual beta barrel topology and an effective EC50 concentration against HIV of 12 ± 2 nM (24). Since binding of 3α,6α-mannopentaose by individual BOA binding sites occurs with Kd values between 20 and 60 µM, as measured by NMR, the high apparent inhibitory activity of BOA is avidity driven, through multiple interactions with gp120 (24).
Here, we demonstrate that BOA interacts with the SARS-CoV-2 spike protein and inhibits cellular entry of SARS-CoV-2 at concentrations less than 10 nM. We further characterized the mechanism of inhibition demonstrating that BOA interacts with the glycans on the SARS-CoV-2 spike protein and is inhibitory through interactions outside of the receptor binding domain.
Results
Lectins as Inhibitors of Viral Entry.
Three different lectins, griffithsin (GRFT), OAA, and BOA, were evaluated for their ability to inhibit infection of Vero E6 cells by the Munich SARS-CoV-2 authentic virus isolate. All three lectins have been reported to inhibit HIV-1 infection, although GRFT and OAA/BOA differ significantly in structure and glycan binding specificities (22, 24, 25). No inhibition of SARS-CoV-2 infectivity was observed for GRFT (at 80 nM), partial inhibition was seen for OAA (at 75 nM), and complete inhibition was achieved using BOA (at 115 nM) (SI Appendix, Fig. S1).
Since BOA was the most potent lectin tested, we evaluated its activity in three different virus neutralization systems. First, using plaque assays, we found that BOA blocked the infectivity of the SARS-CoV-2 Munich isolate in Vero E6 cells with an EC50 value of 9 ± 2 nM (Fig. 1C). Second, using replication-defective, luciferase-expressing HIV-1 pseudotyped with spike from the Wuhan-Hu-1 isolate containing the D614G substitution, BOA reduced infection of Vero E6 cells, as measured by luciferase activity, with an EC50 value of 1.6 ± 0.3 nM (Fig. 1D). Note, the D614G substitution increases the spike protein density on virions resulting in improved transduction (27–29). Finally, infection of HEK293F cells expressing human ACE2 (hACE2) with replication-incompetent GP-expressing HIV-1 pseudotyped with Wuhan-Hu-1 D614G spike generated an EC50 value of 48 ± 16 nM (SI Appendix, Fig. S2). All three assays demonstrate that BOA inhibits SARS-CoV-2 cellular entry at nanomolar concentrations.
Fig. 1.
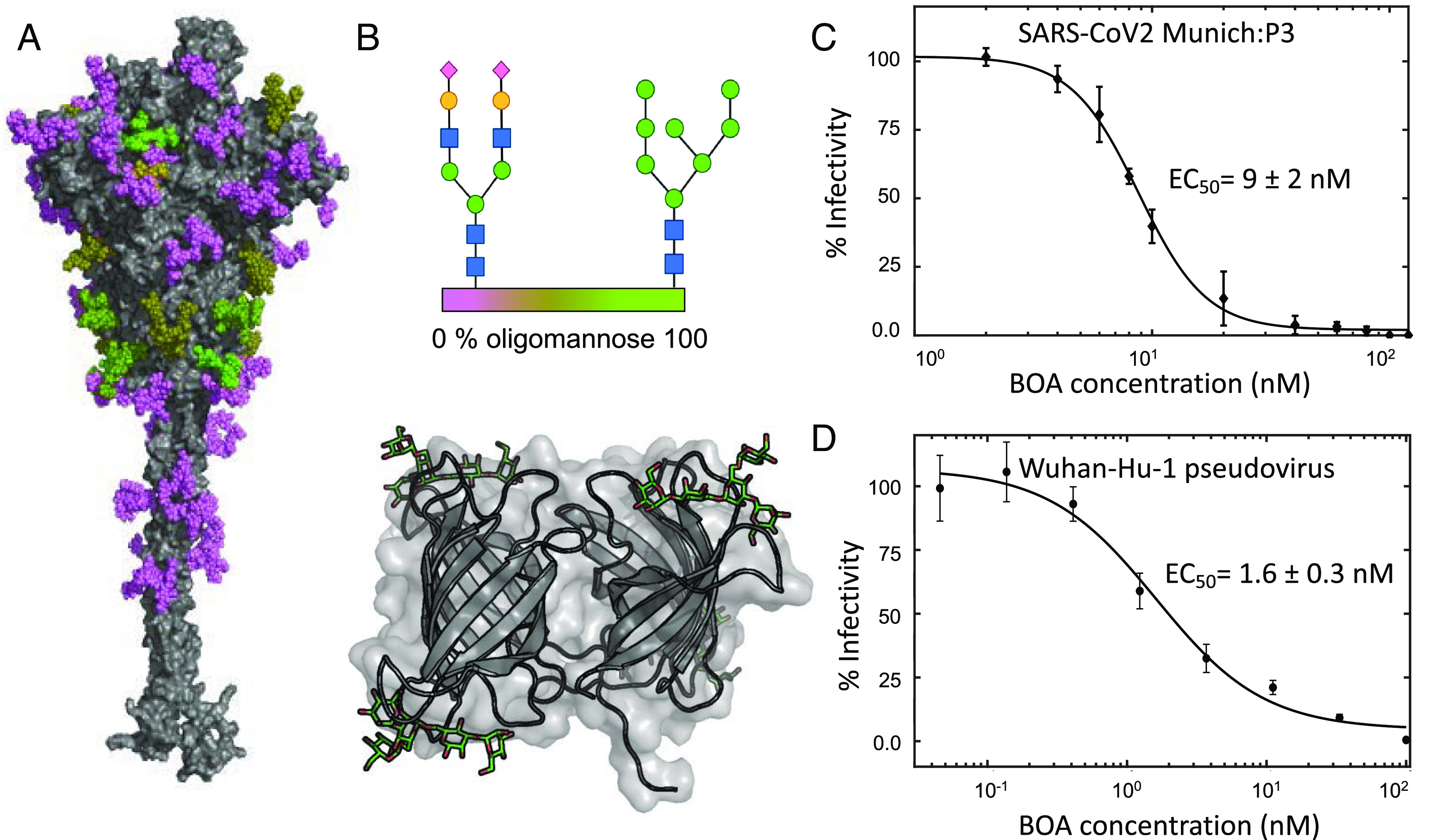
Inhibition of SARS-CoV-2 infection by BOA. (A) Surface representation of the SARS-CoV-2 spike protein (gray) with glycans colored according to glycotype (>90% oligomannose occupancy (lime), 50 to 75% oligomannose occupancy (olive), or complex (violet)). (B) Schematic representation of a complex glycan and high mannose glycan (Top) and the BOA-mannopentaose structure (Bottom). Each sugar is denoted by shape and color (green circles, mannose; blue squares, N-acteyl glucosamine; yellow circles, galactose; purple diamonds, N-acetyl-neuraminic acid). The structure of BOA is shown in ribbon representation and 3α,6α-mannopentaose in green stick representation (PDB ID: 4GK9). (C) Concentration-dependent inhibition of the SARS-CoV-2 Munich isolate by BOA, as measured by a plaque reduction assay and following normalization, yielding an EC50 value of 9 ± 2 nM. (D) Concentration-dependent inhibition of HIV-1 pseudotyped with Wuhan-Hu-1 spike protein by BOA, as measured by using a luciferase reporter, yielding an EC50 value of 1.6 ± 0.3 nM.
