Abstract
Since its isolation in 1988, Afipia felis has been associated with cat scratch disease (CSD) in only one report and its role in CSD has been questioned. We have cultured A. felis from a lymph node of a patient with CSD. 16S rRNA gene sequencing, DNA relatedness studies, fatty acid analysis, and PCR of the A. felis ferredoxin gene showed that the isolate is identical to the previously reported A. felis isolate. To determine the role of A. felis in CSD, PCR of the 16S rRNA gene followed by hybridizations with specific probes were performed with lymph node specimens from CSD patients. All 32 specimens tested positive for Bartonella henselae and negative for A. felis. We conclude that A. felis is a rare cause of CSD. Diagnostic tests not conducive to the identification of A. felis might cause the diagnosis of CSD due to A. felis to be missed.
Since its first description in 1950 (11) cat scratch disease (CSD) was suspected to be an infectious process. In 1988, English et al. (14) successfully cultured the Cat Scratch Disease bacillus, later designated Afipia felis, from the lymph nodes of 10 patients with CSD. Three additional isolates of A. felis, cultured from the lymph nodes of two pediatric patients at one medical center, were described by Brenner et al. (9) in 1991. Since these publications, there have been no reports of the isolation of A. felis from CSD patients; however, numerous reports from various countries have identified Bartonella henselae (formerly Rochalimaea henselae) as the major etiologic cause of CSD. B. henselae has been cultured from the lymph nodes and pus of patients with CSD as well as from cat blood, anti-B. henselae immunoglobulin M and immunoglobulin G antibodies have been detected in the serum of CSD patients, and B. henselae DNA has been identified by PCR in the lymph nodes of patients with clinical CSD and in pus used for the preparation of the CSD skin test (3, 4, 7, 12, 18, 25, 28). As a result, recent studies have focused their diagnostic efforts on the identification of B. henselae. In this report, a patient with CSD from whom A. felis was cultured and characterized is described.
(This study was presented in part at the Infectious Diseases Society of America 34th Annual Meeting, 18 to 20 September 1996, New Orleans, La.)
MATERIALS AND METHODS
Bacterial strains.
A. felis ATCC 53690T was provided by D. J. Wear (Armed Forces Institute of Pathology, Washington, D.C.). The Afipia strains A. clevelandensis ATCC 49720T, A. broomeae ATCC 49717T, and Afipia genomospecies 1 to 3 (strains ATCC 49721, ATCC 49722, and ATCC 49723, respectively), whose DNAs were used for the purpose of comparison with A. felis, have been described previously (9). B. henselae 87-66T (ATCC 49793T) was described by Welch et al. (27). BhTA-2 and BhTA-3 are B. henselae isolates cultured at Tel Aviv Medical Center from patients with CSD and have been described previously (6). Staphylococcus aureus and Pseudomonas aeruginosa are clinical isolates from the microbiology laboratory at the Tel Aviv Medical Center. Borrelia burgdorferi B31 was obtained from M. L. Lovett (University of California at Los Angeles School of Medicine, Los Angeles, Calif.).
Skin test.
Pus was aspirated from the lymph nodes of patients with CSD and was prepared as described previously (6). A positive CSD skin test was defined by induration of ≥5 mm at 48 to 72 h.
Extraction of DNA and PCR amplification.
DNA was extracted from bacteria and clinical samples as described previously (6). Primers corresponding to the ferredoxin and the 16S rRNA genes were synthesized (Tal Ron Scientific Products Ltd., Rehovot, Israel) according to described for previously published sequences (Table 1) and were used for PCR amplification as described previously (7, 26).
TABLE 1.
