Abstract
Primary supranuclear palsy (PSP) is a rare neurodegenerative disease that perturbs body movement, eye movement, and walking balance. Similar to Alzheimer’s disease (AD), the abnormal aggregation of tau fibrils in the central neuronal and glial cells is a major hallmark of PSP disease. In this study, we use multiple approaches, including docking, molecular dynamics, and metadynamics simulations, to investigate the binding mechanism of 10 first- and second-generations of PET tracers for PSP tau and compare their binding in cortical basal degeneration (CBD) and AD tauopathies. Structure–activity relationships, binding preferences, the nature of ligand binding in terms of basic intermolecular interactions, the role of polar/charged residues, induced-fit mechanisms, grove closures, and folding patterns for the binding of these tracers in PSP, CBD, and AD tau fibrils are evaluated and discussed in detail in order to build a holistic picture of what is essential for the binding and also to rank the potency of the different tracers. For example, we found that the same tracer shows different binding preferences for the surface sites of tau fibrils that are intrinsically distinct in the folding patterns. Results from the metadynamics simulations predict that PMPBB3 and PBB3 exhibit the strongest binding free energies onto the Q276[I277]I278, Q351[S352]K353, and N368[K369]K370 sites of PSP than the other explored tracers, indicating a solid preference for vdW and cation−π interactions. Our results also reproduced known preferences of tracers, namely, that MK6240 binds better to AD tau than CBD tau and PSP tau and that CBD2115, PI2620, and PMPBB3 are 4R tau binders. These findings fill in the well-sought-after knowledge gap in terms of these tracers’ potential binding mechanisms and will be important for the design of highly selective novel PET tracers for tauopathies.
Keywords: 4R tau fibrils, metadynamics, positron emission tomography tracer, free energy surface, Alzheimer disease, molecular dynamics
Introduction
Proteinopathies is a broad umbrella term used to represent neurodegenerative disorders that are characterized by different target proteins and featured by the formation of abnormal protein aggregates in the brain, such as amyloid-β (Aβ), tau, and α-synuclein.1−5 Alzheimer’s disease (AD) and Parkinsonian syndromes are the most representative neurodegenerative disorders that are broadly spreading over the world.6 Cortical basal degeneration (CBD) and primary supranuclear palsy (PSP) represent atypical forms of Parkinsonian syndromes and are considered as primary tauopathies with an underlying brain aggregation of tau proteins.7,8 It has been reported that most of these abnormal protein aggregates are present in the brain much in advance of the appearance of dementia symptoms.9 It has therefore become of high relevance to visualize these harmful aggregations of the tau protein in the brains of living patients to detect the disease at an early stage.2,3,10,11 However, the research in this area has been hampered by the fact that imaging of tau fibrils is much more complicated than imaging of Aβ amyloids because the tau proteins are embedded inside the neuronal cells, and there are different isoforms of the tau protein with different folding pathological subtypes. Tau fibrils from AD are comprised of mixed 3/4-repeat (3/4R) microtube binding domains, while only 4-repeat tau (4R tau) aggregates are identified in CBD and PSP.12−16 Additionally, these fibrils are distributed in different sites/regions in the AD/CBD/PSP brains, and the distributions are also related to the progress of dementia.17
Positron emission tomography (PET) has been widely used as a noninvasive imaging tool for neuro disorder diseases.18 Compared to other imaging techniques, PET has a higher sensitivity in detecting the γ rays emitted from positron–electron annihilation, where the positrons and electrons originate from the isotope decay of the labeled small tracer molecules and the atoms of the surrounding brain tissue, respectively. The first generation of tau PET tracers including Flortaucipir (also named AV1451 or T807),19,20TKH5351,21 and PBB3,22 show binding both in vitro and in vivo to both AD (3/4R tau) as well as PSP/CBD (4R tau).11 Among the second-generation tau PET tracers, RO948(23) and MK6240(24) selectively show high binding in AD and no binding in non-AD tauopathies, while PI2620(25) and PMPBB3 (also named APN1607)26 show high binding in the AD brain tissue, but also in the PSP/CBD brain tissue. The in vitro binding data in AD and PSP brain tissues for PI2620, PMPBB3, and CBD2115 seem to be comparable with Kd values in the nM range.27−31
In silico modeling is a powerful aid in structural-based tracer design.32 Several in silico binding studies with different tracers have been reported for AD, CBD, and PSP tau protofibrils.33−37 Some interior tracer binding sites in the protofibrils (e.g., the e1 site in CBD tau) usually exhibit much stronger binding free energies than the fully exposed surface sites.35 However, the interior sites can be difficult to access in a real tissue with many long-range fibrils36 because the entrance to these sites of each real fibril may just interact with the body of other fibrils. Our previous simulations for PI2620 on AD tau protofibrils have proved a high energy barrier for the binding of the tracer into the core site C1.34 Interestingly, there are also partially solvent-exposed surface sites for AD tau fibrils due to its special V-shape (groove) folding pattern.14 This leads to a configuration of a small molecule binding pocket that is closer to the traditional binding pocket on AD tau fibrils.28 Compared to the fully solvent-exposed surface sites on CBD and PSP tau fibrils, the tracer can be less exposed to the solvent environment in these groove sites.37 This may explain why most of the first and second generations of tau tracers show less (or no) binding to 4R tau than to AD tau fibrils.
We use a series of computational tools to investigate the 10 selected first and second generations of tau PET tracers with different scaffolds. Extensive docking studies were first performed to initially rank the binding sites throughout all of the surface sites for PSP, CBD, and AD tau protofibrils. The binding of these tracers to the top-ranked sites is presented and discussed. From molecular dynamics (MD) simulations, we observed large movements on some tracers that show weak binding affinities on some specific sites from docking studies. The probability of tracer binding to new sites is also observed. Finally, we performed metadynamics (metaD) simulations to evaluate the binding free energy profiles for 6 tracers on the surface sites of PSP tau protofibrils. We believe that the mechanistic insight at the atomic level, as provided in this in silico work, may be used to guide the further development of 4R-specific tracers.
Results and Discussion
Determination of Surface Binding Sites for Tracers from Docking Studies
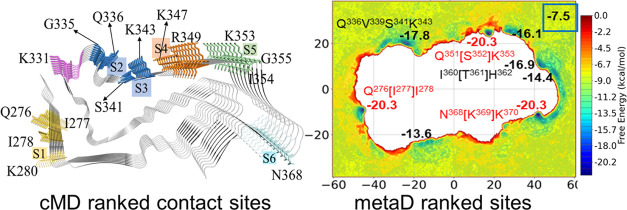
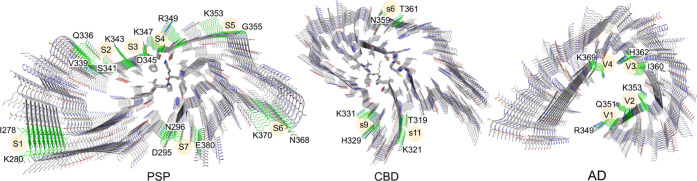
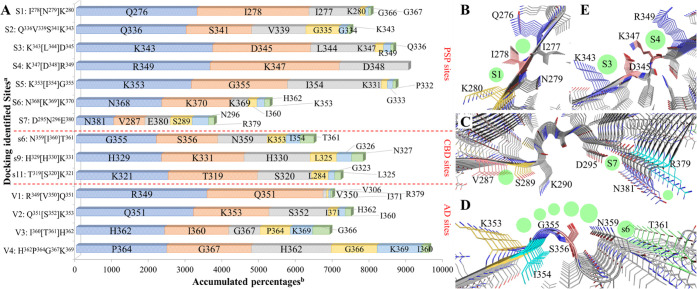
A large amount of compounds with different scaffolds have been reported to be potential PET tracers for the imaging of tau fibrils.11 However, tracers exhibiting high selective binding toward 4R tau fibrils have not yet been reported, especially the PSP tau fibril. To start, we selected from literature data 10 compounds with various scaffolds, including nitrogen-embedded polycyclic aromatic rings (Flortaucipir, RO948, PI2620, and GTP1), pyridine-indole (CBD2115), buta-1,3-dienyl benzothiazole (PBB3, PMPBB3), naphthyridine (JNJ311), pyrrolo-pyridine (MK6240), and pyridine-quinoline (THK5351), for the docking studies on the surface binding sites (Figure 1) on the 9-chain CBD, AD, and PSP tau protofibrils.
Figure 1.
Structures of and ranked surface binding sites for the 9-chain PSP (numbering after “S”), CBD (numbering after “s”), and AD (numbering after “V”) tau protofibrils.
By comparing the Cryo-EM structures of PSP,15 CBD,38 and AD tau protofibrils,28 the volumes of the interior cavities were calculated and follow the order of PSP > CBD > AD (Figure S1). Künze et al. evaluated the associated binding constant of PI2620 to the cavity sites of CBD and PSP tau protofibril by Brown dynamics simulations,37 indicating a very slow intercalation of the tracer to the interior cavity sites due to the low kassoc values. In this study, we focus on comparing the binding on the surface sites of different tracers on PSP, CBD, and AD tau protofibrils. Based on the results from molecular docking and a previous study on CBD and AD protofibrils,35 we selected out 7 and 3 surface sites for PSP and CBD tau, respectively, and 4 concave sites located in the groove of the AD tau protofibril for MD simulations.