Biophysical Characterization of Lectin Spike Interactions.
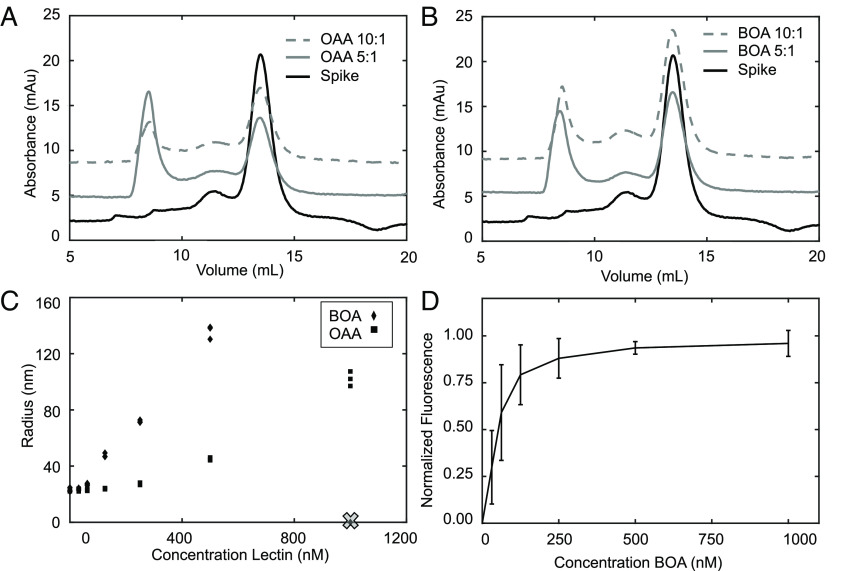
To explore whether BOA and OAA binding to spike was responsible for inhibition of infection, in vitro binding was evaluated using purified spike, OAA, and BOA proteins. Mixing equimolar amounts of lectin and spike proteins, at micromolar concentrations, resulted in coprecipitation (SI Appendix, Fig. S3). Size exclusion chromatography (SEC) generated data consistent with this observation. Specifically, spike protein alone eluted at a volume of 13 mL (Fig. 2 A and B), consistent with its molecular mass of ~500 kDa and previous reports (30, 31). In the presence of OAA or BOA, each in either 5:1 or 10:1 molar excess, additional elution peaks were observed, at ~8 mL (Fig. 2 A and B), the exclusion limit of the column, suggesting that large molecular mass complexes are formed. Visualization of the material in the peak fractions by negative stain electron microscopy revealed large amorphous aggregates (SI Appendix, Fig. S4). Building on these observations, we characterized the concentration dependence of OAA- and BOA-induced aggregation of spike protein by dynamic light scattering (DLS). At concentrations as low as 60 nM BOA or 125 nM OAA, a significant increase in the hydrodynamic radius of the spike protein complex was observed. This increase in hydrodynamic radius continued with increasing lectin concentration until the detector limit was reached (Fig. 2C, X represents the detection limit). Finally, we used SeTau-405 fluorescently labeled BOA in fluorescence titration experiments with spike. In agreement with the above results, saturable binding was observed, with a half-saturation point between 30 and 60 nM (Fig. 2D). This value is of the same order of magnitude as the measured inhibitory concentration of BOA in infectivity assays.
Fig. 2.
OAA and BOA interact with and aggregate spike protein in vitro at nM concentrations. (A and B) Size exclusion chromatography of spike protein without or with OAA (A) or BOA (B) at 5:1 (gray) and 10:1 (gray dotted) molar excess of lectin. (C) Hydrodynamic radius of particles monitored by DLS of spike protein in the presence of increasing concentrations of OAA or BOA (X represents the detection limit). (D) Binding isotherm of BOA binding to spike protein extracted from fluorescence measurements. Normalized fluorescence is plotted as a function of BOA concentrations.
Mechanism of Inhibition.
The high mannose glycan on N234 of the spike protein has been implicated in regulating the position of the RBD (13, 16). We therefore considered the possibility that BOA binding to this particular glycan plays a role in its inhibition of viral entry. To test this idea, we generated pseudoviruses expressing spike protein variants N234A and as a control N165A. While the N165A variant infected Vero E6 cells, the N234A variant was noninfectious (SI Appendix, Fig. S5), confirming that the identity of N234 and/or the glycan at this position is important for viral entry. As N165 is a complex glycan and thus not targeted by BOA, deletion of this glycan by the N165A mutation had minimal impact on BOA’s ability to inhibit viral entry, resulting in an EC50 value of 10 ± 2 nM (SI Appendix, Fig. S5D). Given that the N234A pseudovirus was not infectious, N234A spike and BOA interactions were assessed in vitro using size exclusion chromatography. Similar to what was observed with WT spike protein, BOA and N234A spike eluted at ~8 mL, the exclusion limit of the column, consistent with the formation of soluble aggregates. To corroborate our experimental observations, we computationally evaluated the solvent accessibility of the glycans on the spike protein structure using probes of different sizes, ranging in radius from 1.4 Å, comparable to a water molecule, to 20 Å, comparable to the size of BOA (SI Appendix, Fig. S6). The data indicate that the N234 glycan is buried in the WT spike structure and that BOA interacts with the N234A spike variant through multiple other glycans. Thus, it is likely that the N234 glycan is inaccessible to BOA and does not play an important role in BOA-mediated inhibition.
The role of multivalency in the interaction between BOA and spike was tested by removing three of the four glycan binding sites on BOA. Key arginine, glutamic acid, and tryptophan residues were substituted by alanine. The resulting BOAd3WRE1 variant (W18A:R101A:E64A:W218A:R170AE197A:W152A:R261A:E236A) is a monovalent version of BOA (Fig. 3A) that retains the wild-type (WT) BOA fold, is capable of sugar binding as evidenced by NMR (SI Appendix, Fig. S7), and binds spike as demonstrated by size exclusion chromatography (SI Appendix, Fig. S8). DLS measurements revealed that the BOAd3WRE1 variant fails to aggregate spike at nanomolar concentrations (Fig. 3B). Similarly, SEC of spike in the presence of BOAd3WRE1 shows that the elution volume of the complex is only slightly changed from that of spike alone, indicating that no large soluble aggregates are formed (Fig. 3C). In agreement with the above in vitro data, BOAd3WRE1 failed to interfere with infectivity of the authentic SARS-CoV-2 or pseudovirus (Fig. 3 D and E), demonstrating the importance of multivalency in the interaction between OAA and spike for antiviral activity.
Fig. 3.