Primers and probes used for PCR and hybridization assays
| Oligonucleotide name | Oligonucleotide sequence (5′→3′) | Target organism | Target gene | Nucleotide position | Product size (bp) | Reference |
|---|---|---|---|---|---|---|
| p93E | CCGCACAAGCGGTGGAGCA | Eubacteria | 16S rRNA | 931–949a | 480 | 26 |
| p13B | AGGCCCGGGAACGTATTCAC | Eubacteria | 16S rRNA | 1390–1371a | ||
| AF 1013F | CTCGGAAACCGCAAGGACTG | A. felis | Ferredoxin | 41–60b | 632 | 7 |
| AF 1013R | GATGTTGGGCCAAGTCTTAG | A. felis | Ferredoxin | 635–672b | ||
| Bhens probe | TGCGAGCATTTGGTTGGGCA | B. henselae | 16S rRNA | 1050–1069c | 1 | |
| Afelis probe | TGCTACCATTTAGTTGAGCA | A. felis | 16S rRNA | 1073–1092d | 1 |
Positions within Escherichia coli 16S rRNA gene sequence (GenBank accession no. Eco 16s).
Positions within A. felis ferredoxin gene sequence (GenBank accession no. X81826).
Positions within B. henselae 16S rRNA gene sequence (GenBank accession no. M73229).
Positions within A. felis 16S rRNA gene sequence (GenBank accession no. M65248).
Dot blot hybridization assay.
The 16S rRNA PCR products were spotted, in duplicate, onto two identical membranes and were hybridized with either A. felis- or B. henselae-specific probes (Table 1). Labelling of the probes and hybridization assay were performed as described previously (5). Tetramethylammonium chloride (Sigma Chemical Co., St. Louis, Mo.) was used in the posthybridization washes to eliminate disparities in the melting temperatures for the oligonucleotide probes (17), thus enabling the hybridization of B. henselae- and A. felis-specific probes under identical conditions.
Cellular fatty acid analysis.
Whole-cell fatty acid analysis was performed with cultures harvested from plates containing heart infusion agar supplemented with 5% rabbit blood and incubated at 30°C for 5 days. The preparation and analysis of fatty acid methyl esters were done by the techniques developed by Miller and Berger (19).
Sequencing of the 16S rRNA gene.
The 16S rRNA gene from strain AfTA-1 was amplified and sequenced as described previously (10). Sequence analysis for AfTA-1 and A. felis ATCC 53690T was performed with the Phylogeny Inference Package (Phylip), version 3.51c (14a).
DNA relatedness.
The cultivation of bacteria, purification and radiolabelling of DNA, and the hydroxyapatite method used for DNA hybridization have been described previously (8). Hybridization reactions were performed at 65°C. Levels of divergence were calculated to the nearest 0.5%, as described previously (8).
Nucleotide sequence accession number.
The 16S rRNA gene sequence of strain AfTA-1 was deposited in the GenBank database under accession no. AF003937.
RESULTS
Case history.
A 21-year-old white female presented to the otorhinolaryngology service with right supraclavicular lymphadenopathy of 3 weeks’ duration. She was afebrile, felt well, and had no other complaints. The patient owned a kitten which she hugged and kissed and was often scratched during play. Physical examination was unremarkable except for bilateral upper-extremity scratches and a right supraclavicular enlarged lymph node. The node was minimally tender without erythema or local warmth and measured 2.0 by 1.5 cm. No inoculation papule was identified. Complete blood count, chemistry panel, sedimentation rate, and chest radiogram were within normal limits. No blood was left for serological testing, and the patient refused additional venipuncture. A skin test for CSD resulted in 2 mm of a palpable induration and erythema and was interpreted as negative. Fine-needle aspiration of the node was nondiagnostic. Four days later the puncture site began to drain purulent material. An excisional biopsy was performed. Histopathological examination showed reactive lymphadenitis with focal necrosis but without granulomata or microabscesses. Staining with Warthin-Starry silver stain was negative. Standard aerobic, anaerobic, mycobacterial, and fungal cultures were negative. The patient had an uneventful postoperative course and remained asymptomatic thereafter.
Isolation and characterization of A. felis.