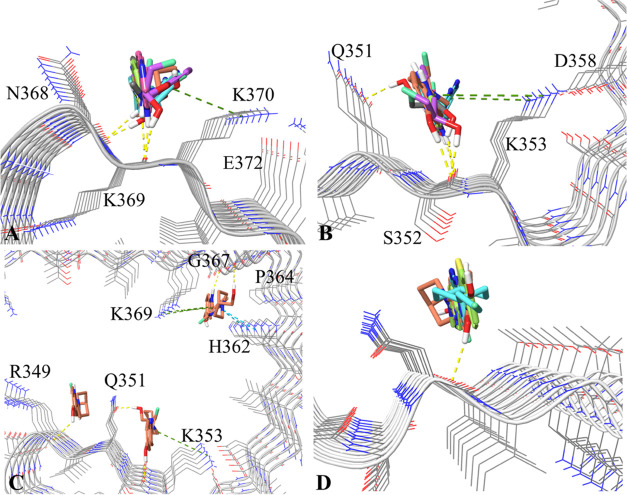
From the docking calculations, we found that site S6 (Figure 1) exhibits the highest binding affinities for the binding of 6 selected tracers (PI2620, CBD2115, PBB3, PMPBB3, and THK5351) on the surface of the PSP tau protofibrils. Site S6 is constituted by the side-chain/backbone atoms of N368 and K370 and the backbone atoms of K369 (denoted as N368[K369]K370; the residue in the bracket has very few side-chain interactions with the tracer, and this will used throughout the text). The backbone of K369 and N368 can serve as hydrogen bond acceptors to contact the hydrogen bond donors in the tracers (Figure 2A). Due to the adjacent E372, the distance between the positive amine group and aromatic rings of the tracers is somehow too far for suitable cation−π interactions. It is notable that the concave sites V2 of AD tau protofibrils also have a similar configuration of residues (Q351[S352]K353, Figure 2B). The docking scores between the V2 and S6 sites are generally similar for the tracers THK5351, PBB3, GTP1, PI2620, CBD2115, and PMPBB3.
Figure 2.
(A) Superimposed binding modes of PI2620, GTP1, CBD2115, PBB3, PMPBB3, and THK5351 at the S6 site of PSP tau. (B) Superimposed binding modes of PI2620, GTP1, CBD2115, PBB3, PMPBB3, and THK5351 at the V2 site of AD tau. (C) Superimposed binding modes of CBD2115 at the V1, V2, and V4 sites of AD tau. (D). Superimposed binding modes of CBD2115, PBB3, and GTP1 on the s6 site of CBD tau. The hydrogen bonds, cation-π, and π–π interactions are depicted as yellow, green, and cyan dash lines, respectively.
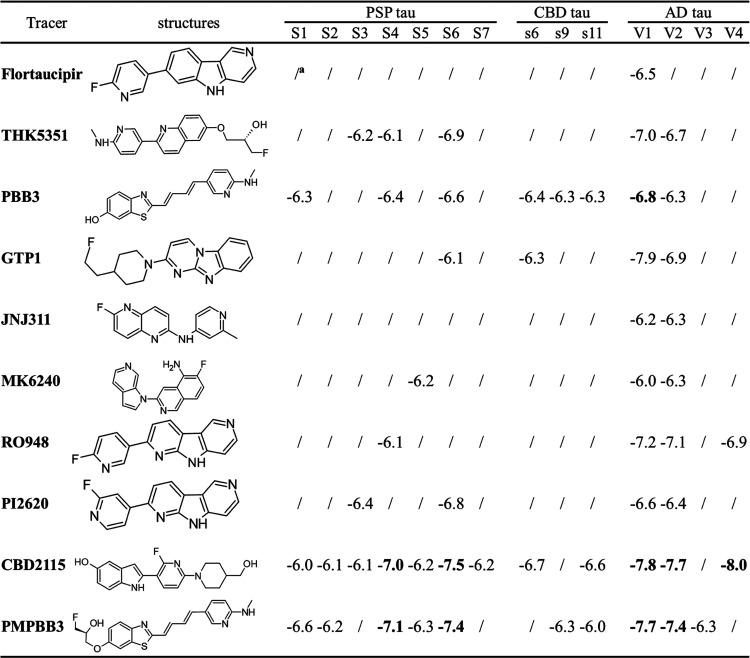
We observed that CBD2115 and PMPBB3 can show high binding affinities in that the docking score could reach −7 to −8 kcal/mol for some sites in both AD and PSP tau fibrils. The docking scores at AD tau are better than those on PSP tau fibrils. Both of CBD2115 and PMPBB3 show moderate binding affinities (−6.0 to −6.7 kcal/mol) to the surface sites of CBD tau protofibrils. CBD2115 exhibits very high binding affinity at the AD tau site V4 with H362P364[G367N368]L369 (Figure 2C). PBB3 also shows many binding sites with docking scores in the range of −6 to −7 kcal/mol on the surface of PSP (S1, S4, and S6) and CBD (s6, s9, and s11) tau with comparable docking scores to the AD tau fibrils (Table 1).
Table 1. Docking Scores of 10 Selected Tracers on Different Surface Sites of PSP, CBD, and AD tau Protofibrils.
Low binding, docking score worse than −6.0 kcal/mol.
The s6 site (N359[I360]T361) of CBD tau ranked from blind docking is a unique site without a side-chain of basic residues. For the site-specific CBD docking studies, s6 exhibits higher binding affinities for GTP1, CBD2115, and PBB3 than the s9 and s11 sites (Figure 2D). Through docking studies, we identified the initial binding modes of the 10 selected tracers to the surface sites of PSP, CBD, and AD tau fibrils. The AD tau sites are located in the concave groove due to their folding pattern, which provides more contact to the tracers. The site S6 in the PSP tau protofibril is similar to the V2 site in AD tau and also fits well with the binding of most of the tracers.
The docking-derived 140 complexes were then subjected to MD simulations. Here, we also performed more docking studies using Autodock439 with a similar grid box setting in our previous study.34 The internal cavity sites were much better ranked than the surface sites that are generally scored in the range of −5.0 to −6.0 kcal/mol and partially overlapped with the Glide docking results (Figure S2).
Mobility of Tracer Binding on the Surface Sites Evaluated by Molecular Dynamics Simulations
From the docking-derived initial binding modes of these tracers, the compensation of the solvent effect and protein flexibility was introduced by performing a 100-ns MD simulation for each compound at each site of the three protofibrils. The time scale of 100-ns is often insufficient to observe the unbinding of small molecules from an enclosed binding site; however, the folding pattern of pathological tau proteins exposes the ligand binding sites to the solvent, and dissociation can, therefore, be observed due to a smaller number of contacts between the ligand and its surface binding sites.37 For example, the binding and unbinding of 60 bTVBT4 molecules onto a full periodic model of the AD tau fibril could be easily observed in the simulations performed by Todarwal et al.36
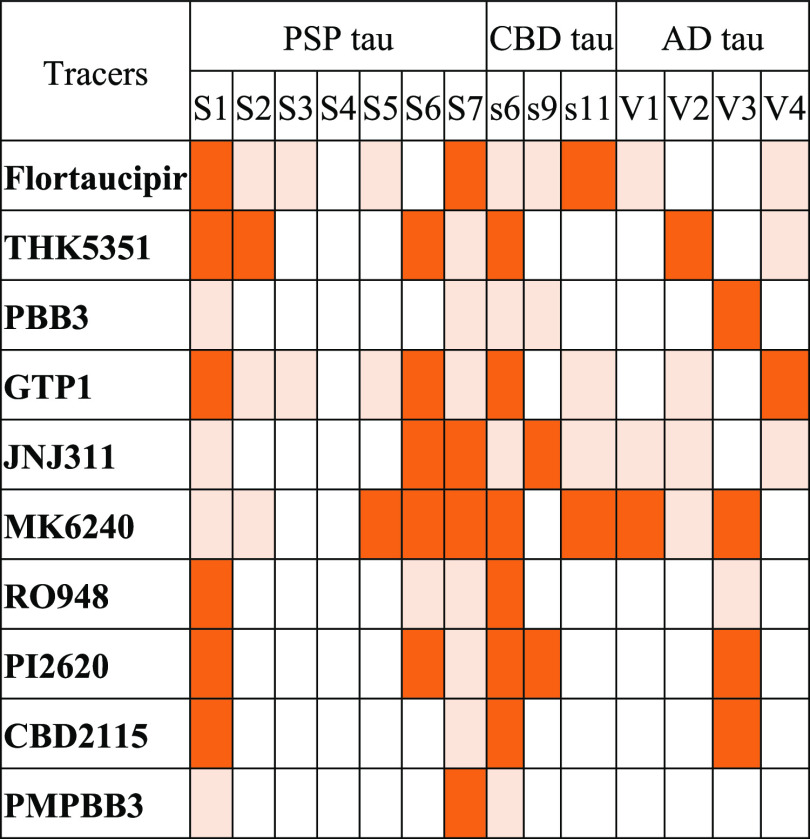
We inspected all of the trajectories to check if the tracers were stable in the docking-derived binding sites (Table 2). For the PSP protofibril, sites S1 and S7 seem not to be suitable for the binding of all tracers, while site S4 can enclose all of the tracers. Site s6 on the CBD protofibril is unstable for all of the tracers, in which all of the tracers shifted to neighbor regions. The shift may be induced by the flat shapes of the nearby residues of site s6 (Figure 1). For the AD protofibrils, V3, the site neighboring the V4 site, is the most unstable site for tracer binding, whereas V4 is a small groove-shaped site gated by H362 and N368 (Figure 1). We can see at a gross level that the binding modes of PBB3, RO948, PI2620, CBD2115, and PMPBB3 are relatively stable in most of the docking identified binding sites.