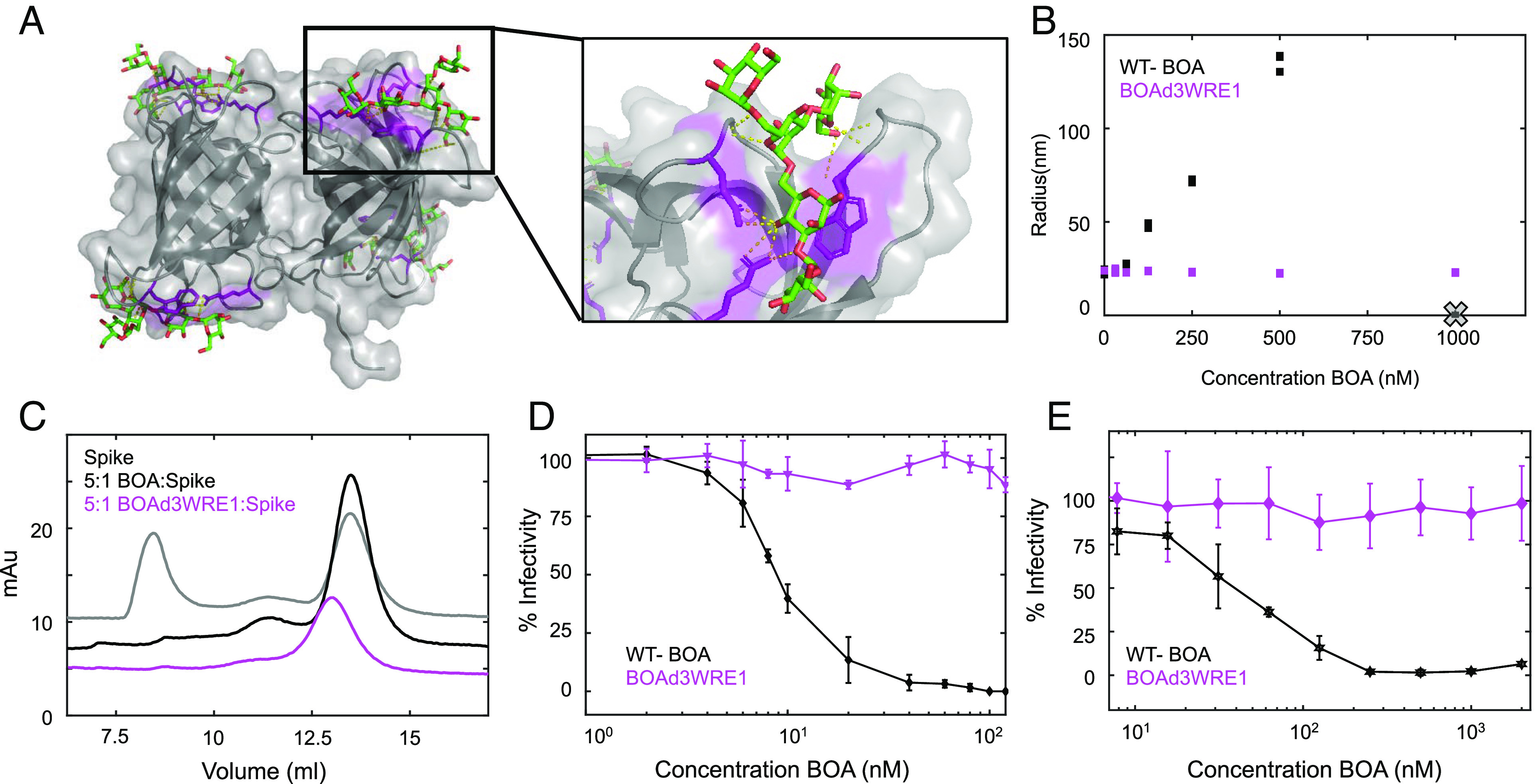
Glycan binding sites in BOA and their importance for avidity-mediated interactions with the spike protein. (A) X-ray structure of the BOA-mannopentaose complex in transparent surface and embedded ribbon representation (PDBID 4GK9). Key Arg, Glu, and Trp residues that are important for glycan recognition in each binding site are highlighted in purple stick representation with the glycan shown in green stick representation. (B) Comparison of spike protein aggregation in the presence of increasing concentrations of WT BOA or BOAd3WRE1, which lacks three of its four glycan binding sites (X represents the detection limit). No aggregation was observed for the variant with a single binding site. (C) SEC of spike protein alone (black) with WT-BOA (gray trace) or with BOAd3WRE1 (magenta trace) at a 5:1 lectin:spike protein ratio. (D and E) Infectivity assays in the presence of WT BOA or BOAd3WRE, using Vero E6 cells infected with the SARS-CoV-2 Munich isolate (D) or hACE2-expressing HEK293F cells infected with HIV-1 pseudotyped with D614G spike (E).
Effectiveness Against Variants of Concern.
As new variants of SARS-CoV-2 emerge, mutations that facilitate transmission and/or immune escape are classified by the World Health Organization (WHO) as variants of concern (7, 32–36). (Fig. 4A). For all variants of concern, decreased recognition by several antibodies has been reported, and symptomatic infections in fully vaccinated individuals with these variants have been observed (7, 35–37). Numerous mutations in the virus, including in the spike gene, have been mapped, especially for regions in the protein under selective pressure, such as position 484 in the RBD or position 681 in the furin cleavage site of the spike protein (38, 39). In contrast, all 22 sites of Asn-linked glycans have remained invariant since the original SARS-CoV-2 isolates in 2019 (Fig. 4B), suggesting that spike retains its original glycan shield. Therefore, BOA should be able to interact with these sugars and retain its ability to inhibit cellular entry of these viral variants. This was tested for the Munich isolate, Delta, and Omicron variants, and only negligible changes in EC50 were observed (Fig. 4C and SI Appendix, Table S1). BOA-mediated inhibition was also tested using replication-defective lentiviruses that were pseudotyped with the spike protein from several variants of concern. Our data show that BOA inhibits Alpha, Beta, Gamma, Delta, and Omicron variants of concern with EC50 values similar to that of the ancestral virus (Fig. 4D and SI Appendix, Table S1).
Fig. 4.