A portion of the excised lymph node was ground, cultured with brain heart infusion broth and agar, and incubated at 32°C without CO2 and at 35°C with 5% CO2. Two weeks later, fine growth was demonstrated in the broth incubated at 32°C, and grey colonies were noted on the agar plate. Subculture was performed on brain heart infusion and charcoal yeast extract agar plates. After several passages, the time to detection of growth was reduced to 5 days. Gram staining revealed small, thin, curved, gram-negative bacilli which were motile on wet mount. The biochemical profile of this microorganism included positive oxidase, urease, and nitrate reactions and negative catalase reactions. This biochemical profile was identical to that of the type strain of A. felis. The catalase and nitrate reactions distinguished this isolate from other Afipia species (Table 2).
TABLE 2.
Phenotypic characterization of the patient’s isolate (isolate AfTA-1) and other Afipia strains
| Strain | Test result
|
||||
|---|---|---|---|---|---|
| Oxidase | Catalase | Urease | Nitrate | Motility | |
| AfTA-1 | + | − | + | + | + |
| A. felis ATCC 53690T | + | − | + | + | + |
| A. clevelandensis ATCC 49720Ta | + | − | + | − | + |
| A. broomeae ATCC 49717T, ATCC 49718, and ATCC 49719a | + | wb | + | − | + |
| Afipiagenomospecies 1 to 3c | + | w | + | − | + |
Results from Brenner et al. (9).
w, weakly positive.
Strains ATCC 49721, ATCC 49722, and ATCC 49723, respectively.
PCR of the ferredoxin and the 16S rRNA genes.
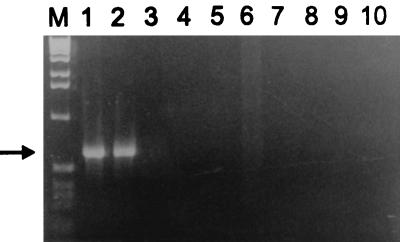
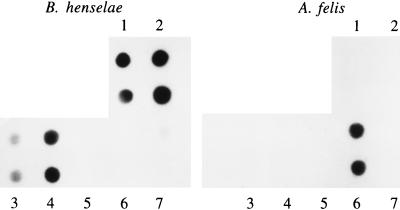
PCR amplification of the isolate’s DNA with primers corresponding to the A. felis ferredoxin gene yielded, as expected, a single 632-bp band (Fig. 1). The identity of the isolate was also confirmed by PCR amplification of the 16S rRNA gene and hybridization with an A. felis-specific probe (Fig. 2, sample 6).
FIG. 1.
Agarose gel electrophoresis of the ferredoxin gene PCR products. Lane M, 1-kb ladder size marker; lane 1, A. felis AfTA-1; lane 2, A. felis ATCC 53690T; lane 3, B. henselae BhTA-2; lane 4, B. henselae BhTA-3; lane 5, B. henselae ATCC 49793T; lane 6, sterile pus; lane 7, S. aureus; lane 8, B. burgdorferi; lane 9, P. aeruginosa; lane 10, water. The arrow indicates a 632-bp band of the PCR product.
FIG. 2.
The 16S rRNA PCR products were spotted, in duplicate, onto two identical membranes which were hybridized with either A. felis- or B. henselae-specific probes. 1, B. henselae DNA; 2 to 4, representative samples from CSD patients; 5, pus from a non-CSD patient; 6, A. felis AfTA-1 DNA; 7, water.
Cellular fatty acid analysis.
Cellular fatty acid analysis showed the following composition: C18:1ω7C (50%), C16:1ω7C (6%), C16:0 (4%), C17:0cyc (6%), C16:02OH (4%), C18:1ω9C (2%), C18:0 (11%), C19:0cyc ω8C (14%), and C12:03OH (1%) and also a component with an equivalent chain length of 18.08, an unsaturated branched-chain fatty acid methyl ester (11-methyloctadec-12-enoic acid). The latter is a unique fatty acid found in the type strain of A. felis and characterized by gas-liquid chromatography and chromatography-mass spectrometry (21).