Table 2. Overview of the Tracer Mobilities during MD Simulationsa,b,c.
Blank: The tracer remained stable in the initial binding pocket.
Light orange: The tracer left the initial site after 50-ns.
Orange: The tracer left the initial site quickly within 50-ns.
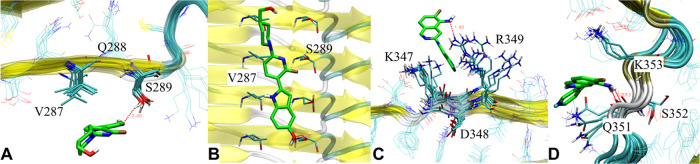
In many cases, though the unbinding to the initial docked sites is observed, the tracer can either stay in the solvent or bind to the other sites. So, we performed contact analysis for all of the trajectories by counting the snapshots with the non-hydrogen atoms of the tracer and protein being closer than 4 Å (Figure 3A). In the MD simulations for these sites, tracers shifted the major contacts with neighboring Q276[I277]I278, V287[Q288]S289, and G355S356K353[I354] for the docked S1, S7, and s6 sites, respectively (Figure 3B–D). The side-chain atoms of I278 became the wall between site S1 and the new site (Figure 3B). Site S7 on the PSP protofibril exhibits the least capacity for enclosing the tracers (Figure 3A). The major tracer binding site starting from S7 shifted to the neighboring R379N381 and a middle-distance site V287[Q288]S289, which is separated by a small groove between D295 and K290 (Figure 3C). However, this groove is found to close up due to the formation of the D295-K290 salt bridges, laying down the conformation of K290’s side-chain atoms (Figure 4). Such dynamical structural bias explains why V287[Q288]S289 becomes a competitive binding site (845% accumulated portion) to S7 (700%, Figure 3A) because the space barrier between V287[Q288]S289 and S7 was flattened. Site S7, located at β13, was also found by Künze et al.37 to be unstable for the binding of PI2620 with a positive binding free energy. For the CBD protofibril, site s6 was found to be extended to more neighboring residues, K353[I354]G355S356, indicating an orientation of tracers that is perpendicular to the axis of the protofibril (Figures 3D and S3). In contrast to S1, S7, and S6, site S4 (K347[D348]R349) is found to fit all of the tracers with the accumulated portion of more than 3500% and without other contacts (Figure 3A,E). Moreover, from the row of site S3 (Figure 3A), we can see that ∼200% of accumulated contacts shifted to the neighboring S4 site. Interestingly, S4 is mainly configurated by the side-chain atoms of two basic residues, K347 and R349, with the lysine forming a salt bridge with D345. This site is also validated by the free energy surfaces (FES) obtained by Künze et al.,37 which indicates a deep local minimum for the binding of PI2620. The CBD protofibril also exhibits a similar surface site s9 that is constituted by two basic residues and shows good stability for tracer binding. For the AD tau protofibril, the concave sites are the most identified sites for the PET tracer binding from experimental research.28,40 The V4 site has two basic residues on the two sides; however, H362 and K269 are separated by 7 residues. Looking into the sites ranked between the top 3 and top 6 from the simulations starting from V3 and V4, we can identify a certain amount of exchange of tracer binding between the two sites (Figure 3A). The structural and dynamical features of these sites will also play different roles in the binding profiles of these tracers to 4R- and 3R/4R tau fibrils.
Figure 3.
Overview of the surface residues that come into contact with tracers during the 100-ns MD simulations. (A) Statistical data of the contacts. A threshold distance of 4.0 Å between the non-hydrogen atoms of the protein residues and tracers was used to count the contacts. The top 6 most contacting residues are depicted. aConfiguration of each docking identified site is given for comparison. bPercentages are accumulated from the MD simulations of each tracer over all of the protofibril chains. (B), (C), and (D) Configurations of new sites (green spheres) in the MD simulations of docked sites (green spheres with labels) S1, S7, and s6, respectively. (E) Comparison of the sites S4 and S3 on the PSP protofibril.
Figure 4.
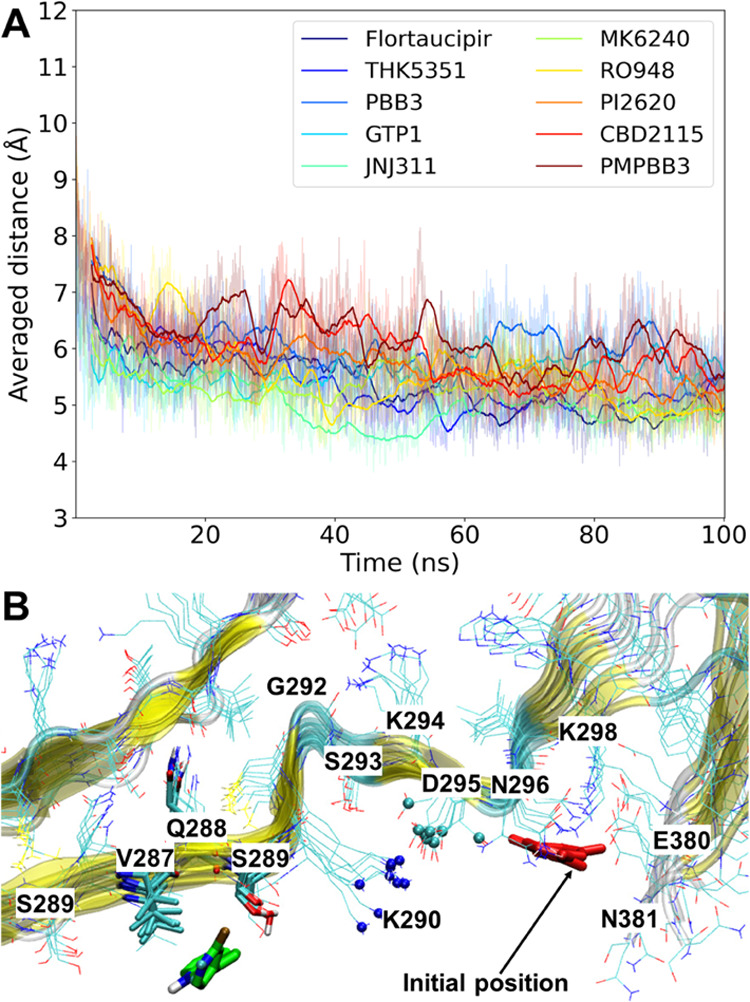
(A) Time course of the averaged distance of the “NZ” atoms of K290 and the “CG” atoms of D295 for all of the 9 chains during the 10 MD simulations with different tracers at site S7. (B) Representative snapshot of PI2620 falling into site V287[Q288]S289 from the simulation starting from PSP site S7. The “NZ” atoms of K290 and “CG” atoms of D295 are depicted in blue and cyan spheres, respectively. PI2620 and the residues within 4.0 Å of PI2620 are rendered as cyan and green sticks, respectively.
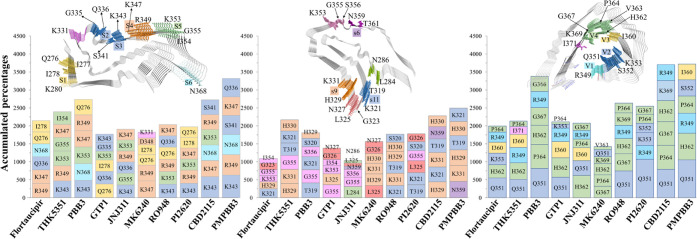
We further summarize the overall contacts for ranking the top 6 most contacting residues for each tracer (Figure 5). Although the basic residues are included in the initial sites, the binding pattern of tracers on these protofibrils shows different preference to the basic residue sites, in which PSP shows the highest portion (K343 from site S2 or S3 and R349 from site S4 for all of the tracers except GTP1), followed by the CBD (K321 from site s11, H329 from site s6, and R349 from a new site for Flortaucipir, THK5351, RO948, and CBD2115) and AD protofibrils (H362 for JNJ311). Interestingly, all of the four “V” sites of AD protofibrils contain at least one basic residue (Figure 1). In the V1 and V2 sites, the distances of R349-Q351 and K353-Q351 pairs are slightly enlarged after MD simulations (Figure S4A), which may be caused by the repulsive interactions between R349 and K375 on the other side of the groove. Such repulsive interaction also slightly enlarges the mouth of this groove (Figure S4B), providing a reasonable explanation for the better preference for contacting the basic residues of the PSP and CBD protofibrils. The accumulated portion of contacts for each top 6 ranked residue on CBD seems to be lower than those on the PSP and AD protofibrils, indicating a larger mobility of tracers on the CBD protofibril. For the binding of PI2620, the top 6 most contacting residues are involved in 5 sites (Q276[I278]K280 near S1, S2, S3, S4, and S5) and 3 sites (s11, extended s6, and L325[G326], Figure 5) for the PSP and CBD protofibrils.