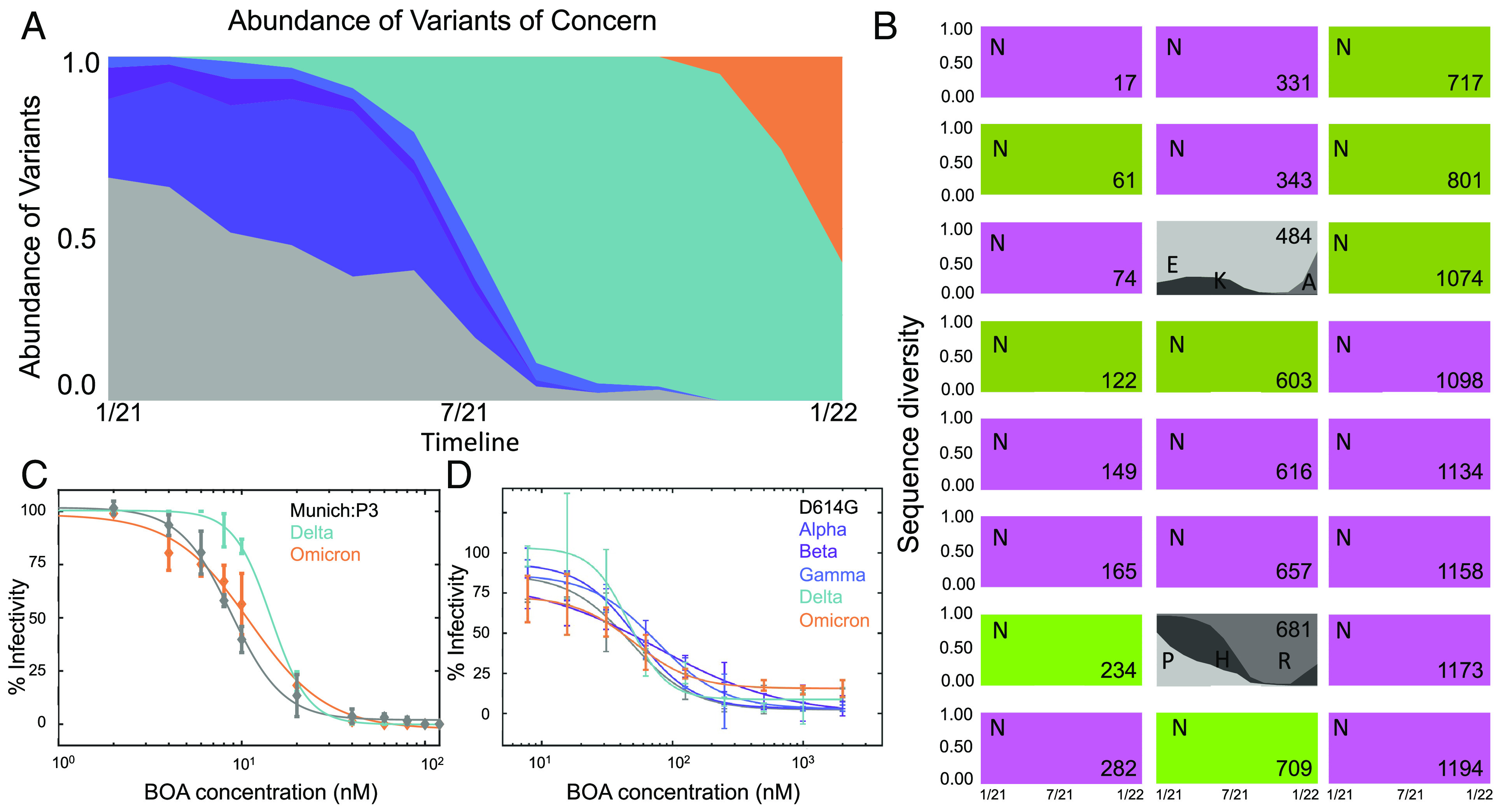
Assessment of BOA activity against emerging variants of concern. (A) World-wide prevalence of ancestral SARS-CoV-2 (gray) and Alpha (indigo), Beta (purple), Gamma (blue), Delta (teal), and Omicron BA.1 (orange) variants of concern in 2021 [Data from Nextstrain database (40)]. (B) Sequence conservation and diversity across variants at positions that possess glycosylated Asn residues. The boxes are colored by glycotype, >90% oligomannose occupancy (lime), 50 to 75% oligomannose occupancy (olive), and complex (violet), with positions shown by numbers in the bottom right corner. In addition to Asn residues, amino acids at positions 484 in the RBD and 681 in the furin cleavage site are shown in gray boxes, with amino acids that are changed in variants of concern labeled in one letter code. (C and D) Infectivity data for several variants of concern using both authentic SARS-CoV-2 (C; Delta and Omicron) and pseudotyped viruses (D; Alpha, Beta, Gamma, Delta, and Omicron).
BOA Is a Potential Pan-CoV Inhibitor.
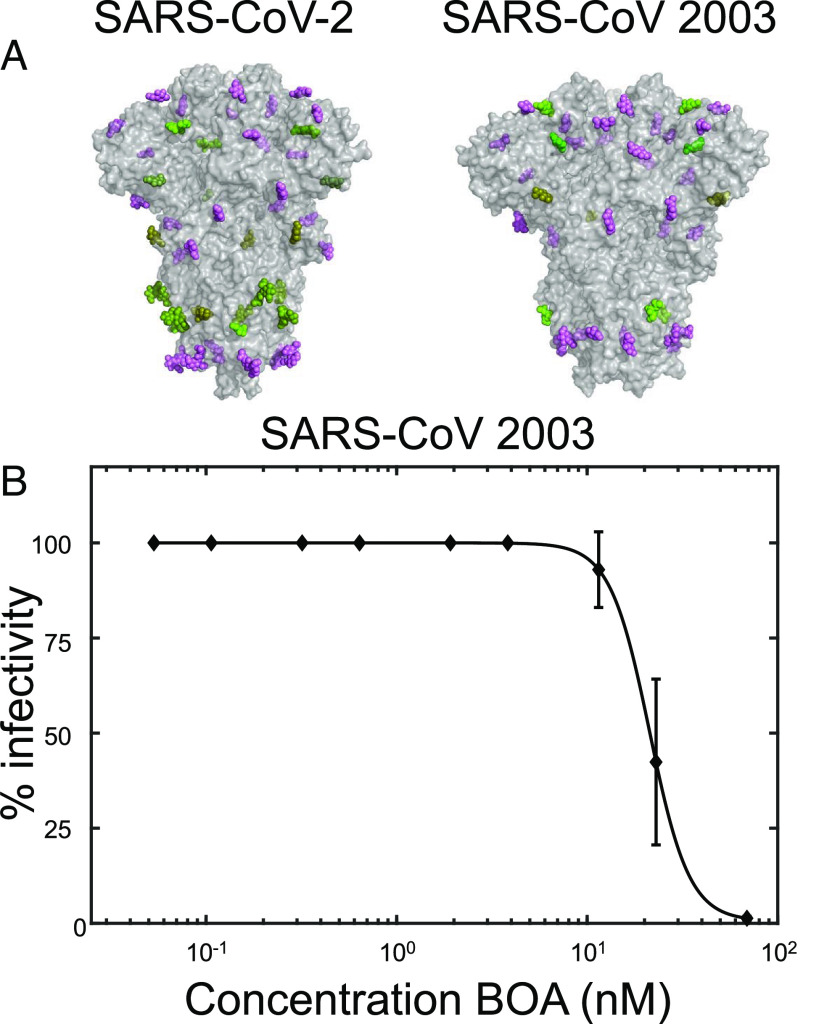
Since SARS-CoV-2 and SARS-CoV 2003 are closely related coronavirus with similar spike protein structures (Fig. 5A), exhibiting 70% amino acid sequence identity, we evaluated whether BOA can also inhibit SARS-CoV 2003. Importantly, both spike proteins possess 22 sites of N-linked glycosylation, with 28% and 32% oligomannose content found in the spike proteins of SARS-CoV-2 and SARS-CoV 2003, respectively (8, 9). Gratifyingly, BOA inhibits infection by SAR-CoV 2003 with an EC50 value of 26 ± 0.3 nM (Fig. 5B), the same order of magnitude as seen for the SARS-CoV-2 variants (cf. Figs. 1 C and D and 4 C and D).