Sequencing of the 16S rRNA gene.
In a pairwise comparison of the 16S rRNA gene sequence from the A. felis type strain (GenBank accession no. M65248) and AfTA-1, the two sequences were 99.86% similar (2 nucleotide differences of 1,391 nucleotides). When the type strain and AfTA-1 sequences were included in a multiple sequence alignment, edited to remove the variable regions at the 5′ and 3′ ends, and used as input for Phylip, A. felis ATCC 53690T and AfTA-1 were found to be identical.
DNA relatedness studies.
Labelled DNA from AfTA-1 was reacted with unlabelled DNAs from the type strains of A. felis, A. clevelandensis, and A. broomeae and reference strains for Afipia genomospecies 1, 2 and 3, as shown in Table 3. The relatedness of AfTA-1 to the A. felis type strain was 99%, with 0.5% divergence within the related sequences.
TABLE 3.
DNA relatedness of strain AfTA-1 to type and reference strains of Afipia species
| Source of unlabelled DNA | % Relatedness to labelled AfTA-1 DNA | % Divergencea |
|---|---|---|
| AfTA-1 | 100 | 0.0 |
| A. felis ATCC 53690T | 99 | 0.5 |
| A. clevelandensis ATCC 49720T | 26 | |
| A. broomeae ATCC 49717T | 21 | |
| Afipia genomospecies 1 (ATCC 49721) | 25 | |
| Afipia genomospecies 2 (ATCC 49722) | 22 | |
| Afipia genomospecies 3 (ATCC 49723) | 20 |
A blank space indicates that the reaction was not performed.
Detection of A. felis and B. henselae DNAs in clinical specimens.
In a recent study, we described 32 specimens (28 samples of pus and 4 samples of lymph node tissue) from 29 patients with typical clinical cases of CSD. These specimens were subjected to PCR for amplification of the 16S rRNA gene. All patients had regional lymphadenopathy and a history of contact with a cat(s). Hybridization identified B. henselae in all 32 specimens (6). To rule out dual infection with A. felis in the current study, aliquots of the PCR products were spotted in duplicate onto two identical membranes, and each membrane was hybridized with either a B. henselae- or an A. felis-specific probe. All specimens were positive for B. henselae DNA but negative for A. felis DNA. Representative results are presented in Fig. 2.
DISCUSSION
The phenotypic and genotypic characterizations as well as the DNA relatedness determination reported here leave no doubt that AfTA-1, isolated from a lymph node of a patient with clinical CSD, is a A. felis.
The isolation and characterization of A. felis from CSD patients have been extremely rare. English et al. (14) isolated the first A. felis strain (ATCC 53690T) by direct culture on bacteriological media. Three other strains were isolated at the Centers for Disease Control and Prevention by a tissue culture method (9). To the best of our knowledge, AfTA-1 is the first A. felis strain cultured on standard bacteriological media since the first report of English et al. (14) in 1988 and the third report of the isolation of A. felis from the lymph nodes of CSD patients.
Despite its rare isolation, indirect evidence suggests that A. felis may be more commonly linked to CSD than is currently appreciated. Drancourt et al. (13) reported on a patient with acute meningoencephalitis and a history of close cat contact who seroconverted to positivity for A. felis antibodies. In a serological study conducted in Italy among 80 patients with suspected classical or atypical CSD, 24 (30%) patients had antibodies against B. henselae, 10 (12%) had antibodies against A. felis, and 4 (5%) had antibodies against both organisms, as determined by an immunofluorescence assay (IFA) (15). In another study which examined the binding capacity of B. henselae and A. felis to peripheral blood lymphocytes of patients with clinical CSD, one of the five patients studied had elevated anti-A. felis antibody titers without anti-B. henselae antibodies, as determined by IFA (24). By a microagglutination test with heat-killed whole-cell antigen, 5 to 7% of 430 serum samples were reported to be positive for A. felis in one survey which studied the seroprevalence of antibodies against various Afipia strains (22). Amerein et al. (2) used an IFA to study serum samples from 35 CSD patients, 123 control patients without lymphadenopathy, 57 control patients with lymphadenopathy, and 102 nonpatient controls. Although no difference in A. felis antibody titers was found among these groups (contrary to the findings for anti-B. henselae antibody titers, which were significantly higher in the CSD group), 3 patients demonstrated high A. felis antibody titers. Two of these three patients belonged to the CSD group (representing 6% of the 35 CSD patients) and, in addition to anti-A. felis antibodies, also had high antibody titers to B. henselae.