Figure 5.
Top 6 most contacted residues for each tracer on PSP (left), CBD (middle), and AD (right) protofibrils throughout 140 (10 tracers * 14 initial sites) 100-ns MD simulations.
The protein–tracer contact information has proven that when a tracer leaves the docked surface sites (within or after 50-ns), the tracer could shortly remain at other surface sites that are poorly ranked by the docking. For example, in the last 20-ns simulation of CBD2115 starting from PSP site S1, the tracer is found to stay in a site V287[N288]S289 (Figure 6A,B) with the accumulated portion of contact of 210% (Figure 3A), which is a competitive site in the simulations starting from site S7. Hydrogen bonding between CBD2115 and V287[N288]S289 can be observed. The simulations for PBB3, CBD2115, PI2620, and PMPBB3 are found to be relatively stable in most of the PSP surface sites (Table 2), while three of them are second-generation tau tracers. For the experimentally characterized 3R/4R tau binder MK6240, although it also exhibits a strong preference for contact with K347 and R349 on the PSP protofibril (Figure 4), we find that its orientation is actually vertical to the axis of the fibril, exposing the large benzopyridine moiety to the solvent environment (Figure 6C). The binding mode of MK6240 on PSP is, therefore, different in all of the sites on the AD protofibril (Figure 6D), which is also observed in a previous small section of autoradiography.31
Figure 6.
Examples of binding modes. (A) and (B) Representative snapshots of CBD2115 on the V287[Q288]S289 site of PSP protofibril from the front and bottom view, respectively. (C) Vertical binding mode of MK6240 on the S4 site of the PSP protofibril (front view). (D) Parallel binding mode of MK6240 in the V2 site of the AD protofibril.
The time length of simulations may impact the distribution of the tracer most contacted sites. Additionally, we performed 70 (starting from sites S1-S7 for each of the 10 tracers) 500-ns MD simulations for the tracers on PSP protofibril with new seeds generating initial velocities. Compared to the simulations with a length of 100-ns, the top-1 and top-2 most contacted residues changes for all of the systems, but they mostly belong to sites S2 or S3 for THK5351, PBB3, JNJ311, PI2620, CBD2115, and PMPBB3 (Figure S5). For the top 6 most contacted sites, the scope of contacted residues is similar to the distribution derived from the simulation length of 100-ns. We believe that the simulation length of 100-ns starting from multiple sites is generally sufficient for studying the mobility of tracers.
In the docking top-ranked CBD surface site without basic residues, s6, the simulations were unstable for all of the tracers that moved to the neighboring sites K353[I354]G355S356 or T361[H362]V363 (Figure 3D). Only CBD2115 moved to T361[H362]V363, while the other 9 tracers fell into K353[I354]G355S356. For site s9, H329[H330]K331, RO948, CBD2115, PBB3, PMPBB3, and MK6240 were found to be stable. Since MK6240 is known as a 3R/4R tau binder (more than 500-fold selectivity over 4R tau in KD),24,41 site s9 may not be considered as a good site to distinguish the binding of different tau tracers for CBD protofibrils. Site s11 is close to the concave groove of CBD tau, which folds more tightly than that in AD tau. Simulations starting from this site were stable for the binding of Flortaucipir, PI2620, CBD2115, PBB3, PMPBB3, and JNJ311. From the simulation results, the bias of selecting only a few sites (especially for the CBD protofibril) was compensated to some extent because new surface sites were observed during the MD simulations.
A very recent paper40 has shown a novel and interesting binding mode that multiple GTP1 molecules can be stacked along the axis of the fibril in the groove site of the AD tau fibril with a very high concentration of tracer, indicating that ligand cooperativity can compensate for the instability of these half (the V2 and V3 sites in the AD protofibril) solvent-exposed binding sites. This also implies a further consideration of the possibility of ligand cooperativity in the fully solvent-exposed surface sites for the 4R tau fibrils.
Free Energy Profiles of Tracer Binding Derived from Metadynamics Simulations
A typical feature of the surface sites for the pathological tau fibril is that they usually contain only 2 to 4 residues that are repeated 4 to 6 times on the axis of the protofibril.33,34,42 Through the MD simulations, the dynamics of both tracers and protofibrils can be well captured, such as the new ionic interactions within the protofibril in PSP and the unbinding and rebinding of tracers. However, a qualitative comparison of binding modes of tracers on tau protofibrils often needs further evaluation, such as that given by binding free energy calculations.
To gain energy insight into the binding of these tracers to the surface sites of PSP protofibrils, we further performed well-tempered metadynamics simulations for the tracers of CBD2115, PI2620, Flortaucipir, PMPBB3, PBB3, and MK6240. Reweighting the biased potential obtained from the metadynamics simulations to recover the full FES of the rare events has shown great advantages over the end-point free energy calculations implemented with implicit solvent models.43−45 Here, by using the X and Y coordinates of the center of the tracer as collective valuables (CVs) for the bias potential to act on, the conformational sampling of the tracer is enhanced, and thereby, one can evaluate the favorable sites and strength for the binding of different tracers from the FESs.
The setting of CVs allows the interactions between the tracer and solvent molecules and ions with the strength of −6 to −9 kcal/mol, the averaged energies can be treated as the solvation free energies for each tracer (Figure 4), which should be canceled out for measuring the real binding free energy of the tracer on a specific surface site. There are many strong hydrogen bonds and salt bridges between the folded chains; therefore, the ligand will not crash the inherent structure of the tau fibrils, which leaves a blank area (nonbinding area) on the FES shapes exactly the same as the protofibril viewed from the z-axis. It is possible to adopt other sets of CVs to enable the tracer to enter the interior cavities during metadynamics simulations; previous studies have indicated that the entrance of tracers into the interior cavities of the tau protein along other CVs requires relatively high energy barriers.34,37 Hence, compared to the conventional MD simulations for which the dissociation on a particular site takes place occasionally, setting CVs along the X and Y dimensions of the ligand centroid allows us to sample more thoroughly throughout all of the surface residues without crashing the structure of the protofibril and to randomly place the tracers at the beginning.
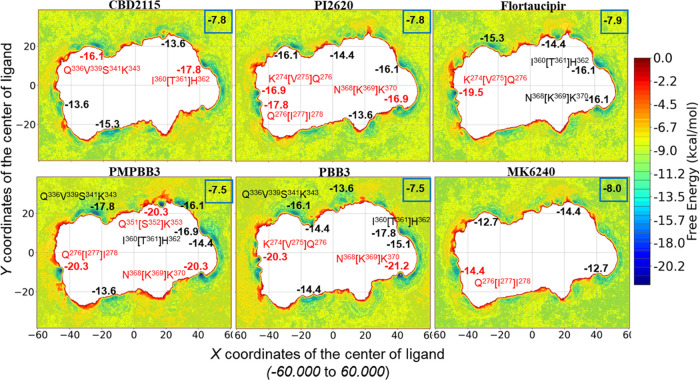
From the distributions of the local minima of the simulated systems, all six tracers are not found to bind to the small groove near docking site S7. This agrees with the MD simulations that the ionic interactions between K290 and D295 close the groove formed by residues K290 to D295 (Figure 4). This site was found to be a local minimum for the binding of PI2620 from the metadynamics performed by Künze et al. (site 7).37 As discussed above, in our settings of CVs, the dynamics of protein are not greatly perturbated by the bias potential. CBD2115, PI2620, Flortaucipir, and PMPBB3 are the know tracers that can bind both 3R/4R and 4R tau fibrils,25−27,31,46 while MK6240 shows few bindings to the 4R tau fibrils. At an overall level, the well depth and numbers of local minima on these FESs also indicate the better PSP binding preference for PMPBB3, CBD2115, Flortaucipir, and PI2620 than MK6240 (Figure 7). CBD2115 exhibits the highest binding affinity of −10.0 kcal/mol (with the cancelation of solvent free energy) to the site I360[T361]H362, followed by a local minimum (−16.1 kcal/mol in Figure 7) at the site Q336V339S341K343, which is also the site S2 identified by the docking study. Despite the contribution of more possibility for hydrogen bonding interactions from the hydroxyl group on the indole and piperidine rings, we find that the cation−π interaction or hydrogen bonding is not the leading force for binding compared to PI2620. This is, maybe, caused by the piperidine moiety in CBD2115 that affects the orientations of both hydroxyl groups and the indole and pyridine moieties. In our previous metadynamics simulation for CBD2115 on the CBD tau protofibril, a similar effect of CBD2115’s piperidine ring was observed.35 Two neighboring local minima (Q276[I277]I278 and K274[V275]Q276) near the docking identified site S1 show strong preference for the binding for PI2620, PMPBB3, and Flortaucipir. For PI2620, both sites can fit the ligand with a slightly higher preference on Q276[I277]I278 (−17.8 kcal/mol). For Flortaucipir, the binding is much more, preferring the K274[V275]Q276 site with deeper minima (−19.5 kcal/mol). We speculate that the more linear scaffold of Flortaucipir can have better cation-π interactions than PI2620. However, on an overall level, the number of deep local minima (<−13.0 kcal/mol) for PI2620 (7) is larger than for Flortaucipir (5), which means more possibility for PI2620 binding. It may compensate for the overall binding affinity for PI2620 measure at the tissue level for the real PSP tau fibrils.