Fig. 5.
Comparison of SARS-CoV-2 and SARS-CoV 2003 spike proteins and BOA inhibition of SARS-CoV 2003. (A) Surface representation of the X-ray structures of SARS-CoV-2 (PDBID: 6VXX) and SARS-CoV 2003 (PDBID: 5X58) spike proteins. Amino acids are shown in gray and sugars are colored by glycotype: >90% oligomannose occupancy (lime), 50 to 75% oligomannose occupancy (olive), and complex (violet). (B) Inhibition assay of the SARS-CoV 2003 Urbani isolate modified with a luciferase reporter reveals an EC50 of 26 ± 0.3 nM.
Discussion
The ability of three lectins (BOA, OAA, and GRFT) to interfere with SARS-CoV-2 infectivity was evaluated employing different assays. While GRFT exhibited no activity in all assays, both BOA and OAA possess potent inhibitory activity. BOA and OAA belong to the same structural family and bind to identical sugar substructures of high mannose glycans (22, 24), whereas GRFT is structurally very different and binds to the terminal sugars in the D2 and D3 arms of high mannose glycans (25). Unfortunately, conflicting reports on GRFT’s activity toward SARS-CoV-2 infection exist, describing very different EC50 values of 6 nM (41) or 63 nM (42). Since we did not observe any inhibition at 80 nM GRFT concentration, our results are at odds with both of these values (41, 42). Variation in antiviral assays has been well documented (43). However, whether the observed disparities are due to different cellular glycan processing, variations in viral isolates, or specific details in the assays remains to be evaluated.
Similarly to GRFT and BOA, inhibitory interactions with the SARS-CoV-2 spike protein have been reported for additional lectins. Muñoz et al. (44) demonstrated that Cyanovirin-N (CV-N) binds to the S1 region of spike of the D614G ancestral virus, with an EC50 of 100 nM. Similarly, for H84T BanLec, characterized by Chan et al. (45), an EC50 of 6 nM was reported, and this lectin was hypothesized to interact with multiple glycans on the same spike molecule. GRFT, H84T BanLec, and CV-N all are assumed to interact with spike via a similar mechanism, namely binding to glycans on the S1 region of the same spike molecule and thereby interfering with the attachment of spike RBD to ACE2.
The lectins studied here function via a different mechanism. Both, BOA and OAA are structural homologs, and the key structural differences between them are the number of beta-barrel domains and the number of glycan binding sites: OAA consists of a single beta-barrel with two binding sites, while BOA contains two domains and therefore four glycan binding sites. The amino acids in the binding sites for glycans are identical; thus, they exhibit the same glycan specificities. As a result, the mechanisms of inhibition mediated via glycan binding are also the same. Given that the antiviral activity toward SARS-CoV2 is associated with glycan binding, BOA, with twice the glycan binding sites compared to OAA, is a more potent inhibitor. Likewise, removing three of the four sugar binding sites on BOA (BOAd3WRE1, Fig. 3) obliterates activity, confirming that multisite interactions are essential. Considering our biophysical assays that report the formation of soluble aggregates at nanomolar concentrations, the multisite interaction between BOA and the spike protein most likely accounts for the antiviral activity of the lectin.
Since one or more glycans on the spike protein are likely targeted by BOA and given the reported importance of the N234 glycan for the conformation of the spike RBD that engages the cellular receptor ACE2, we aimed to test whether the N234A spike glycan deletion variant could be inhibited by BOA. However, because pseudovirus carrying the N234A spike modification was noninfectious, a specific role for the glycan in BOA-mediated viral inhibition could not be ascertained. Instead, the inability of N234A virus to infect cells suggests that N234 and/or its glycan is essential for viral entry. The latter possibility is supported by the in silico and biophysical observations made by Casilano et al. (13), identifying the 234 glycan as essential for spike function. Since N234A spike protein and BOA still interact in vitro, as seen by our SEC results, glycans other than the one attached at N234 must play a role in the interaction.
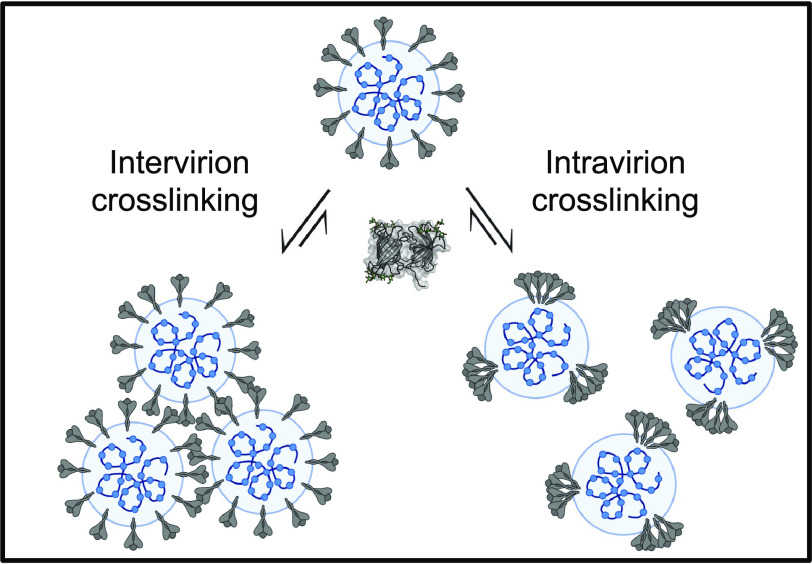
What picture emerges from our observations? Our in vitro data suggest that BOA binds sugars on multiple spike proteins, possibly cross-linking different spike proteins on a single virion or cross-linking spike proteins on different virions (Fig. 6). Such a mechanism is compatible with results obtained for the 2G12 antibody, which binds to glycans at positions 709, 717, and 801 of the spike protein but fails to inhibit viral infection (46, 47). Because 2G12 is a monovalent glycan-binding antibody (47), it is not able to cross-link the spike protein, in contrast to BOA and OAA that can bind in a multisite mode. Therefore, we posit that BOA inhibition of SARS-CoV-2 infection is associated with simultaneous binding to multiple spike proteins. This mechanistic model is further supported by our finding that a monovalent BOA variant binds the spike protein but does not form soluble aggregates in vitro and fails to inhibit viral entry.