Since both A. felis and B. henselae are members of the alpha-2 group of proteobacteria it would be reasonable to assume that common epitopes among these bacteria are responsible for cross-antigenicity. However, previous studies failed to demonstrate cross-reactivity between these two microorganisms, which differ both phenotypically and genotypically. Regnery et al. (25) have shown that high-titer human anti-B. henselae sera did not cross-react with A. felis antigen in the IFA and that hyperimmune rabbit antisera against A. felis did not react with B. henselae whole-cell antigen (25). Amerein et al. (2) could not demonstrate cross-reactivity between each of two hyperimmune rabbit antiserum samples against A. felis and B. henselae and the heterologous antigen. Due to the apparent lack of cross-reactivity between A. felis and B. henselae, the demonstration of simultaneous seropositivity to both organisms suggests that coinfection with both A. felis and B. henselae may occur in at least a small minority of patients with CSD. Although A. felis and B. henselae have never been cocultivated from the same clinical specimen, they were coidentified by PCR in 2 of 12 lymph node specimens from CSD patients (1). We did not identify A. felis DNA in any of the 32 specimens which were PCR positive for B. henselae DNA. This observation is in accordance with that of other studies (4, 7, 16), suggesting that if coinfection does occur, it is rare.
Also, when the diagnosis of CSD is based solely on histopathology, some cases could be wrongly interpreted as being caused by B. henselae instead of A. felis since both microorganisms are morphologically indistinguishable and both stain well with the Warthin-Starry silver stain (14, 20).
In conclusion, evidence based on serological studies or DNA amplification suggest that A. felis may play a role in the pathogenesis of CSD, either as a single pathogen or in conjunction with B. henselae. These data are difficult to interpret and are inconclusive. However, the culture-confirmed cases of CSD caused by A. felis described by English et al. (14) and Brenner et al. (9), together with the case of CSD caused by A. felis in the patient described in this report, implicate A. felis as a rare cause of CSD. If attempts at the diagnosis of CSD continue to be directed only at the identification of B. henselae, as was the case in several recent studies (4, 20, 23, 28), and not at A. felis as well, the diagnosis of CSD might be missed for those patients with CSD caused by A. felis.