Figure 7.
Binding sites and free energy profiles from the 4.5-μs well-tempered metadynamics simulations for 6 selected tracers in PSP protofibrils. CV1: the X coordinates of the ligand centroid; CV2: the Y coordinates of the ligand centroid. The averaged free energy of the tracer in the solvent and the lowest free energy of some local minima are given in bold numbers with and without a blue frame box, respectively. The free energies of some top-ranked local minima are colored in red, with the label of their binding site configuration marked also in red. The orientation of the PSP protofibril is identical to the one depicted in Figure 1.
The binding of PMPBB3 and PBB3 shows three and two highly strong binding sites (<−20.0 kcal/mol) on the FESs, respectively. None of these sites overlaps with the strongest binding site identified for CBD2115 (I360[T361]H362), which clearly reveals the distinct binding preferences of PMPBB3 and CBD2115 and may support the results from a current displacement study by Graham et al.47 Our previous metadynamics simulations for CBD and AD tau protofibrils also indicated very high binding for PMPBB3.35 Recently, Qi et al. identified that PMPBB3 has the top rank of hydrophobic contacts against other tracers to the 3R/4R chronic traumatic encephalopathy (CTE) tau protofibrils.48 These results indicated that the combination of buta-1,3-dienyl and benzothiazole in the scaffold of PMPBB3 and PBB3 is a good strategy for binding to the surface sites on both 3R/4R and 4R tau fibrils. The use of 1,3-butadiene to connect the benzothiazole and pyridine rings leads to a longer linear scaffold compared to CBD2115, which is believed to be beneficial for contacting with more fibril chains. However, the 1,3-butadiene moiety is also photon-sensitive, which can lead to undesired photoisomerization. Further investigations and improvements for the potential and selective 4R tau PET tracers are still ongoing.
Conclusions
Motivated by the great need to develop biomarkers for early diagnosis of neurodegenerative diseases and by the outstanding possibilities offered by the rapid development of in silico hard- and software, in this work, we have carried out a computational investigation of first- and second-generation PET tracers for 4R tau fibrils based on fundamental atomic principles. We illustrated the binding of 10 first- and second-generations of PET tracers for PSP tau protofibrils by different levels of methodology representing different rigor, accuracy, and efficiency. The binding sites and energies are also compared to CBD and AD tau by docking and conventional MD simulations. Most of these sites contain at least one polar/charged residue. This feature is present for all of the sites identified by the docking, MD, and metadynamics simulations in this study. The docking studies indicate that CBD2115, PBB3, and PMPBB3 exhibit higher binding probabilities and affinities for many surface sites in PSP tau than other tracers, in contrast to the stronger binding for the concave sites on AD tau fibrils. Our MD simulations clearly showed that due to the differences between the folding patterns between PSP/CBD tau and AD tau, one and the same tracer behaves differently on the surface sites of these tau fibrils. For the identified unstable docking sites, tracers can occasionally move to other sites that are low-ranked in the docking. For example, the closure and enlargement for the small groove (K290[C291][G292]S293[K294]D295 on PSP) and the big groove of the AD protofibril, respectively. The results may stress the importance of the induced-fit mechanism where the tracer and fibril mutually and dynamically perturb the structures of the counterpart. This process may also affect the so-called kon and koff reactivity constants for the binding of tracers. From metadynamics simulations, the preferred surface binding sites on PSP tau were investigated for CBD2115, PI2620, Flortaucipir, PMPBB3, PBB3, and MK6240. PMPBB3 and PBB3 exhibited stronger binding to the Q276[I277]I278, Q351[S352]K353, and N368[K369]K370 sites than the other tracers.
Our results can be divided into general and specific outcomes: (1) general in the sense of showing that, indeed, in silico modeling can act as a valuable trendsetter and give structure-property relation predictors providing tracer–fibril interaction patterns beneficial for further chemical radio labeling and in vitro/in vivo development; (2) specific because we could rank potent 4R tau PET tracers both with respect to effectivity and selectivity. We believe that exploring the binding characteristics of the tracers to PSP, CBD, and AD tau protofibrils can be valuable to establish and understand the structure–activity relationships between the known tracers and tau fibrils, as well as help to design novel high-affinity and selectivity PET tracers targeting a specific tau fibril.
Methods
Preparation of the Protofibrils
All of the initial structures of AD, CBD, and PSP protofibrils were taken from the Cryo-EM structures stored in the Protein Data Bank (PDB) with the PDB codes 7NRV,286VHA,38 and 7P65,15 respectively. For CBD tau, we chose 6VHA because of its smaller buried cavity compared to 6TJX (Figure S6).16 For AD tau, 7NRV was selected because it was fitted with the existence of PMPBB3.28 For PSP tau, 7P65 is selected because it is from the canonical tissue with Richardson’s syndrome.15 Considering the movement along the principal axis of fibrils and the consumption of computational resources, a nine-chain protofibril was constructed for each Cryo-EM structure of PSP, CBD, and AD using the method mentioned in our previous study.35,49 The automated preparation protocol implemented in the Protein Preparation Workflow of the Schrödinger Suite (Version 2021-4) was used,50,51 which assigns the atom types, bond orders, hydrogen atoms, and protonation states of ionizable residues at the pH of 7.4 and minimizes the system by the convergence of heavy atoms to 0.3 Å of root-mean-square deviations (RMSD) with the OPLS4 force field.52 The assigned protonation states of the ionizable residues for each prepared protofibrils were visually inspected.53
Molecular Docking
The structures of the selected tracer molecules were either downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/) or manually depicted in the Maestro interface of the Schrödinger Suite, followed by the preparation with the LigPrep module for the assignment of atom types, bond orders, and atomic partial charges.54 All of the docking runs were accomplished by the Glide module in the Schrödinger Suite,55,56 with the same protocol as in our previous study.35
Molecular Dynamics Simulations
For the tracer binding sites identified by docking, molecular dynamics simulations were carried out using the Desmond package (version 2021-4) with an OPLS4 force field.52,57 A box with an orthorhombic shape was used to place each initial protofibril–tracer complex with a minimum buffering area of 10 Å to the boundary of the filled TIP3P water molecules.58 Sodium or/and chloride ions were added to neutralize the system and to increase the concentration of salt to 0.15 M. The default energy minimization schemes were applied, followed by the relaxations using the Nose–Hoover thermostat and Martyna–Bobias–Klein barostat for 10.0 and 20.0 ps, respectively.59,60 After that, a 100-ns production simulation was performed in the NPT ensemble (300 K and 1 atm) for each system. The trajectories were inspected by Visual Molecular Dynamics (VMD, version 1.9.4a57) software with the in-house Tcl scripts to calculate the contacts.61 The accumulated percentage of contacts was calculated by counting the total number of protein–ligand atom pairs within 4 Å over all of the snapshots in the simulations for each ligand.
Metadynamics Simulations
To thoroughly explore the binding and unbinding of the tracer on the surface sites, we further performed well-tempered metadynamics simulations for each tracer on PSP protofibrils with a randomly placed position.62 The simulation protocols were adopted as our previous studies35,49 using PLUMED (version 2.8.0) patched with GROMACS (version 2021.4).63−65 Briefly, the tracers were parameterized with the general Amber force field with the charges fitted by the restrained electrostatic potential procedure, in which the electrostatic charges were firstly calculated by Gaussian 16 (rev. C01) at the Hartree–Fock level using the 6–31G(d) basis set.66 The Amber 99SB-ildn force field and TIP3P model were applied for the protein and solvent atoms, respectively.58,67,68 A 4.5-μs production run was performed for each tracer in the NVT ensemble (T = 300 K), using the 2D (x and y) coordinates of the center of mass of the tracer as collective variables (CVs), with a ±5 Å restraint on the z coordinates (along the principal axis of the protofibril) of the tracer’s center of mass. To calculate the biased potential along CVs, the initial Gaussian height and the bias factor were set at 0.2 kcal/mol and 6 with an interval of 1000 steps, respectively.69 The free energy surface (FES) was obtained by reweighting the biased potential for the sampled CV space with 1000 bins.45
Acknowledgments
The computations were enabled by resources provided by the National Academic Infrastructure for Supercomputing in Sweden (NAISS) at the National Supercomputer Centre at Linköping University (Sweden), partially funded by the Swedish Research Council through Grant Agreement 2022-3-34.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschemneuro.3c00437.