Fig. 6.
Model of BOA-mediated cross-linking of spike proteins on SARS-CoV-2 (created with BioRender.com)
SARS-CoV-2 variants of concern possess minimal changes with respect glycosylation (40). For this reason and because BOA inhibition of viral entry is mediated through interactions with glycans on the spike protein, BOA is a potent inhibitor against variants of concern. Our finding that BOA also inhibits an evolutionarily similar virus, SARS-CoV 2003, demonstrates that the inhibitory activity of BOA is minimally dependent on protein sequence. Since BOA inhibits viral entry of all tested SARS-CoV-2 variants, as well as SARS-CoV 2003, our findings may open the way to develop and exploit multivalent glycan binders in the quest to generate pan-coronavirus inhibitors.
Conclusions
The potent inhibitory activity of the BOA lectin toward SARS-CoV-2 was demonstrated and mechanistically characterized. This activity persists against variants of concern and SARS-CoV 2003, illustrating the tantalizing potential to develop multisite glycan binders as pan-coronavirus inhibitors.
Materials and Methods
Protein Expression and Purification.
BOA.
Escherichia coli BL21(DE3) cells were transformed with a pET15b plasmid harboring the gene for BOA; the protein was expressed with an N-terminal hexa-His tag and purified as previously described (24). In short, 100 mL of Luria-Bertani (LB) broth containing 100 µg/mL of carbenicillin was inoculated with a single colony and incubated overnight at 37 °C. 10 mL of the overnight culture was added to 0.5 L of LB containing 100 µg/mL carbenicillin, followed by incubation at 37 °C until an optical density at 600 nm of 0.6 was achieved. Protein expression was then induced with 1 mM IPTG, followed by incubation at 18 °C for 16 h. Cells were harvested by centrifugation at 3,000 × g, and the cell pellet was resuspended in 50 mL of 20 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole, and 1 mM TCEP. The cells were lysed using a microfluidizer, and cell debris was removed by centrifugation for 45 min at 18,000 × g. The supernatant was loaded onto a Cytiva HisTrap HP column and eluted using an imidazole gradient from 0 to 100% (20 mM NaH2PO4 pH 8.0, 300 mM NaCl, 500 mM imidazole, and 1 mM TCEP). Fractions containing BOA were dialyzed overnight at 4 °C against 50 mM Tris–HCL pH 8.0, 375 µM EDTA, and 1 mM TCEP, and the His-tag was cleaved by overnight incubation with tobacco etch virus protease (1:20 TEV to BOA ratio) and removed by passage over a HisTrap HP column. The flow-through containing BOA was concentrated to ~2 mL and loaded onto a S75 size-exclusion column and separated in 20 mM NaPO4 pH 7.5. Fractions containing BOA were collected, concentrated, and frozen at −20 °C. The purity and identity of the BOA protein was assessed by SDS-PAGE and LC-MS.
OAA.
E. coli BL21DE3 cells were transformed with a pET26b plasmid containing the gene for OAA, and the protein was expressed following the same procedure as was used for BOA. OAA was purified as previously described (22). Briefly, cells were pelleted at 3000 × g and resuspended in 20 mM Tris–HCl pH 8.5, lysed using a microfluidizer, and centrifuged at 18,000 × g for 45 min to remove cellular debris. This lysate was dialyzed overnight into 20 mM Tris-HCl pH 8.5 (at 25 °C) and loaded onto a Q HP anion exchange column and eluted using a 0-1 M gradient of NaCl + 20 mM Tris-HCl. Fractions were analyzed via SDS-PAGE and fractions containing OAA were concentrated using an Amicon spin concentrator. OAA was further purified by SEC chromatography using 20 mM Tris–HCl and 100 mM NaCl, pH 8.5 (at 25 °C). Fractions containing OAA were collected, concentrated, and frozen at −20 °C until further use.
Spike protein.
The paH vector harboring the gene for Hexapro S was obtained from Addgene (Plasmid #166856) and used to transfect expi293F cells as previously described (30). Cells were induced for expression with 3.5 mM valproic acid, and media containing the secreted spike protein were collected after 3 d and clarified by centrifugation. The clarified supernatant was passed over a HisTrap HP column, and spike protein was eluted using a 0 to 100% gradient of elution buffer (20 mM NaH2PO4, pH 8.0, 150 mM NaCl, and 500 mM imidazole). Fractions were analyzed for the presence of spike protein on SDS-PAGE, and spike protein-containing fractions were pooled and concentrated to <1 mL. Further purification was carried out by size exclusion chromatography over a Superose-6 10/300 GL column in 20 mM NaPO4, 150 mM NaCl, pH 7.5, and spike protein-containing fractions were pooled, concentrated, and flash frozen in 100 µL aliquots. The purity and identity of the spike protein was confirmed by SDS-PAGE. Negative stain electron microscopy was used to confirm the spike protein was in the prefusion conformation.
Analytical Size Exclusion Chromatography.
100 mL samples of purified spike protein (1 µM) in the absence or presence of 5 or 10 µM BOA were passed over a Superose-6 10/300 GL column in 20 mM NaPO4 and 150 mM NaCl, pH 7.5, using a flowrate of 0.8 mL/min over one column volume. Data were exported and plotted in MatLab.
DLS.
Light scattering data were acquired at 298 K using a DynaPro Plate Reader III (Wyatt Technology, Santa Barbara, CA) in a 384-well plate format. Samples (60 µL total volume in 20 mM NaPO4 and 150 mM NaCl, pH 7.5) contained 2 µM spike protein and increasing BOA concentrations (32 nM to 2 µM). For each sample, ten sets of scattering data were collected and averaged in Dynamics 7.1.7. Data for each concentration were collected in triplicate. Hydrodynamic radii were extracted, and the data were analyzed in MatLab. The errors are given as the SD of the mean for three replicates.
Negative Stain Electron Microscopy.
Carbon-coated grids (Electron Microscopy Sciences, Hatfield, PA) were glow discharged for 30 s at 10 mA using the PELCO system (Ted Pella, USA). Grids were prepared by applying a 2 mL protein sample to the glow-discharged grid, followed by removal of excess liquid after 30 s, and staining with 2% uranyl acetate. The grids were allowed to dry for 15 min before use. All negative stain EM images were obtained on a Tecnai TF20 microscope (FEI Company) equipped with a TVIPS CMOS camera.
Fluorescence Binding Experiments.
BOA was incubated with 2 × molar excess of Setau-405-maleimide (Seta biomedicals; Cat# K7-548) in 20 mM NaPO4, 150 mM NaCl for 12 to 16 h. The reaction was quenched by filtering through a 10-kDa cutoff spin concentrator, with three buffer exchanges into fresh 20 mM NaPO4 and 150 mM NaCl. Labeling was confirmed by LC-MS. Binding was followed by incubating 125 nM BOA with increasing amounts of spike protein (31, 62, 125, 250, 500, and 1,000 nM). Fluorescence intensity was normalized to the maximum fluorescence. Errors are given as the SD of the mean for triplicate measurements. Since no 1:1 or defined stoichiometry of binding exists, the data were not fit to a model, and only the half point of the saturation was extracted.