REFERENCES
- 1.Alkan S, Morgan M B, Sandin R L, Moscinski L C, Ross C W. Dual role for Afipia felis and Rochalimaea henselae in cat scratch disease. Lancet. 1995;345:385. doi: 10.1016/s0140-6736(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 2.Amerein M P, De Briel D, Jaulhac B, Meyer P, Monteil H, Piemont Y. Diagnostic value of the indirect immunofluorescence assay in cat scratch disease with Bartonella henselae and Afipia felis. Clin Diagn Lab Immunol. 1996;3:200–204. doi: 10.1128/cdli.3.2.200-204.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson B E, Kelly C, Threlkel R, Edwards K. Detection of Rochalimaea henselae in cat scratch disease skin test antigens. J Infect Dis. 1993;168:1034–1036. doi: 10.1093/infdis/168.4.1034. [DOI] [PubMed] [Google Scholar]
- 4.Anderson B E, Slims K, Regnery R L, Robinson I, Schmidt M J, Goral S, Hager C, Edwards K. Detection of Rochalimaea henselae DNA in specimens from cat scratch disease patients by PCR. J Clin Microbiol. 1994;32:942–948. doi: 10.1128/jcm.32.4.942-948.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avidor B, Zakut H, Keren B. Simple, rapid nonradioactive method to detect major cystic fibrosis mutations in Ashkenazi Jews. Clin Chem. 1996;42:103–105. [PubMed] [Google Scholar]
- 6.Avidor B, Kletter Y, Abulafia S, Golan Y, Ephros M, Giladi M. Molecular diagnosis of cat scratch disease: a two-step approach. J Clin Microbiol. 1997;35:1924–1930. doi: 10.1128/jcm.35.8.1924-1930.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergmans A M, Groothedde J W, Schellekens J F P, Embden J D A, Ossewaarde J M, Schouls L M. Etiology of cat scratch disease: comparison of polymerase chain reaction detection of Bartonella (formerly Rochalimaea) and Afipia felis DNA with serology and skin tests. J Infect Dis. 1995;171:916–923. doi: 10.1093/infdis/171.4.916. [DOI] [PubMed] [Google Scholar]
- 8.Brenner D J, McWhorter A C, Leete-Knutson J K, Steigerwalt A G. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol. 1982;15:1133–1140. doi: 10.1128/jcm.15.6.1133-1140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner D J, Hollis D G, Moss C W, English C K, Hall G S, Vincent J, Radosevic J, Birkness K A, Bibb W F, Quinn F D, Swaminathan B, Weaver R E, Reeves M W, O’Connor S P, Hayes P S, Tenover F C, Steigerwalt A G, Perkins B A, Daneshvar M I, Hill B C, Washigton J A, Woods T C, Hunter S B, Hadfield T L, Ajello G W, Kaufmann A F, Wear D J, Wenger J D. Proposal of Afipia gen. nov., with Afipia felis sp. nov. (formerly the Cat Scratch Disease bacillus), Afipia clevelandensis sp. nov. (formerly the Cleveland Clinic Foundation strain), Afipia broomeae sp. nov., and three unnamed genospecies. J Clin Microbiol. 1991;29:24250–2460. doi: 10.1128/jcm.29.11.2450-2460.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen J J, Whitney A M, Teixeira L M, Steigerwalt A G, Facklam R R, Korner B, Brenner D J. Aerococcus urinae: intraspecies genetic and phenotypic relatedness. Int J Syst Bacteriol. 1997;47:28–32. doi: 10.1099/00207713-47-1-28. [DOI] [PubMed] [Google Scholar]
- 11.Debre R, Lamy M, Jammet M L, Costil L, Mozziconacci P. La maladie des griffes de chat. Bull Mem Soc Med Hos Paris. 1950;66:76–79. [PubMed] [Google Scholar]
- 12.Dolan M J, Wong M T, Regnery L, Jorgensen J H, Garcia M, Peters J, Drehner D. Syndrome of Rochalimaea henselae adenitis suggesting cat-scratch disease. Ann Intern Med. 1993;118:331–336. doi: 10.7326/0003-4819-118-5-199303010-00002. [DOI] [PubMed] [Google Scholar]
- 13.Drancourt M, Donnet A, Pelletier J, Raoult D. Acute meningoencephalitis associated with seroconversion to Afipia felis. Lancet. 1992;340:558. doi: 10.1016/0140-6736(92)91761-v. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 14.English C K, Wear D J, Margileth A M, Lissner C R, Walsh G P. Cat scratch disease. Isolation and culture of the bacterial agent. JAMA. 1988;259:1347–1352. doi: 10.1001/jama.259.9.1347. [DOI] [PubMed] [Google Scholar]