Inner cavities of selected tau structures; Autodock4 results; binding modes of tracers in site s6 after MD simulations; mobility of PI2620 in the V2 site; most contact residue profile of PSP tau derived from 100- and 500-ns MD simulations; and Cryo-EM structures of CBD tau protofibrils (PDF)
Author Contributions
H.Å., J.L., and A.N. designed the research; J.L. performed the research and analyzed the data; J.L. wrote the first draft of the manuscript, while H.Å and A.K. participated in updating it; and B.L. actively joined the discussion of the project. All of the authors are involved in the finalization of the manuscript and revision, while J.L. incorporated all of the changes.
This study was financially supported by the Michael J. Fox Foundation, MJFF-019728 (H.Å. and A.N.), the Swedish Science Research Council, VR2017-06086 (A.N.), and the Rainwater Charitable Foundation for the Tau consortium project: In silico development and in vitro characterization of PET tracers for 4R tau. A.K. would like to acknowledge the funding from the Alzheimer Association USA (AARF-21-848395).
The authors declare no competing financial interest.
Supplementary Material
References
- Dugger B. N.; Dickson D. W. Pathology of Neurodegenerative Diseases. Cold Spring Harbor Perspect. Biol. 2017, 9, a028035 10.1101/cshperspect.a028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M.; Yamaguchi Y.; Mishra S. K.; Higuchi M.; Sahara N. Tau Filaments and the Development of Positron Emission Tomography Tracers. Front. Neurol. 2018, 9, 70 10.3389/fneur.2018.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer L.; Brendel M. Imaging of Tau Pathology in Neurodegenerative Diseases: An Update. Semin. Nucl. Med. 2021, 51, 253–263. 10.1053/j.semnuclmed.2020.12.004. [DOI] [PubMed] [Google Scholar]
- Twohig D.; Nielsen H. M. α-synuclein in the Pathophysiology of Alzheimer’s Disease. Mol. Neurodegener. 2019, 14, 25 10.1186/s13024-019-0320-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles T. P. J.; Vendruscolo M.; Dobson C. M. The Amyloid State and its Association with Protein Misfolding Diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- DeKosky S. T.; Duara R. The Broad Range of Research in Alzheimer’s Disease and Related Dementias. Neurotherapeutics 2022, 19, 1–7. 10.1007/s13311-022-01245-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlden H.; Baker M.; Morris H. R.; MacDonald N.; Pickering–Brown S.; Adamson J.; Lees A. J.; Rossor M. N.; Quinn N. P.; Kertesz A.; Khan M. N.; Hardy J.; Lantos P. L.; George–Hyslop P. S.; Munoz D. G.; Mann D.; Lang A. E.; Bergeron C.; Bigio E. H.; Litvan I.; Bhatia K. P.; Dickson D.; Wood N. W.; Hutton M. Corticobasal Degeneration and Progressive Supranuclear Palsy Share a Common tau Haplotype. Neurology 2001, 56, 1702. 10.1212/WNL.56.12.1702. [DOI] [PubMed] [Google Scholar]
- Santos-Santos M. A.; Mandelli M. L.; Binney R. J.; Ogar J.; Wilson S. M.; Henry M. L.; Hubbard H. I.; Meese M.; Attygalle S.; Rosenberg L.; Pakvasa M.; Trojanowski J. Q.; Grinberg L. T.; Rosen H.; Boxer A. L.; Miller B. L.; Seeley W. W.; Gorno-Tempini M. L. Features of Patients With Nonfluent/Agrammatic Primary Progressive Aphasia With Underlying Progressive Supranuclear Palsy Pathology or Corticobasal Degeneration. JAMA Neurol. 2016, 73, 733–742. 10.1001/jamaneurol.2016.0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K.; del C Alonso A.; Chen S.; Chohan M. O.; El-Akkad E.; Gong C.-X.; Khatoon S.; Li B.; Liu F.; Rahman A.; Tanimukai H.; Grundke-Iqbal I. Tau Pathology in Alzheimer Disease and Other Tauopathies. Biochim. Biophys. Acta, Mol. Basis Dis. 2005, 1739, 198–210. 10.1016/j.bbadis.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Wang Y. T.; Edison P. Tau Imaging in Neurodegenerative Diseases Using Positron Emission Tomography. Curr. Neurol. Neurosci. Rep. 2019, 19, 45 10.1007/s11910-019-0962-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Liu T.; Cui M. Recent Development in Selective Tau Tracers for PET Imaging in the Brain. Chin. Chem. Lett. 2022, 33, 3339–3348. 10.1016/j.cclet.2022.03.024. [DOI] [Google Scholar]
- Goedert M.; Spillantini M. G.; Jakes R.; Rutherford D.; Crowther R. A. Multiple Isoforms of Human Microtubule-associated Protein tau: Sequences and localization in Neurofibrillary Tangles of Alzheimer’s Disease. Neuron 1989, 3, 519–526. 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- Goedert M. Tau Filaments in Neurodegenerative Diseases. FEBS Lett. 2018, 592, 2383–2391. 10.1002/1873-3468.13108. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick A. W. P.; Falcon B.; He S.; Murzin A. G.; Murshudov G.; Garringer H. J.; Crowther R. A.; Ghetti B.; Goedert M.; Scheres S. H. W. Cryo-EM Structures of tau Filaments from Alzheimer’s Disease. Nature 2017, 547, 185–190. 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.; Zhang W.; Yang Y.; Murzin A. G.; Falcon B.; Kotecha A.; van Beers M.; Tarutani A.; Kametani F.; Garringer H. J.; Vidal R.; Hallinan G. I.; Lashley T.; Saito Y.; Murayama S.; Yoshida M.; Tanaka H.; Kakita A.; Ikeuchi T.; Robinson A. C.; Mann D. M. A.; Kovacs G. G.; Revesz T.; Ghetti B.; Hasegawa M.; Goedert M.; Scheres S. H. W. Structure-based Classification of Tauopathies. Nature 2021, 598, 359–363. 10.1038/s41586-021-03911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Tarutani A.; Newell K. L.; Murzin A. G.; Matsubara T.; Falcon B.; Vidal R.; Garringer H. J.; Shi Y.; Ikeuchi T.; Murayama S.; Ghetti B.; Hasegawa M.; Goedert M.; Scheres S. H. W. Novel tau Filament Fold in Corticobasal Degeneration. Nature 2020, 580, 283–287. 10.1038/s41586-020-2043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan S.; Jing L. G.; Lakshmi C.; Anna S.; Jennifer D. M.; Luisa V. S.; Zhuohao H.; Bin Z.; Ronald J. G.; John Q. T.; Virginia M. Y. L. Pathological Tau Strains from Human Brains Recapitulate the Diversity of Tauopathies in Nontransgenic Mouse Brain. J. Neurosci. 2017, 37, 11406. 10.1523/JNEUROSCI.1230-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G. M.; Knopman D. S.; Chertkow H.; Hyman B. T.; Jack C. R.; Kawas C. H.; Klunk W. E.; Koroshetz W. J.; Manly J. J.; Mayeux R.; Mohs R. C.; Morris J. C.; Rossor M. N.; Scheltens P.; Carrillo M. C.; Thies B.; Weintraub S.; Phelps C. H. The Diagnosis of Dementia due to Alzheimer’s Disease: Recommendations from the National Institute on Aging-Alzheimer’s Association Workgroups on Diagnostic Duidelines for Alzheimer’s Disease. Alzheimer’s Dementia 2011, 7, 263–269. 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C.-F.; Arteaga J.; Chen G.; Gangadharmath U.; Gomez L. F.; Kasi D.; Lam C.; Liang Q.; Liu C.; Mocharla V. P.; Mu F.; Sinha A.; Su H.; Szardenings A. K.; Walsh J. C.; Wang E.; Yu C.; Zhang W.; Zhao T.; Kolb H. C. [18F]T807, A Novel tau Positron Emission Tomography Imaging Agent for Alzheimer’s Disease. Alzheimer’s Dementia 2013, 9, 666–676. 10.1016/j.jalz.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Ossenkoppele R.; Rabinovici G. D.; Smith R.; Cho H.; Schöll M.; Strandberg O.; Palmqvist S.; Mattsson N.; Janelidze S.; Santillo A.; Ohlsson T.; Jögi J.; Tsai R.; La Joie R.; Kramer J.; Boxer A. L.; Gorno-Tempini M. L.; Miller B. L.; Choi J. Y.; Ryu Y. H.; Lyoo C. H.; Hansson O. Discriminative Accuracy of [18F]flortaucipir Positron Emission Tomography for Alzheimer Disease vs Other Neurodegenerative Disorders. JAMA 2018, 320, 1151–1162. 10.1001/jama.2018.12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada R.; Okamura N.; Furumoto S.; Furukawa K.; Ishiki A.; Tomita N.; Tago T.; Hiraoka K.; Watanuki S.; Shidahara M.; Miyake M.; Ishikawa Y.; Matsuda R.; Inami A.; Yoshikawa T.; Funaki Y.; Iwata R.; Tashiro M.; Yanai K.; Arai H.; Kudo Y. 18F-THK5351: A Novel PET Radiotracer for Imaging Neurofibrillary Pathology in Alzheimer Disease. J. Nucl. Med. 2016, 57, 208–214. 10.2967/jnumed.115.164848. [DOI] [PubMed] [Google Scholar]