NMR Spectroscopy.
All NMR spectra were collected at 37 °C using a Bruker AVANCE 600 MHz spectrometer equipped with a triple resonance cryoprobe. 2D 1H-15N HSQC spectra were collected on 250 µM samples of BOAd3WRE1 in 20 mM NaAcetate pH 5.0 with 10% D2O. Glycan binding was assessed by addition of 250 µM 3α,6α-mannopentaose. All spectra were processed in NMRFAM-Sparky (48).
Generation of Wuhan-Hu-1 Pseudoviruses.
Replication-defective pseudotyped HIV-1 encoding firefly luciferase was produced by transfection of 293T cells with plasmids encoding the SARS-CoV-2 spike (Wuhan-Hu-1 with a 21 amino acid C-terminal deletion and the D614G substitution (HDM-SARS2-S-del21-D614G) (49) and the HIV-1 provirus (NLdE-luc) (50) as previously described. The N165A and N234A variants of HDM-SARS2-S-del21-D614G were generated by site-directed PCR mutagenesis using the QuickChange II kit (Agilent), and changes were confirmed by sequencing. Specific infectivity, the luciferase actvity normalized to the amount of virus, was determined by measuring the luciferase activity of virus stocks pseudotyped with WT, N165A, or N234A spike in Vero E6 cells and quantifying HIV-1 capsid (p24) concentrations by ELISA (XpressBio).
Pseudotyped replication-defective lentivirus encoding a GFP reporter gene and expressing D614G SARS-CoV-2 spike protein or the corresponding spike proteins of the Alpha, Beta, Gamma, Delta, and Omicron variants of concern were purchased from Integral Molecular (catalog #s RVP-702, RVP-706, RVP-724, RVP-708, RVP-763, and RVP-768G).
Pseudovirus Inhibition Assays.
Infections of Vero E6 cells with pseudoviruses encoding luciferase were performed in triplicate for each condition in two independent experiments in 96-well plates. Eight dilutions of BOA, or no BOA (100% infectivity), were used, as previously described (51). For GFP encoding pseudovirus infection assays, HEK293F cells expressing a clonally stable hACE2 were obtained from Integral Molecular (cat# c-HA102) and cultured with puromycin to induce hACE2 expression. Infection assays were performed in 96-well plates with BOA concentrations ranging from 15 nM to 2 µM, generated by serial dilution with Dulbecco’s Modified Eagle Media (DMEM). Each well contained 50 µL of BOA in DMEM, to which 20 µL of virus and 30 µL of DMEM were added; these along with infection control wells (20 µL of virus to which 80 µL of DMEM was added) were incubated at 37 °C for 1 h. HEK293F cells were cultured to 80% confluency and lifted from the plate by trypsinization for 3 min, followed by addition of 20 mL DMEM, and centrifugation at 90 × g for 10 min. The cell pellet was resuspended in fresh DMEM. Cell concentrations were determined using nucleocounter NC-100, and stock solutions of 2 × 105 cells per mL were prepared. To each virus-containing well, 100 µL of cell stock solution was added. Plates were incubated for 72 h at 37 °C to allow for infection and expression of GFP. After 72 h, the supernatant in each well was removed by gentle aspiration, and 200 µL of PBS was added. The plate was subsequently imaged using a bottom reading plate reader with 488 nm excitation and 535 nm emission filters. All variants were imaged in triplicate, and the imaging data were exported to MatLab. The fluorescence signal was normalized to control cells without BOA and fit to a sigmoidal function. The error bars represent SD from the mean of triplicate or duplicate experiments.
SARS-CoV-2 Live-Virus Neutralization Assays.
BOA was diluted to 240, 200, 160, 120, 80, 40, 20, 16, 12, 8, or 4 nM in Opti-MEM. One hundred microliters of BOA at the different concentrations was mixed with 100 μL of SARS-CoV-2 inoculum (50 plaque-forming units of virus in Opti-MEM) to yield final concentrations of 120, 100, 80, 60, 40, 20, 10, 8, 6, 4, and 2 nM BOA. These mixes were used in inhibition assays as previously described (52) in Vero E6 cells for the Munich isolate or Vero-hACE2-TMPRSS2 cells for Delta and Omicron variants. In short, virus and lectin mixtures were incubated for 1 h at 37 °C and added dropwise onto a confluent monolayer of Vero-E6 cells in a six-well plate. After incubation at 37 °C for 1 h, 2 mL of 0.1% (w/v) immunodiffusion agarose in DMEM containing 10% FBS and 1× penstrep were added to each well and incubated at 37 °C for 72 h. After incubation, the agarose overlay was removed, and the cells were fixed with 1 mL/well of formaldehyde (37% w/v) stabilized in 10% methanol for 20 min at room temperature. Fixative was discarded, and 1 mL/well of crystal violet in 10% methanol was added. Plates were incubated at room temperature for 20 min and rinsed with water, and plaques were enumerated.
SARS-CoV 2003 Live-Virus Neutralization Assays.
Full-length SARS-CoV 2003 urbani expressing nanoluciferase (nLuc) was used as described previously (53, 54). One day prior to the assay, Vero E6 USAMRID cells were plated at 20,000 cells per well in clear-bottom black-walled plates. BOA was tested at a starting concentration of 2 μg/mL and was serially diluted in equal volume of diluted virus and incubated at 37 °C with 5% CO2 for 1 h. After incubation, BOA and viral complexes were added at 800 plaque-forming units (PFU) at 37 °C with 5% CO2. Twenty-four hours later, plates were read by measuring luciferase activity using a Nano-Glo Luciferase Assay System (Promega) kit. Luminescence was measured by a Spectramax M3 plate reader (Molecular Devices). Virus neutralization titers were defined as the sample dilution at which a 50% reduction in relative light units was observed relative to the average of the virus control wells.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Teresa Bronsenitsch for her critical reading and editing of this manuscript, Mike Delk for NMR support, and Kanako Mori for PCR mutagenesis to produce spike mutants. HDM_SARS2_spike_del21_D614G was a gift from Jesse Bloom (Addgene plasmid # 158762; http://n2t.net/addgene:158762; RRID:Addgene_158762). SARS-CoV-2 S HexaPro was a gift from Jason McLellan (Addgene plasmid # 154754; http://n2t.net/addgene:154754; RRID:Addgene_154754). This work was supported by an NIH grant P50AI150481 (A.M.G.) and the University of Pittsburgh Clinical and Translational Science Institute and the DSF Charitable Foundation (Z.A.). A.J.G is supported by K99GM145970. A.J.G. was a Merck Fellow of the Life Science Research Foundation. R.S.B. was supported by U54 CA260543. D.R.M. is funded by a Hanna H. Gray Fellowship from the Howard Hughes Medical Institute. A.J.G. and D.R.M. hold Postdoctoral Enrichment Program Award from the Burroughs Wellcome Fund.