- 14a.Felsenstein J. Phylogeny Inference Package, version 3.51c. Seattle: University of Washington; 1993. [Google Scholar]
- 15.Fumarola D, Petruzzelli R, Giuliani G, Partipilo M R, Pece S. Cat scratch disease in Italy: a serological approach. Microbiology. 1994;17:255–258. [PubMed] [Google Scholar]
- 16.Goral S, Anderson B E, Hager C, Edwards K M. Detection of Rochalimaea henselae DNA by polymerase chain reaction from suppurative nodes of children with cat scratch disease. Pediatr Infect Dis J. 1994;13:994–997. doi: 10.1097/00006454-199411000-00011. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs K A, Rudersdorf R, Neil S D, Douherty J P, Brown E L, Fritsch E F. The thermal stability of oligonucleotide duplexes is sequence independent in tetraalkylammonium salt solution: application to identifying recombinant DNA clones. Nucleic Acids Res. 1988;16:4637–4649. doi: 10.1093/nar/16.10.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koehler J E, Glaser C E, Tappero W. Rochalimaea henselae infection: new zoonosis with the domestic cat as reservoir. JAMA. 1994;271:531–535. doi: 10.1001/jama.271.7.531. [DOI] [PubMed] [Google Scholar]
- 19.Miller L, Berger T. Bacterial identification by gas chromatography of whole cell fatty acids. Hewlett-Packard application note 228–41. Avondale, Pa: Hewlett-Packard; 1985. [Google Scholar]
- 20.Min K W, Reed J A, Welch D F, Slater L N. Morphologically variable bacilli of cat scratch disease are identified by immunocytochemical labeling with antibodies to Rochalimaea henselae. Am J Clin Pathol. 1994;101:607–610. doi: 10.1093/ajcp/101.5.607. [DOI] [PubMed] [Google Scholar]
- 21.Moss C W, Holzer G, Wallace P L, Hollis D G. Cellular fatty acid compositions of an unidentified organism and a bacterium associated with cat scratch disease. J Clin Microbiol. 1990;28:1071–1074. doi: 10.1128/jcm.28.5.1071-1074.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller, H. E. Detection of antibodies to Afipia felis by microagglutination test. Eur. J. Clin. Microbiol. Infect. Dis. 12:951–954. [DOI] [PubMed]
- 23.Nadal D, Zbinden R. Serology to Bartonella (Rochalimaea) henselae may replace traditional diagnostic criteria for cat scratch disease. Eur J Pediatr. 1995;154:906–908. doi: 10.1007/BF01957503. [DOI] [PubMed] [Google Scholar]
- 24.Pece S, Maffione A B, Petruzzelli R, Greco B, Giuliani G, Partipilo M R, Amarri S, Schettini F, Jirillo E, Fumarola D. Rochalimaea henselae organisms possess an elevated capacity of binding to peripheral blood lymphocytes from patients with cat scratch disease. Microbios. 1994;77:95–100. [PubMed] [Google Scholar]
- 25.Regnery R L, Olson J G, Perkins B A, Ribb W. Serological response to “Rochalimaea henselae” antigen in suspected cat scratch disease. Lancet. 1992;339:1443–1445. doi: 10.1016/0140-6736(92)92032-b. [DOI] [PubMed] [Google Scholar]
- 26.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 27.Welch D F, Hensel D M, Pickett D A, San-Joaquin V H, Robinson A, Slater L N. Bacteremia due to Rochalimaea henselae in a child: practical identification of isolates in the clinical laboratory. J Clin Microbiol. 1993;31:2381–2386. doi: 10.1128/jcm.31.9.2381-2386.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zangwill K M, Hamilton D H, Perkins B A, Regnery R L, Plikaytis B D, Hadler J L, Cartter M L, Wenger J D. Cat scratch disease in Connecticut: epidemiology, risk factors and evaluation of a new diagnostic test. N Engl J Med. 1993;329:8–13. doi: 10.1056/NEJM199307013290102. [DOI] [PubMed] [Google Scholar]