- Maruyama M.; Shimada H.; Suhara T.; Shinotoh H.; Ji B.; Maeda J.; Zhang M.-R.; Trojanowski John Q.; Lee Virginia M. Y.; Ono M.; Masamoto K.; Takano H.; Sahara N.; Iwata N.; Okamura N.; Furumoto S.; Kudo Y.; Chang Q.; Saido Takaomi C.; Takashima A.; Lewis J.; Jang M.-K.; Aoki I.; Ito H.; Higuchi M. Imaging of Tau Pathology in a Tauopathy Mouse Model and in Alzheimer Patients Compared to Normal Controls. Neuron 2013, 79, 1094–1108. 10.1016/j.neuron.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbi L. C.; Knust H.; Körner M.; Honer M.; Czech C.; Belli S.; Muri D.; Edelmann M. R.; Hartung T.; Erbsmehl I.; Grall-Ulsemer S.; Koblet A.; Rueher M.; Steiner S.; Ravert H. T.; Mathews W. B.; Holt D. P.; Kuwabara H.; Valentine H.; Dannals R. F.; Wong D. F.; Borroni E. Identification of Three Novel Radiotracers for Imaging Aggregated Tau in Alzheimer’s Disease with Positron Emission Tomography. J. Med. Chem. 2017, 60, 7350–7370. 10.1021/acs.jmedchem.7b00632. [DOI] [PubMed] [Google Scholar]
- Malarte M.-L.; Nordberg A.; Lemoine L. Characterization of MK6240, a tau PET Tracer, in Autopsy Brain Tissue from Alzheimer’s Disease Cases. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1093–1102. 10.1007/s00259-020-05035-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel M.; Barthel H.; van Eimeren T.; Marek K.; Beyer L.; Song M.; Palleis C.; Gehmeyr M.; Fietzek U.; Respondek G.; Sauerbeck J.; Nitschmann A.; Zach C.; Hammes J.; Barbe M. T.; Onur O.; Jessen F.; Saur D.; Schroeter M. L.; Rumpf J.-J.; Rullmann M.; Schildan A.; Patt M.; Neumaier B.; Barret O.; Madonia J.; Russell D. S.; Stephens A.; Roeber S.; Herms J.; Bötzel K.; Classen J.; Bartenstein P.; Villemagne V.; Levin J.; Höglinger G. U.; Drzezga A.; Seibyl J.; Sabri O. Assessment of 18F-PI-2620 as a Biomarker in Progressive Supranuclear Palsy. JAMA Neurol. 2020, 77, 1408–1419. 10.1001/jamaneurol.2020.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng C.-C.; Hsiao I.-T.; Yang Q.-F.; Yao C.-H.; Tai C.-Y.; Wu M.-F.; Yen T.-C.; Jang M.-K.; Lin K.-J. Characterization of 18F-PM-PBB3 (18F-APN-1607) Uptake in the rTg4510 Mouse Model of Tauopathy. Molecules 2020, 25, 1750. 10.3390/molecules25071750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg A.; Knight A. C.; Sohn D.; Rakos L.; Tong J.; Radelet A.; Mason N. S.; Stehouwer J. S.; Lopresti B. J.; Klunk W. E.; Sandell J.; Sandberg A.; Hammarström P.; Svensson S.; Mathis C. A.; Vasdev N. Radiosynthesis, In Vitro and In Vivo Evaluation of [18F]CBD-2115 as a First-in-Class Radiotracer for Imaging 4R-Tauopathies. ACS Chem. Neurosci. 2021, 12, 596–602. 10.1021/acschemneuro.0c00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.; Murzin A. G.; Falcon B.; Epstein A.; Machin J.; Tempest P.; Newell K. L.; Vidal R.; Garringer H. J.; Sahara N.; Higuchi M.; Ghetti B.; Jang M.-K.; Scheres S. H. W.; Goedert M. Cryo-EM Structures of tau Filaments from Alzheimer’s Disease with PET Ligand APN-1607. Acta Neuropathol. 2021, 141, 697–708. 10.1007/s00401-021-02294-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.; Beyer L.; Kaiser L.; Barthel H.; van Eimeren T.; Marek K.; Nitschmann A.; Scheifele M.; Palleis C.; Respondek G.; Kern M.; Biechele G.; Hammes J.; Bischof G.; Barbe M.; Onur Ö.; Jessen F.; Saur D.; Schroeter M. L.; Rumpf J.-J.; Rullmann M.; Schildan A.; Patt M.; Neumaier B.; Barret O.; Madonia J.; Russell D. S.; Stephens A. W.; Mueller A.; Roeber S.; Herms J.; Bötzel K.; Danek A.; Levin J.; Classen J.; Höglinger G. U.; Bartenstein P.; Villemagne V.; Drzezga A.; Seibyl J.; Sabri O.; Boening G.; Ziegler S.; Brendel M. Binding Characteristics of [18F]PI-2620 Distinguish the Clinically Predicted tau Isoform in Different Tauopathies by PET. J. Cereb. Blood Flow Metab. 2021, 41, 2957–2972. 10.1177/0271678X211018904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagai K.; Ono M.; Kubota M.; Kitamura S.; Takahata K.; Seki C.; Takado Y.; Shinotoh H.; Sano Y.; Yamamoto Y.; Matsuoka K.; Takuwa H.; Shimojo M.; Takahashi M.; Kawamura K.; Kikuchi T.; Okada M.; Akiyama H.; Suzuki H.; Onaya M.; Takeda T.; Arai K.; Arai N.; Araki N.; Saito Y.; Trojanowski J. Q.; Lee V. M. Y.; Mishra S. K.; Yamaguchi Y.; Kimura Y.; Ichise M.; Tomita Y.; Zhang M.-R.; Suhara T.; Shigeta M.; Sahara N.; Higuchi M.; Shimada H. High-Contrast In Vivo Imaging of Tau Pathologies in Alzheimer’s and Non-Alzheimer’s Disease Tauopathies. Neuron 2021, 109, 42–58e48. 10.1016/j.neuron.2020.09.042. [DOI] [PubMed] [Google Scholar]
- Malarte M.-L.; Gillberg P.-G.; Kumar A.; Bogdanovic N.; Lemoine L.; Nordberg A. Discriminative Binding of tau PET Tracers PI2620, MK6240 and RO948 in Alzheimer’s Disease, Corticobasal Degeneration and Progressive Supranuclear Palsy Brains. Mol. Psychiatry 2023, 28, 1272–1283. 10.1038/s41380-022-01875-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabe V. T.; Ntombela T.; Jhamba L. A.; Maguire G. E. M.; Govender T.; Naicker T.; Kruger H. G. Current Trends in Computer Aided Drug Design and a Highlight of Drugs Discovered via Computational Techniques: A Review. Eur. J. Med. Chem. 2021, 224, 113705 10.1016/j.ejmech.2021.113705. [DOI] [PubMed] [Google Scholar]
- Murugan N. A.; Nordberg A.; Ågren H. Different Positron Emission Tomography Tau Tracers Bind to Multiple Binding Sites on the Tau Fibril: Insight from Computational Modeling. ACS Chem. Neurosci. 2018, 9, 1757–1767. 10.1021/acschemneuro.8b00093. [DOI] [PubMed] [Google Scholar]
- Kuang G.; Murugan N. A.; Zhou Y.; Nordberg A.; Ågren H. Computational Insight into the Binding Profile of the Second-Generation PET Tracer PI2620 with Tau Fibrils. ACS Chem. Neurosci. 2020, 11, 900–908. 10.1021/acschemneuro.9b00578. [DOI] [PubMed] [Google Scholar]
- Zhou Y.; Li J.; Nordberg A.; Ågren H. Dissecting the Binding Profile of PET Tracers to Corticobasal Degeneration Tau Fibrils. ACS Chem. Neurosci. 2021, 12, 3487–3496. 10.1021/acschemneuro.1c00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todarwal Y.; Gustafsson C.; Thi Minh N. N.; Ertzgaard I.; Klingstedt T.; Ghetti B.; Vidal R.; König C.; Lindgren M.; Nilsson K. P. R.; Linares M.; Norman P. Tau Protein Binding Modes in Alzheimer’s Disease for Cationic Luminescent Ligands. J. Phys. Chem. B 2021, 125, 11628–11636. 10.1021/acs.jpcb.1c06019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künze G.; Kümpfel R.; Rullmann M.; Barthel H.; Brendel M.; Patt M.; Sabri O. Molecular Simulations Reveal Distinct Energetic and Kinetic Binding Properties of [18F]PI-2620 on Tau Filaments from 3R/4R and 4R Tauopathies. ACS Chem. Neurosci. 2022, 13, 2222–2234. 10.1021/acschemneuro.2c00291. [DOI] [PubMed] [Google Scholar]
- Arakhamia T.; Lee C. E.; Carlomagno Y.; Duong D. M.; Kundinger S. R.; Wang K.; Williams D.; DeTure M.; Dickson D. W.; Cook C. N.; Seyfried N. T.; Petrucelli L.; Fitzpatrick A. W. P.; et al. Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains. Cell 2020, 180, 633–644e612. 10.1016/j.cell.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M.; Huey R.; Lindstrom W.; Sanner M. F.; Belew R. K.; Goodsell D. S.; Olson A. J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz G. E.; Chalkley M. J.; Tan S. K.; Tse E.; Lee J.; Prusiner S. B.; Paras N. A.; DeGrado W. F.; Southworth D. R. Stacked Binding of a PET Ligand to Alzheimer’s tau Paired Helical Filaments. Nat. Commun. 2023, 14, 3048 10.1038/s41467-023-38537-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascoal T. A.; Therriault J.; Benedet A. L.; Savard M.; Lussier F. Z.; Chamoun M.; Tissot C.; Qureshi M. N. I.; Kang M. S.; Mathotaarachchi S.; Stevenson J.; Hopewell R.; Massarweh G.; Soucy J.-P.; Gauthier S.; Rosa-Neto P. 18F-MK-6240 PET for Early and Late Detection of Neurofibrillary Tangles. Brain 2020, 143, 2818–2830. 10.1093/brain/awaa180. [DOI] [PubMed] [Google Scholar]