Author contributions
A.J.G., L.J.R., S.N., Z.A., W.P.D., and A.M.G. designed research; A.J.G., L.J.R., S.N., C.N.R., D.R.M., D.T.Y., F.B., S.V., and A.S. performed research; A.J.G., F.B., Z.A., and W.P.D. contributed reagents/analytic tools; A.J.G., L.J.R., S.N., C.N.R., D.R.M., D.T.Y., F.B., S.V., R.S.B., Z.A., W.P.D., and A.M.G. analyzed data; and A.J.G., R.S.B., Z.A., W.P.D., and A.M.G. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Dong E., Du H., Gardner L., An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinreich D. M., et al. , REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 384, 238–251 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes C. O., et al. , SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottlieb R. L., et al. , Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: A randomized clinical trial. JAMA 325, 632–644 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walls A. C., et al. , Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrapp D., et al. , Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Beltran W. F., et al. , Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184, 2372–2383.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe Y., Allen J. D., Wrapp D., McLellan J. S., Crispin M., Site-specific glycan analysis of the SARS-CoV-2 spike. Science 369, 330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe Y., et al. , Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 11, 2688 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe Y., Bowden T. A., Wilson I. A., Crispin M., Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta Gen. Subj. 1863, 1480–1497 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen J. D., et al. , Site-specific steric control of SARS-CoV-2 spike glycosylation. Biochemistry 60, 2153–2169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao H., et al. , Molecular architecture of the SARS-CoV-2 virus. Cell 183, 730–738.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casalino L., et al. , Beyond shielding: The roles of glycans in the SARS-CoV-2 spike protein. ACS Central Sci. 6, 1722–1734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sztain T., et al. , A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat. Chem. 13, 963–968 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Q., et al. , Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. ELife 9, e61552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson R., et al. , Glycans on the SARS-CoV-2 spike control the receptor binding domain conformation. bioRxiv [Preprint] (2020). 10.1101/2020.06.26.173765 (Accessed 1 June 2021). [DOI]
- 17.Mitchell C. A., Ramessar K., O’Keefe B. R., Antiviral lectins: Selective inhibitors of viral entry. Antiviral Res. 142, 37–54 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown G. D., Willment J. A., Whitehead L., C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 18, 374–389 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Koharudin L. M. I., Gronenborn A. M., Antiviral lectins as potential HIV microbicides. Curr. Opin. Virol. 7, 95–100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millet J. K., et al. , Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin. Antiviral Res. 133, 1–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Férir G., et al. , Broad anti-HIV activity of the Oscillatoria agardhii agglutinin homologue lectin family. J. Antimicrob. Chemother. 69, 2746–2758 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koharudin L. M. I., Furey W., Gronenborn A. M., Novel fold and carbohydrate specificity of the potent anti-HIV cyanobacterial lectin from Oscillatoria agardhii*. J. Biol. Chem. 286, 1588–1597 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato Y., Okuyama S., Hori K., Primary structure and carbohydrate binding specificity of a potent anti-HIV lectin isolated from the filamentous cyanobacterium Oscillatoria agardhii*. J. Biol. Chem. 282, 11021–11029 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Whitley M. J., Furey W., Kollipara S., Gronenborn A. M., Burkholderia oklahomensis agglutinin is a canonical two-domain OAA-family lectin: Structures, carbohydrate binding and anti-HIV activity. FEBS J. 280, 2056–2067 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziółkowska N. E., Wlodawer A., Structural studies of algal lectins with anti-HIV activity. Acta Biochim. Pol. 53, 617–626 (2006). [PubMed] [Google Scholar]
- 26.Sato Y., Murakami M., Miyazawa K., Hori K., Purification and characterization of a novel lectin from a freshwater cyanobacterium, Oscillatoria agardhii. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 125, 169–177 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Daniloski Z., et al. , The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. ELife 10, e65365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L., et al. , SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 11, 6013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yurkovetskiy L., et al. , Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 183, 739–751.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh C.-L., et al. , Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henderson R., et al. , Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 27, 925–933 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geers D., et al. , SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci. Immunol. 6, eabj1750 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gobeil S. M., et al. , Effect of natural mutations of SARS-CoV-2 on spike structure, conformation and antigenicity. bioRxiv [Preprint] (2021). 10.1101/2021.03.11.4350 (Accessed 15 June 2021). [DOI] [PMC free article] [PubMed]
- 34.McCarthy K. R., et al. , Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 371, 1139 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becker M., et al. , Immune response to SARS-CoV-2 variants of concern in vaccinated individuals. Nat. Commun. 12, 3109 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krause P. R., et al. , SARS-CoV-2 variants and vaccines. N. Engl. J. Med. 385, 179–186 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mannar D., et al. , SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex. Science 375, 760–764 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harvey W. T., et al. , SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19, 409–424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.López-Cortés G. I., et al. , The spike protein of SARS-CoV-2 is adapting because of selective pressures. Vaccines 10, 864 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hadfield J., et al. , Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahan R. E., et al. , A highly potent SARS-CoV-2 blocking lectin protein. ACS Infect. Dis. 8, 1253–1264 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai Y., et al. , Griffithsin with a broad-spectrum antiviral activity by binding glycans in viral glycoprotein exhibits strong synergistic effect in combination with a pan-coronavirus fusion inhibitor targeting SARS-CoV-2 Spike S2 subunit. Virologica Sinica 35, 857–860 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahita J., et al. , Coronavirus immunotherapeutic consortium database. Database 2023, baac112 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muñoz-Basagoiti J., et al. , Cyanovirin-N binds to select SARS-CoV-2 spike oligosaccharides outside of the receptor binding domain and blocks infection by SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 120, e2214561120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan J.F.-W., et al. , A molecularly engineered, broad-spectrum anti-coronavirus lectin inhibits SARS-CoV-2 and MERS-CoV infection in vivo. Cell Rep. Med. 3, 100774 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mannar D., Leopold K., Subramaniam S., Glycan reactive anti-HIV-1 antibodies bind the SARS-CoV-2 spike protein but do not block viral entry. Sci. Rep. 11, 12448 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams W. B., et al. , Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies. Cell 184, 2955–2972.e25 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee W., Tonelli M., Markley J. L., NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crawford K. H. D., et al. , Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12, 513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee K., et al. , Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7, 221–233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melody K., et al. , Low frequency of drug-resistant variants selected by long-acting rilpivirine in macaques infected with simian immunodeficiency virus containing HIV-1 reverse transcriptase. Antimicrob. Agents Chemother. 59, 7762–7770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klimstra W. B., et al. , SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J. Gen. Virol. 101, 1156–1169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez D. R., et al. , A broadly cross-reactive antibody neutralizes and protects against sarbecovirus challenge in mice. Sci. Transl. Med. 14, eabj7125 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez D. R., et al. , Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science 373, 991–998 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.