- Ono M.; Sahara N.; Kumata K.; Ji B.; Ni R.; Koga S.; Dickson D. W.; Trojanowski J. Q.; Lee V. M.-Y.; Yoshida M.; Hozumi I.; Yoshiyama Y.; van Swieten J. C.; Nordberg A.; Suhara T.; Zhang M.-R.; Higuchi M. Distinct Binding of PET Ligands PBB3 and AV-1451 to tau Fibril Strains in Neurodegenerative Tauopathies. Brain 2017, 140, 764–780. 10.1093/brain/aww339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H.; Chen H.; Blazhynska M.; Goulard Coderc de Lacam E.; Szczepaniak F.; Pavlova A.; Shao X.; Gumbart J. C.; Dehez F.; Roux B.; Cai W.; Chipot C. Accurate Determination of Protein:ligand Standard Binding Free Energies from Molecular Dynamics Simulations. Nat. Protoc. 2022, 17, 1114–1141. 10.1038/s41596-021-00676-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden S.; Ryde U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-binding Affinities. Expert Opin. Drug Discovery 2015, 10, 449–461. 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwary P.; Parrinello M. A Time-Independent Free Energy Estimator for Metadynamics. J. Phys. Chem. B 2015, 119, 736–742. 10.1021/jp504920s. [DOI] [PubMed] [Google Scholar]
- Tezuka T.; Takahata K.; Seki M.; Tabuchi H.; Momota Y.; Shiraiwa M.; Suzuki N.; Morimoto A.; Nakahara T.; Iwabuchi Y.; Miura E.; Yamamoto Y.; Sano Y.; Funaki K.; Yamagata B.; Ueda R.; Yoshizaki T.; Mashima K.; Shibata M.; Oyama M.; Okada K.; Kubota M.; Okita H.; Takao M.; Jinzaki M.; Nakahara J.; Mimura M.; Ito D. Evaluation of [18F]PI-2620, a Second-generation Selective tau Tracer, for Assessing Four-repeat Tauopathies. Brain Commun. 2021, 3, fcab190 10.1093/braincomms/fcab190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham T. J. A.; Lindberg A.; Tong J.; Stehouwer J. S.; Vasdev N.; Mach R. H.; Mathis C. A. In Silico Discovery and Subsequent Characterization of Potent 4R-Tauopathy Positron Emission Tomography Radiotracers. J. Med. Chem. 2023, 66, 10628–10638. 10.1021/acs.jmedchem.3c00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi B.; Tan J.; Sun Y.; Cao M.; Lin J.; Zhang Q.; Zou Y. Mechanistic Insights into the Binding of Different Positron Emission Tomography Tracers to Chronic Traumatic Encephalopathy Tau Protofibrils. ACS Chem. Neurosci. 2023, 14, 1512–1523. 10.1021/acschemneuro.3c00061. [DOI] [PubMed] [Google Scholar]
- Uzuegbunam B. C.; Li J.; Paslawski W.; Weber W.; Svenningsson P.; Ågren H.; Yousefi B. H. Toward Novel [18F]Fluorine-Labeled Radiotracers for the Imaging of α-Synuclein Fibrils. Front. Aging Neurosci. 2022, 14, 830704 10.3389/fnagi.2022.830704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger Release 2021-4Jaguar, version 11.3; Schrödinger, LLC: New York, NY, 2021. [Google Scholar]
- Madhavi Sastry G.; Adzhigirey M.; Day T.; Annabhimoju R.; Sherman W. Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments. J. Comput.-Aided Mol. Des. 2013, 27, 221–234. 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Lu C.; Wu C.; Ghoreishi D.; Chen W.; Wang L.; Damm W.; Ross G. A.; Dahlgren M. K.; Russell E.; Von Bargen C. D.; Abel R.; Friesner R. A.; Harder E. D. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. 10.1021/acs.jctc.1c00302. [DOI] [PubMed] [Google Scholar]
- Olsson M. H. M.; Søndergaard C. R.; Rostkowski M.; Jensen J. H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- LigPrep, Schrödinger LLC: New York, NY, 2021. [Google Scholar]
- Friesner R. A.; Banks J. L.; Murphy R. B.; Halgren T. A.; Klicic J. J.; Mainz D. T.; Repasky M. P.; Knoll E. H.; Shelley M.; Perry J. K.; Shaw D. E.; Francis P.; Shenkin P. S. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Halgren T. A.; Murphy R. B.; Friesner R. A.; Beard H. S.; Frye L. L.; Pollard W. T.; Banks J. L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Bowers K. J.; Chow E.; Xu H.; Dror R. O.; Eastwood M. P.; Gregersen B. A.; Klepeis J. L.; Kolossvary I.; Moraes M. A.; Sacerdoti F. D.; Salmon J. K.; Shan Y.; Shaw D. E.. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing; Association for Computing Machinery: Tampa, Florida, 2006; p 84. [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Martyna G. J.; Klein M. L.; Tuckerman M. Nosé–Hoover Chains: The Canonical Ensemble via Continuous Dynamics. J. Chem. Phys. 1992, 97, 2635–2643. 10.1063/1.463940. [DOI] [Google Scholar]
- Martyna G. J.; Tobias D. J.; Klein M. L. Constant Pressure Molecular Dynamics Algorithms. J. Chem. Phys. 1994, 101, 4177–4189. 10.1063/1.467468. [DOI] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: Visual Molecular Dynamics. J. Mol. Graphics 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Barducci A.; Bussi G.; Parrinello M. Well-Tempered Metadynamics: A Smoothly Converging and Tunable Free-Energy Method. Phys. Rev. Lett. 2008, 100, 020603 10.1103/PhysRevLett.100.020603. [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. C.; van der Spoel D.; van Drunen R. GROMACS: A Message-Passing Parallel Molecular Dynamics Implementation. Comput. Phys. Commun. 1995, 91, 43–56. 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- Van Der Spoel D.; Lindahl E.; Hess B.; Groenhof G.; Mark A. E.; Berendsen H. J. C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Branduardi D.; Gervasio F. L.; Parrinello M. From A to B in Free Energy Space. J. Chem. Phys. 2007, 126, 054103 10.1063/1.2432340. [DOI] [PubMed] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Petersson G. A.; Nakatsuji H.; Li X.; Caricato M.; Marenich A. V.; Bloino J.; Janesko B. G.; Gomperts R.; Mennucci B.; Hratchian H. P.; Ortiz J. V.; Izmaylov A. F.; Sonnenberg J. L.; Williams-Young D.; Ding F.; Lipparini F.; Egidi F.; Goings J.; Peng B.; Petrone A.; Henderson T.; Ranasinghe D.; Zakrzewski V. G.; Gao J.; Rega N.; Zheng G.; Liang W.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T.; Throssell K.; Montgomery J. A. Jr; Peralta J. E.; Ogliaro F.; Bearpark M. J.; Heyd J. J.; Brothers E. N.; Kudin K. N.; Staroverov V. N.; Keith T. A.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A. P.; Burant J. C.; Iyengar S. S.; Tomasi J.; Cossi M.; Millam J. M.; Klene M.; Adamo C.; Cammi R.; Ochterski J. W.; Martin R. L.; Morokuma K.; Farkas O.; Foresman J. B.; Fox D. J.. Gaussian 16, revision C. 01; Gaussian. Inc.: Wallingford, CT, 2016. [Google Scholar]
- Cornell W. D.; Cieplak P.; Bayly C. I.; Gould I. R.; Merz K. M.; Ferguson D. M.; Spellmeyer D. C.; Fox T.; Caldwell J. W.; Kollman P. A. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995, 117, 5179–5197. 10.1021/ja00124a002. [DOI] [Google Scholar]
- Lindorff-Larsen K.; Piana S.; Palmo K.; Maragakis P.; Klepeis J. L.; Dror R. O.; Shaw D. E. Improved Side-chain Torsion Potentials for the Amber ff99SB Protein Force Field. Proteins: Struct., Funct., Bioinf. 2010, 78, 1950–1958. 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsson O.; Tiwary P.; Parrinello M. Enhancing Important Fluctuations: Rare Events and Metadynamics from a Conceptual Viewpoint. Annu. Rev. Phys. Chem. 2016, 67, 159–184. 10.1146/annurev-physchem-040215-112229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.